- Esophageal and Lung Institute, Allegheny Health Network, Pittsburgh, PA, United States
Esophageal adenocarcinoma (EAC) is a lethal cancer requiring improved screening strategies and treatment options due to poor detection methods, aggressive progression, and therapeutic resistance. Emerging circulating tumor DNA (ctDNA) technologies may offer a unique non-invasive strategy to better characterize the highly heterogeneous cancer and more clearly establish the genetic modulations leading to disease progression. The presented review describes the potential advantages of ctDNA methodologies as compared to current clinical strategies to improve clinical detection, enhance disease surveillance, evaluate prognosis, and personalize treatment. Specifically, we describe the ctDNA-targetable genetic markers of prognostic significance to stratify patients into risk of progression from benign to malignant disease and potentially offer cost-effective screening of established cancer. We also describe the application of ctDNA to more effectively characterize the heterogeneity and particular mutagenic resistance mechanisms in real-time to improve prognosis and therapeutic monitoring strategies. Lastly, we discuss the inconsistent clinical responses to currently approved therapies for EAC and the role of ctDNA to explore the dynamic regulation of novel targeted and immunotherapies to personalize therapy and improve patient outcomes. Although there are clear limitations of ctDNA technologies for immediate clinical deployment, this review presents the prospective role of such applications to potentially overcome many of the notable hurdles to treating EAC patients. A deeper understanding of complex EAC tumor biology may result in the progress toward improved clinical outcomes.
Introduction
Esophageal cancer (EC) is currently the 7th deadliest cancer in the United States with an estimated 17,290 newly diagnosed cases and 15,850 deaths in 2018, with an overall 5-years survival rate under 20% (1). For locally advanced disease, standard of care treatment currently includes neoadjuvant chemoradiotherapy plus surgery (2–4). However, ~40% of patients present with unresectable late-stage disease at diagnosis, reducing their 5-years survival to < 5%, and further underscoring the urgent need for earlier detection and improved treatment strategies (5, 6).
Circulating tumor DNA (ctDNA) is fragmented tumor-derived DNA in the blood stream that serves as a non-invasive diagnostic and prognostic tool for a number of cancer types (7, 8). Current standard cancer liquid protein biomarkers, such as PSA, CA 125, CEA, CA 19-9, AFP, etc., are limited by poor specificity and are known to be elevated in a variety of benign conditions. In contrast, ctDNA offers increased specificity of dysregulated genetic tumor markers, with levels of detection correlating well with premalignant to malignant progression (9). Moreover, ctDNA has a shorter half-life than protein biomarkers, and the real-time information that may be gathered from such “liquid biopsies” may provide unique insight to improve screening methodologies, therapeutic monitoring, and personalized therapy to improve outcomes in a cost-effective widely accessible manner (7, 10). Recently, Park et al. demonstrated that deep sequencing of ctDNA KRAS mutations sensitively detects pancreatic ductal adenocarcinoma and correlates with therapeutic response and disease progression (11). Additionally, ctDNA has shown promise for the detection of post-surgical recurrence of colon, breast, and lung cancers (12–14). The Cancer Genome Atlas (TCGA) recently characterized numerous deregulated genes in esophagogastric adenocarcinoma such as TP53, CDKN2A, SMAD4, ARID1A, ERBB2, VEGFA, CCNE1, GATA4, and GATA6 (15). Many of the identified markers overlapped with significant genes in gastric cancer pathogenesis; however, EAC DNA was consistently more hyper-methylated (11). ctDNA technologies may differentiate these important variances through the detection of not only point mutations, but also copy number variations, chromosomal rearrangements, epigenetic alterations, insertions, and deletions (10). The purpose of this review is to describe the potential utility of ctDNA to improve the detection, monitoring, treatment strategies, and prognosis for EAC patients.
Literature Review Methods
A thorough literature review was performed by searching the PubMed database for all relevant articles through September 1st, 2018. The following search criteria were applied: (“circulating tumor DNA” OR “ctDNA” OR “cell free DNA” OR “tumor DNA”) AND (“esophageal” OR “esophageal” OR “gastrointestinal” or “gastroesophageal”) AND (“cancer” OR “tumor” OR “malignancy”). The search produced 165 items, and two independent reviewers (JK and TP) screened the results. References from relevant articles were screened as an additional source of literature. Exclusions included articles that evaluated GI malignancies other than esophageal cancer and articles that were not in English. Additional specific searches of current EAC epidemiology statistics, TCGA data, ctDNA methodologies, and EAC therapeutics were performed to supplement background information.
ctDNA Methodology
ctDNA detection was first established though Sanger sequencing, however limitations such as high cost and complicated protocols have lead to the development of various methodologies to improve efficiency, cost-effectiveness, sensitivity, and specificity (16). Currently, the two main methods of detection include Digital PCR (dPCR) and Next-Generation Sequencing (NGS) (17). dPCR methods, such as Droplet Digital PCR and BEAMing, carry the advantage of being relatively easy, inexpensive methods with short turn around times, which can be a critical advantage when treating aggressive malignancies (18, 19). Additionally, these PCR methods offer superior precision, sensitivity, and reproducibility (19). Specifically, BEAMing has reported up to 100% sensitivity of multiple markers in a variety of cancers (20–22). Although dPCR methods carry higher sensitivity than NGS, it can only detect mutations within a limited number of loci, usually within a single gene, a disadvantage that can be overcome with NGS (23, 24). ctDNA NGS protocols include tagged-amplicon deep sequencing (Tam-Seq), Safe-Sequencing System (Safe-SeqS), and capture based sequencing (CAPP-seq) (23). CAPP-seq targets only areas of interest and is therefore more cost effective and focused than whole exome or whole genome sequencing (16). It also confers the lowest detection limit and background error rate of any NGS-based method and is therefore considered a superior NGS method for practical implementation (16).
Application of ctDNA technologies offers potential unique advantages to clinical care over tissue biopsy, and the concordance of ctDNA with tissue biopsy has been validated across a variety of cancers (25–29). Primarily, tissue DNA collection requires invasive procedures that many frail and elderly patients may not tolerate well (30). Less than 15% of esophageal cancers are diagnosed before the age of 55, so non-invasive detection and monitoring methods may confer particular benefit to the EAC patient population (1). Additionally, it has been well-established that many cancers, including EAC, are very heterogenic in nature and gain additional mutations throughout progression, treatment, and metastasis (31). Tissue biopsy samples only a small section of primary tumor, so obtained samples may not truly reflect all relevant and targetable mutations (32). Comparatively, ctDNA offers a more global perspective of the entire tumor, including metastases, that may offer improved detection of spatial and temporal tumor heterogeneity, which can carry great value by providing up-to-date information on tumor evolution and mutational status throughout disease course (10, 33). Moreover, unlike tissue DNA, analyzing ctDNA does not require fixation of the sample, which can fragment the DNA and cause sequence artifacts that may be misinterpreted as cancer-associated mutations (34).
Despite the improving technologies, there are current limitations to the clinical applications of ctDNA. First, the use of ctDNA as a reliable mechanism to inform clinical decisions lacks standardization due to limited early data and complex bioinformatics processing (35). To date, the only FDA approved tests include methylation-based test of SEPT9 for colorectal cancer and qPCR-based test for EGFR in non-small cell lung cancer (36, 37). Second, the yield of ctDNA material from plasma is usually quite low, especially in early stage cancers and precursor lesions. In such cases, whole genome amplification has been utilized to improve sample yield, however, future studies are needed to determine if this may compromise the clinical characterization of the tumor (35). Third, the mechanisms and rate of elimination of ctDNA from the bloodstream has not yet been fully explored. There is evidence to suggest ctDNA is cleared through the kidney, liver, and spleen, as well as nuclease degradation and phagocytosis; however, the rate of elimination may be very relevant to elucidate to in the setting of therapeutic monitoring or disease progression (38, 39).
ctDNA Detection of EAC
In an at-risk patient population of over 20 million GERD sufferers, it is estimated that only 10–15% develop Barrett's esophagus (BE), 0.5% of patients with BE progress to EAC per year, and 25% of high-grade dysplasia cases progress to EAC (40). The minimal risk of disease progression based on GERD symptoms alone dilutes the practicality of universal esophageal cancer screening protocols. Current testing for EAC and precursor lesions is primarily triggered by persistent GERD symptoms in a goal to detect early disease before development to malignancy. Despite this strategy, ~93% of diagnosed EAC patients received no prior surveillance (41). Non-invasive screening with ctDNA liquid biopsy may offer a cost-effective non-invasive alternative to identify patients at increased risk for early-stage disease or development of EAC.
Numerous genetic alterations have been identified in association with the development and prognosis of EAC that may serve as ideal ctDNA targets for improved detection. Particularly, the tumor suppressors TP53 and CDKN2A are mutated in ~72 and 12% of EAC cases, respectively (42, 43). Bettegowda et al. utilized ctDNA technology to detect multiple early and late stage malignancies, including gastroesophageal cancer (GEC), and were able to reliably detect ~58% of localized GEC (no evidence of metastasis) and 100% of metastatic disease (44). As expected, sensitivity improved from Stage 1 through IV progression across all malignancies (44). Additionally, a meta-analysis by Creemers et al. suggested ctDNA detection of HER2 and MYC may be useful for diagnosis and therapeutic monitoring of GEC (45). In order to improve early detection of EAC, non-invasive ctDNA testing may be strategically applied to patients with clinical risk factors, such as recurrent GERD symptoms, Caucasian ethnicity, obesity, smoking history, age, and/or male sex.
Many of the described mutations that characterize EAC, such as TP53 and CDKN2A, also occur in precursor lesions such as BE and HGD, lowering the specificity of such testing for established cancer (46). Still, alternative ctDNA markers may be useful in identifying patients with BE or HGD who are at increased risk for progression to EAC. Currently, the American College of Gastroenterology recommends patients with established non-dysplastic BE should receive endoscopic biopsy surveillance to assess possible disease progression every 3–5 years; however, 90–95% of these patients have completely stable disease and will never progress to EAC (47–50). In a recent study by Li et al. patients with BE were stratified into 79 progressors and 169 non-progressors to EAC, and biopsies were classified based on genetic expression. The study demonstrated that non-progressing BE lesions had small localized deletions at fragile sites, such as FHIT, WWOX, CDKN2A, and 9p arm loss/copy neutral loss of heterozygosity (LOH) (51). These samples revealed a low level of genetic heterogeneity and remained stable over years of surveillance. Alternatively, lesions that progressed to EAC developed increasing chromosome instability as early as 48 months before progression to EAC with gains and losses of whole chromosomes or chromosome arms, such as loss of 18q (51). Progressors showed significant mutation of SMAD4 and were universally more heterogenetic with progressive diversity and genome doublings (51). Similarly, a high-powered meta-analysis by Gharahkahni et al. suggests HTR3C and ABCC5 may be specific for progression of BE to EAC (52). Rumiato et al. recently utilized circulating cell-free DNA (cfDNA) to evaluate the neoplastic progression of BE and were able to successfully detect notable LOH predicting progression to dysplasia and/or EAC prior to visualization (9, 53). Moreover, post-intervention cfDNA sampling demonstrated a return to baseline levels of expression, further validating the potential of the technology to aid in the evaluation of treatment efficacy (9). Therefore, ctDNA screening for SMAD4, HTR3C, ABCC5, and increasing genetic variability over time may stratify patients with precancerous lesions for more appropriate endoscopic screening or intervention recommendations.
Prognostic Significance and Therapeutic Monitoring
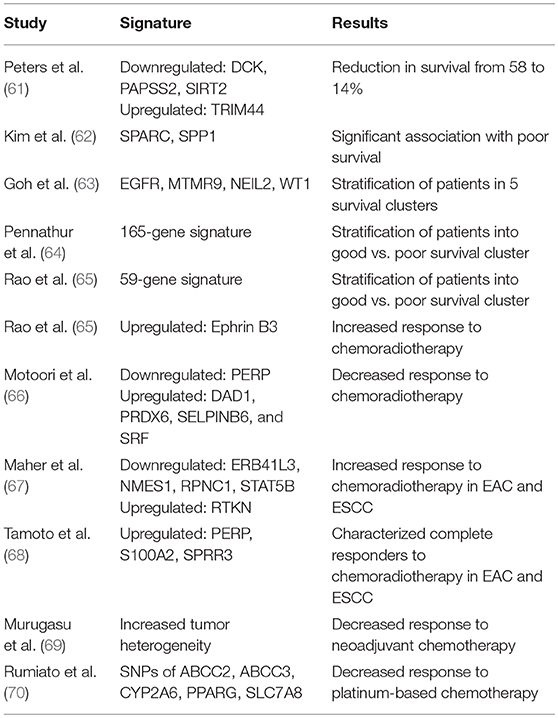
Various previous studies have established ctDNA as a useful tool for the prediction of patient prognosis before and after therapy in a variety of cancer types (54–58). In a recent study evaluating the role of ctDNA for tumor prognosis in breast cancer patients with multiline resistance, Hu et al. revealed unique mutation frequency patterns in those with progression free survival (PFS) <3 months vs. >3 months (59). Additionally, a TP53+PIK3CA mutation pattern successfully predicted progression within 6 months (59). Similarly, in a study investigating the utility of ctDNA to monitor non-small cell lung cancer (NSCLC) patients, tumor and blood samples before and after surgical resection demonstrated that the presence of ctDNA had a higher positive predictive value than six currently utilized clinical tumor biomarkers (60). As EAC is a notably heterozygous malignancy, a variety of diverse studies have reported unique genomic signatures associated with survival and prognostic response to chemoradiotherapy (Table 1) (71). Although, the true clinical utility of these specific genomic profiles has yet to be reliably established, ctDNA may offer an ideal setting for future studies to validate these prognostic indicators, as has been done for other cancers.
Furthermore, 44–61% of patients treated according to the current guidelines of neoadjuvant chemoradiotherapy plus surgery will experience recurrent disease (72). Despite complete resection with pathologically-confirmed clear margins and negative post-therapeutic CT imaging, it is hypothesized that many EAC recurrences are due to minimal residual disease (MRD) or systemic micrometastases (73). Previous studies have demonstrated that ctDNA screening may more sensitively identify small areas of remaining or recurrent disease as compared to standard imaging technology (74, 75). Such sensitive screening methodology may be ideally suited for such an aggressively deadly disease to trigger early intervention and reduce associated mortality. Recently, Chan et al. was able to improve detection of nasopharyngeal carcinoma recurrence by 10 months as compared to standard screening protocols (76). Similarly, ctDNA has also been used to detect MRD and predict recurrence in breast cancer with a mean lead-time of 7.9 months over clinical relapse (12). In a third study for lung cancer, ctDNA mutations predicted recurrence with 94% sensitivity with a median clinical lead-time of 5.2 months (14).
Unfortunately, 50–60% of EAC patients are resistant to standard chemotherapeutic treatment options due to inherent heterogeneity and development of escape mechanisms (69, 77). ctDNA technology provides an additional tool to monitor real-time therapeutic efficacy for more efficient modification of dosing and regimen. Murugasu et al. demonstrated that patients with EAC who had promising response to platinum agents developed decreased C>T mutations and increased C>A mutations through the course of treatment (69). Findlay et al. also confirmed this finding in addition to TT>CT changes, and acquired mutations in SF3B1, TAF1, and CCND2 (78). As neoadjuvant chemotherapy can dramatically and rapidly change the EAC genome profile, real-time monitoring to quickly identify resistance and opportunities for new actionable mutations may lead to clinical benefit. Moreover, ctDNA studies may provide additional insight into disease progression when used in conjunction with clinical imaging. de Figueriredo Barros et al. described an increasing mutation burden correlating with progression in metastatic colorectal cancer, while CT imaging showed stable disease (79).
Personalized Therapeutic Applications
Various targeted therapies have been explored for the treatment of gastroesophageal cancers but have only demonstrated limited efficacy (80, 81). Only trastuzumab has been established as a potential first-line treatment option for advanced GEC in HER2+ patients; however, benefits are minimal with a median overall survival (OS) of 13.8 months vs. 11.1 months (82). Ramucirumab (VEGFR-2) single agent or in combination with paclitaxel are recommended options for second-line treatment demonstrating an OS of 5.2 and 9.6 months, compared to 3.8 and 7.4 months, respectively (83, 84).
Over the last decade, success of immunotherapy across multiple cancer types has prompted exploration of novel immunologic targets for GECs (85). Recently, the late-line KEYNOTE-059 study of pembrolizumab in PD-L1 positive GEC and gastric tumors showed an objective response rate of 13.3% with 58% of the responses lasting 6 months or longer, leading to a third-line FDA approval (86, 87). Similarly, nivolumab has been approved for third-line GEC treatment in Japan after demonstrating an improved median survival of 5.26 months vs. 4.14 months, independent of PD-L1 expression (88, 89). Disappointingly, the most recent KEYNOTE-061 second-line pembrolizumab trial did not demonstrate any significant survival benefit in PD-L1 positive GEC patients (90). Still, pembrolizumab has secured a tumor agnostic approval in microsatellite instability-high or mismatch repair deficient previously treated unresectable or metastatic solid tumors; yet, these criteria only apply to ~3% of GECs (15, 91).
First generation immuno-oncology agents have demonstrated modest activity and potential application for the treatment of EAC; however, better stratification biomarkers and newer immunotherapeutic combination strategies may be required for enhanced durable responses. Only ~40% of EAC patients present with baseline PD-1 positivity, and this expression occurs primarily at the invasive margin (92, 93). Therefore, PD-L1 may not be the ideal predictive marker in EAC due to the inconsistent clinical response and relatively low rate of upregulation. There is evidence that the immune microenvironment as a whole is highly reactive to chemoradiotherapy and significantly increases expression of PD-L1 and other developing targets, such as LAG3, TIM3, and OX40 (94). Therefore, ctDNA technology may be a useful tool to explore the dynamic immunoregulation throughout the course of treatment and to better characterize the immunologic profile of EAC beyond the PD-1/PD-L1 pathway. A number of novel targeted molecular and immunotherapeutic agents are currently under investigation for EAC to explore additional potential treatment alternatives (95). Due to the extreme genomic variability and instability that classically characterize EAC, in addition to the multiple resistance mechanisms that may emerge during treatment, EAC may be an optimal candidate for the application of personalized therapeutics through the use of real-time ctDNA monitoring (96).
Moreover, in patients treated with immunotherapy, radiological evaluation of early disease responsiveness is especially challenging due to immune cell infiltration resulting in pseudo-progression on imaging (97, 98). Cabel et al. showed ctDNA is useful for early monitoring of responsiveness to anti-PD1 immunotherapy and correlated well with PFS and OS (99). Similarly, Xi et al. showed early changes in BRAFV600E ctDNA as early indicator to identify responding from non-responding patients with metastatic melanoma treated with immunotherapy (100). In another study, Raja et al. demonstrated early reduction in ctDNA correlation with survival in lung and bladder cancer after treatment with durvalumab (PD-L1) (101). Lastly, Khagi et al. demonstrated increased ctDNA detection of variants of unknown significance correlated with statistically significant improved PFS and OS in patients with diverse malignancies receiving immunotherapy (102). Exploration into the use of ctDNA technologies for non-invasive therapeutic monitoring of immunotherapy in EAC may be warranted due to the supporting evidence that ctDNA may be a good indicator of immunoresponse and prognosis.
Conclusions
Esophageal adenocarcinoma is a very deadly disease due to the high percentage of patients presenting with late-stage diagnosis and minimally effective treatment strategies. The emergence of non-invasive and cost-effective ctDNA technologies may offer a unique opportunity to improve screening protocols to more effectively monitor benign disease and detect malignancy earlier. Moreover, many patients with EAC are resistant to first line chemoradiotherapy, and therapeutic monitoring of ctDNA mutations throughout the course of treatment may allow for more efficient adjustments of personalized therapy. Despite only minor successes with targeted therapies due to the highly heterogenic nature of EAC, real-time information regarding response and prognosis may allow for more informed clinical decision-making strategies. Additionally, the integration of ctDNA with developing immunotherapeutic options may open the door for improved prognostic outcomes.
Limitations of ctDNA technologies are rooted in the still early development of this new emerging technology and the lack of strongly validated studies characterizing the precise clinical role it may play to truly improve patient care (30). Rigorous future clinical studies will be required to reliably describe specific discoverable changes in ctDNA throughout disease progression before broad implementation. Still, EAC remains an extremely lethal disease, and further investigation into the potential benefits of ctDNA characterization may offer significant benefits to improve early detection, monitor progression, enhance real-time personalization of therapeutics, and evaluate prognosis to improve clinical care.
Author Contributions
This manuscript was drafted by JK, AZ, and TP and was critically reviewed by BJ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Noone AM, Howlader N, Krapcho M, Miller D, Brest A, Yu M, et al. SEER Cancer Statistics Review, 1975-2015. National Cancer Institute (2018). Available online at: https://seer.cancer.gov/csr/1975_2015/
2. Shapiro J, van Lanschot JJB, Hulshof M, van Hagen P, van Berge Henegouwen MI, Wijnhoven BPL, et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol. (2015) 16:1090–8. doi: 10.1016/S1470-2045(15)00040-6
3. Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M, et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Eng J Med. (2006) 355:11–20. doi: 10.1056/NEJMoa055531
4. Walsh TN, Noonan N, Hollywood D, Kelly A, Keeling N, Hennessy TP. A comparison of multimodal therapy and surgery for esophageal adenocarcinoma. N Eng J Med. (1996) 335:462–7. doi: 10.1056/NEJM199608153350702
5. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. (2018) 68:7–30. doi: 10.3322/caac.21442
6. Thrift AP. The epidemic of oesophageal carcinoma: where are we now? Cancer Epidemiol. (2016) 41:88–95. doi: 10.1016/j.canep.2016.01.013
7. Ma M, Zhu H, Zhang C, Sun X, Gao X, Chen G. “Liquid biopsy”-ctDNA detection with great potential and challenges. Ann Transl Med. (2015) 3:235. doi: 10.3978/j.issn.2305-5839.2015.09.29
8. Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer–a survey. Biochim Biophys Acta (2007) 1775:181–232. doi: 10.1016/j.bbcan.2006.10.001
9. Rumiato E, Boldrin E, Malacrida S, Realdon S, Fassan M, Morbin T, et al. Detection of genetic alterations in cfDNA as a possible strategy to monitor the neoplastic progression of Barrett's esophagus. Transl Res. (2017) 190:16–24 e1. doi: 10.1016/j.trsl.2017.09.004
10. Duffy J. Annals express: circulating tumor DNA as a cancer biomarker. Ann Clin Biochem. (2018). doi: 10.1177/0004563218798401. [Epub ahead of print].
11. Park G, Park JK, Son DS, Shin SH, Kim YJ, Jeon HJ, et al. Utility of targeted deep sequencing for detecting circulating tumor DNA in pancreatic cancer patients. Sci Rep. (2018) 8:11631. doi: 10.1038/s41598-018-30100-w
12. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. (2016) 8:346ra92. doi: 10.1126/scitranslmed.aaf6219
13. Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. (2015) 7:302ra133. doi: 10.1126/scitranslmed.aab0021
14. Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. (2017) 7:1394–403. doi: 10.1158/2159-8290.CD-17-0716
15. Cancer Genome Atlas Research N Analysis Working Group: Asan U Agency BCC Brigham Women's H Broad I, et al. Integrated genomic characterization of oesophageal carcinoma. Nature (2017) 541:169–75. doi: 10.1038/nature20805
16. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. (2014) 20:548–54. doi: 10.1038/nm.3519
17. Sawada K, Kotani D, Bando H. The clinical landscape of circulating tumor DNA in gastrointestinal malignancies. Front Oncol. (2018) 8:263. doi: 10.3389/fonc.2018.00263
18. Garcia-Foncillas J, Alba E, Aranda E, Diaz-Rubio E, Lopez-Lopez R, Tabernero J, et al. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: an expert taskforce review. Ann Oncol. (2017) 28:2943–9. doi: 10.1093/annonc/mdx501
19. Hudecova I. Digital PCR analysis of circulating nucleic acids. Clin Biochem. (2015) 48:948–56. doi: 10.1016/j.clinbiochem.2015.03.015
20. Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. (2012) 18:3462–9. doi: 10.1158/1078-0432.CCR-11-2696
21. Diehl F, Schmidt K, Durkee KH, Moore KJ, Goodman SN, Shuber AP, et al. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology (2008) 135:489–98. doi: 10.1053/j.gastro.2008.05.039
22. Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. (2003) 100:8817–22. doi: 10.1073/pnas.1133470100
23. Molina-Vila MA, Mayo-de-las-Casas C, Giménez-Capitán A, Jordana-Ariza N, Garzón M, Balada A, et al. Liquid biopsy in non-small cell lung cancer. Front Med. (2016) 3:69. doi: 10.3389/fmed.2016.00069
25. Husain H, Velculescu VE. Cancer DNA in the Circulation: the liquid biopsy. JAMA (2017) 318:1272–4. doi: 10.1001/jama.2017.12131
26. Yeung KT, More S, Woodward B, Velculescu V, Husain H. Circulating tumor DNA for mutation detection and identification of mechanisms of resistance in non-small cell lung cancer. Mol Diagn Ther. (2017) 21:375–84. doi: 10.1007/s40291-017-0260-5
27. Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. (2017) 9:eaan2415. doi: 10.1126/scitranslmed.aan2415
28. Lipson EJ, Velculescu VE, Pritchard TS, Sausen M, Pardoll DM, Topalian SL, et al. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. J Immunother Cancer (2014) 2:42. doi: 10.1186/s40425-014-0042-0
29. Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. (2014) 4:650–61. doi: 10.1158/2159-8290.CD-13-1014
30. Ignatiadis M, Lee M, Jeffrey SS. Circulating tumor cells and circulating tumor DNA: challenges and opportunities on the path to clinical utility. Clin Cancer Res. (2015) 21:4786–800. doi: 10.1158/1078-0432.CCR-14-1190
31. Noorani A, Bornschein J, Lynch AG, Secrier M, Achilleos A, Eldridge M, et al. A comparative analysis of whole genome sequencing of esophageal adenocarcinoma pre- and post-chemotherapy. Genome Res. (2017) 27:902–12. doi: 10.1101/gr.214296.116
32. Xu H, Chen L, Shao Y, Zhu D, Zhi X, Zhang Q, et al. Clinical application of circulating tumor DNA in the genetic analysis of patients with advanced GIST. Mol Cancer Ther. (2018) 17:290–6. doi: 10.1158/1535-7163.MCT-17-0436
33. Amirouchene-Angelozzi N, Swanton C, Bardelli A. Tumor evolution as a therapeutic target. Cancer Discov. (2017). doi: 10.1158/2159-8290.CD-17-0343. [Epub ahead of print].
34. Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. (2015) 61:64–71. doi: 10.1373/clinchem.2014.223040
35. Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J. (2018) 16:370–8. doi: 10.1016/j.csbj.2018.10.002
36. Song L, Yu H, Jia J, Li Y. A systematic review of the performance of the SEPT9 gene methylation assay in colorectal cancer screening, monitoring, diagnosis and prognosis. Cancer Biomark. (2017) 18:425–32. doi: 10.3233/CBM-160321
37. Brown P. The Cobas(R) EGFR Mutation test v2 assay. Future Oncol. (2016) 12:451–2. doi: 10.2217/fon.15.311
38. Velders M, Treff G, Machus K, Bosnyak E, Steinacker J, Schumann U. Exercise is a potent stimulus for enhancing circulating DNase activity. Clin Biochem. (2014) 47:471–4. doi: 10.1016/j.clinbiochem.2013.12.017
39. Choi JJ, Reich CF III Pisetsky DS. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology. (2005) 115:55–62. doi: 10.1111/j.1365-2567.2005.02130.x
40. Gregson EM, Bornschein J, Fitzgerald RC. Genetic progression of Barrett's oesophagus to oesophageal adenocarcinoma. Br J Cancer (2016) 115:403–10. doi: 10.1038/bjc.2016.219
41. Dong J, Buas MF, Gharahkhani P, Kendall BJ, Onstad L, Zhao S, et al. Determining risk of barrett's esophagus and esophageal adenocarcinoma based on epidemiologic factors and genetic variants. Gastroenterology (2018) 154:1273–81 e3. doi: 10.1053/j.gastro.2017.12.003
42. Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O'Donovan M, et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat Genet. (2015) 47:1038–46. doi: 10.1038/ng.3357
43. Testa U, Castelli G, Pelosi E. Esophageal cancer: genomic and molecular characterization, stem cell compartment and clonal evolution. Medicines (2017) 4:E67. doi: 10.3390/medicines4030067
44. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. (2014) 6:224ra24. doi: 10.1126/scitranslmed.3007094
45. Creemers A, Krausz S, Strijker M, van der Wel MJ, Soer EC, Reinten RJ, et al. Clinical value of ctDNA in upper-GI cancers: a systematic review and meta-analysis. Biochim Biophys Acta (2017) 1868:394–403. doi: 10.1016/j.bbcan.2017.08.002
46. Weaver JMJ, Ross-Innes CS, Shannon N, Lynch AG, Forshew T, Barbera M, et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nature Genet. (2014) 46:837–43. doi: 10.1038/ng.3013
47. Clermont M, Falk GW. Clinical guidelines update on the diagnosis and management of barrett's esophagus. Dig Dis Sci. (2018) 63:2122–8. doi: 10.1007/s10620-018-5070-z
48. Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett's esophagus. N Eng J Med. (2011) 365:1375–83. doi: 10.1056/NEJMoa1103042
49. Macdonald CE, Wicks AC, Playford RJ. Ten years' experience of screening patients with Barrett's oesophagus in a university teaching hospital. Gut (1997) 41:303–7. doi: 10.1136/gut.41.3.303
50. Sikkema M, de Jonge PJ, Steyerberg EW, Kuipers EJ. Risk of esophageal adenocarcinoma and mortality in patients with Barrett's esophagus: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. (2010) 8:235–44. doi: 10.1016/j.cgh.2009.10.010
51. Li X, Galipeau PC, Paulson TG, Sanchez CA, Arnaudo J, Liu K, et al. Temporal and spatial evolution of somatic chromosomal alterations: a case-cohort study of Barrett's esophagus. Cancer Prev Res. (2014) 7:114–27. doi: 10.1158/1940-6207.CAPR-13-0289
52. Gharahkhani P, Fitzgerald RC, Vaughan TL, Palles C, Gockel I, Tomlinson I, et al. Genome-wide association studies in oesophageal adenocarcinoma and Barrett's oesophagus: a large-scale meta-analysis. Lancet Oncol. (2016) 17:1363–73. doi: 10.1016/S1470-2045(16)30240-6
53. Boldrin E, Rumiato E, Fassan M, Balsamo L, Realdon S, Battaglia G, et al. Liquid biopsy as a novel tool to monitor the carcinogenesis of Barrett's esophagus. Transl Res. (2016) 176:127–31. doi: 10.1016/j.trsl.2016.05.001
54. Berger AW, Schwerdel D, Ettrich TJ, Hann A, Schmidt SA, Kleger A, et al. Targeted deep sequencing of circulating tumor DNA in metastatic pancreatic cancer. Oncotarget (2018) 9:2076–85. doi: 10.18632/oncotarget.23330
55. Wei Z, Wang W, Shu Z, Zhou X, Zhang Y. Correlation between circulating tumor DNA levels and response to Tyrosine Kinase Inhibitors (TKI) treatment in non-small cell lung cancer. Med Sci Monit. (2017) 23:3627–34. doi: 10.12659/MSM.902265
56. Imamura F, Uchida J, Kukita Y, Kumagai T, Nishino K, Inoue T, et al. Early responses of EGFR circulating tumor DNA to EGFR tyrosine kinase inhibitors in lung cancer treatment. Oncotarget (2016) 7:71782–9. doi: 10.18632/oncotarget.12373
57. Garlan F, Laurent-Puig P, Sefrioui D, Siauve N, Didelot A, Sarafan-Vasseur N, et al. Early evaluation of circulating tumor DNA as marker of therapeutic efficacy in metastatic colorectal cancer patients (PLACOL Study). Clin Cancer Res. (2017) 23:5416–25. doi: 10.1158/1078-0432.CCR-16-3155
58. Yang X, Zhuo M, Ye X, Bai H, Wang Z, Sun Y, et al. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget (2016) 7:20810–24. doi: 10.18632/oncotarget.8021
59. Hu ZY, Xie N, Tian C, Yang X, Liu L, Li J, et al. Identifying circulating tumor DNA mutation profiles in metastatic breast cancer patients with multiline resistance. EBioMedicine (2018) 32:111–8. doi: 10.1016/j.ebiom.2018.05.015
60. Guo N, Lou F, Ma Y, Li J, Yang B, Chen W, et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci Rep. (2016) 6:33519. doi: 10.1038/srep33519
61. Peters CJ, Rees JR, Hardwick RH, Hardwick JS, Vowler SL, Ong CA, et al. A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology (2010) 139:1995–2004 e15. doi: 10.1053/j.gastro.2010.05.080
62. Kim SM, Park YY, Park ES, Cho JY, Izzo JG, Zhang D, et al. Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome. PloS ONE (2010) 5:e15074. doi: 10.1371/journal.pone.0015074
63. Goh XY, Rees JR, Paterson AL, Chin SF, Marioni JC, Save V, et al. Integrative analysis of array-comparative genomic hybridisation and matched gene expression profiling data reveals novel genes with prognostic significance in oesophageal adenocarcinoma. Gut (2011) 60:1317–26. doi: 10.1136/gut.2010.234179
64. Pennathur A, Xi L, Litle VR, Gooding WE, Krasinskas A, Landreneau RJ, et al. Gene expression profiles in esophageal adenocarcinoma predict survival after resection. J Thorac Cardiovasc Surg. (2013) 145:505–12. doi: 10.1016/j.jtcvs.2012.10.031
65. Rao S, Welsh L, Cunningham D, te-Poele RH, Benson M, Norman A, et al. Correlation of overall survival with gene expression profiles in a prospective study of resectable esophageal cancer. Clin Colorectal Cancer (2011) 10:48–56. doi: 10.3816/CCC.2011.n.007
66. Motoori M, Takemasa I, Yamasaki M, Komori T, Takeno A, Miyata H, et al. Prediction of the response to chemotherapy in advanced esophageal cancer by gene expression profiling of biopsy samples. Int J Oncol. (2010) 37:1113–20. doi: 10.3892/ijo_00000763
67. Maher SG, Gillham CM, Duggan SP, Smyth PC, Miller N, Muldoon C, et al. Gene expression analysis of diagnostic biopsies predicts pathological response to neoadjuvant chemoradiotherapy of esophageal cancer. Ann Surg. (2009) 250:729–37. doi: 10.1097/SLA.0b013e3181bce7e1
68. Tamoto E, Tada M, Murakawa K, Takada M, Shindo G, Teramoto K, et al. Gene-expression profile changes correlated with tumor progression and lymph node metastasis in esophageal cancer. Clin Cancer Res. (2004) 10:3629–38. doi: 10.1158/1078-0432.CCR-04-0048
69. Murugaesu N, Wilson GA, Birkbak NJ, Watkins T, McGranahan N, Kumar S, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. (2015) 5:821–31. doi: 10.1158/2159-8290.CD-15-0412
70. Rumiato E, Boldrin E, Malacrida S, Battaglia G, Bocus P, Castoro C, et al. A germline predictive signature of response to platinum chemotherapy in esophageal cancer. Transl Res. (2016) 171:29–37 e1. doi: 10.1016/j.trsl.2015.12.011
71. Visser E, Franken IA, Brosens LA, Ruurda JP, van Hillegersberg R. Prognostic gene expression profiling in esophageal cancer: a systematic review. Oncotarget (2017) 8:5566–77. doi: 10.18632/oncotarget.13328
72. Neri A, Marrelli D, Voglino C, Di Mare G, Ferrara F, Marini M, et al. Recurrence after surgery in esophago-gastric junction adenocarcinoma: current management and future perspectives. Surg Oncol. (2016) 25:355–63. doi: 10.1016/j.suronc.2016.08.003
73. Koenig AM, Prenzel KL, Bogoevski D, Yekebas EF, Bubenheim M, Faithova L, et al. Strong impact of micrometastatic tumor cell load in patients with esophageal carcinoma. Ann Surg Oncol. (2009) 16:454–62. doi: 10.1245/s10434-008-0169-7
74. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. (2015) 61:112–23. doi: 10.1373/clinchem.2014.222679
75. Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget (2016) 7:48832–41. doi: 10.18632/oncotarget.9453
76. Chan KC, Hung EC, Woo JK, Chan PK, Leung SF, Lai FP, et al. Early detection of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer (2013) 119:1838–44. doi: 10.1002/cncr.28001
77. Davies AR, Gossage JA, Zylstra J, Mattsson F, Lagergren J, Maisey N, et al. Tumor stage after neoadjuvant chemotherapy determines survival after surgery for adenocarcinoma of the esophagus and esophagogastric junction. J Clin Oncol. (2014) 32:2983–90. doi: 10.1200/JCO.2014.55.9070
78. Findlay JM, Castro-Giner F, Makino S, Rayner E, Kartsonaki C, Cross W, et al. Differential clonal evolution in oesophageal cancers in response to neo-adjuvant chemotherapy. Nat Commun. (2016) 7:11111. doi: 10.1038/ncomms11111
79. de Figueiredo Barros BD, Kupper BEC, Aguiar Junior S, de Mello CAL, Begnami MD, Chojniak R, et al. Mutation detection in tumor-derived cell free DNA anticipates progression in a patient with metastatic colorectal cancer. Front Oncol. (2018) 8:306. doi: 10.3389/fonc.2018.00306
80. Pectasides E. Genomic Alterations and targeted therapy in gastric and esophageal adenocarcinoma. Clin Ther. (2016) 38:1589–99. doi: 10.1016/j.clinthera.2016.03.016
81. He YM, Yu C, Li WB, Li ZP, Xu N. Evaluation of short-term effectiveness of eight targeted agents combined with chemotherapy for treating esophageal-gastric junction adenocarcinoma: a network meta-analysis. J Cell Biochem. (2018) 119:1183–92. doi: 10.1002/jcb.26288
82. Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet (2010) 376:687–97. doi: 10.1016/S0140-6736(10)61121-X
83. Wilke H, Muro K, Van Cutsem E, Oh SC, Bodoky G, Shimada Y, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol. (2014) 15:1224–35. doi: 10.1016/S1470-2045(14)70420-6
84. Fuchs CS, Tomasek J, Yong CJ, Dumitru F, Passalacqua R, Goswami C, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet (2014) 383:31–9. doi: 10.1016/S0140-6736(13)61719-5
85. Mizrak Kaya D, Harada K, Amlashi FG, Vasilakopoulou M, Ajani JA. Customization of therapy for gastroesophageal adenocarcinoma patients. Chronic Dis Transl Med. (2018) 4:8–17. doi: 10.1016/j.cdtm.2018.02.003
86. Fuchs CS, Doi T, Jang RW, Muro K, Satoh T, Machado M, et al. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: Phase 2 clinical KEYNOTE-059 trial. JAMA Oncol. (2018) 4:e180013. doi: 10.1001/jamaoncol.2018.0013
87. U.S. Food and Drug Administration: FDA Grants Accelerated Approval to Pembrolizumab for Advanced Gastric Cancer. Available online at: https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm577093.htm (Accessed Sep 22, 2017)
88. Kang YK, Boku N, Satoh T, Ryu MH, Chao Y, Kato K, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet (2017) 390:2461–71. doi: 10.1016/S0140-6736(17)31827-5
89. Boku N, Kang YK, Satoh T. A Phase 3 study of nivolumab (Nivo) in previously treated advanced gastric or gastroesophageal junction (G/GEJ) cancer: updated results and subset analysis by PD-L1 expression (ATTRACTION-02). Ann Oncol. (2017) 28 (Suppl. 5):v209–68. doi: 10.1093/annonc/mdx369.001
90. Shitara K, Ozguroglu M, Bang YJ, Di Bartolomeo M, Mandala M, Ryu MH, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet (2018) 392:123–33. doi: 10.1016/S0140-6736(18)31257-1
91. U.S. Food and Drug Administration: FDA grants accelerated approval to pembrolizumab for first tissue/site agnostic indication. Available online at: https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm560040.htm (Accessed May 30, 2017).
92. Thompson ED, Zahurak M, Murphy A, Cornish T, Cuka N, Abdelfatah E, et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut (2017) 66:794–801. doi: 10.1136/gutjnl-2015-310839
93. Derks S, Nason KS, Liao X, Stachler MD, Liu KX, Liu JB, et al. Epithelial PD-L2 expression Marks Barrett's esophagus and esophageal adenocarcinoma. Cancer Immunol Res. (2015) 3:1123–9. doi: 10.1158/2326-6066.CIR-15-0046
94. Kelly RJ, Zaidi AH, Smith MA, Omstead AN, Kosovec JE, Matsui D, et al. The dynamic and transient immune microenvironment in locally advanced esophageal adenocarcinoma post chemoradiation. Ann Surg. (2017) 268:992–9. doi: 10.1097/SLA.0000000000002410
95. Gaur P, Hunt CR, Pandita TK. Emerging therapeutic targets in esophageal adenocarcinoma. Oncotarget (2016) 7:48644–55. doi: 10.18632/oncotarget.8777
96. Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. (2013) 45:478–86. doi: 10.1038/ng.2591
97. Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Eng J Med. (2015) 373:123–35. doi: 10.1056/NEJMoa1504627
98. Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Eng J Med. (2015) 373:23–34. doi: 10.1056/NEJMoa1504030
99. Cabel L, Riva F, Servois V, Livartowski A, Daniel C, Rampanou A, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol. (2017) 28:1996–2001. doi: 10.1093/annonc/mdx212
100. Xi L, Pham TH, Payabyab EC, Sherry RM, Rosenberg SA, Raffeld M. Circulating tumor DNA as an early indicator of response to T-cell transfer immunotherapy in metastatic melanoma. Clin Cancer Res. (2016) 22:5480–6. doi: 10.1158/1078-0432.CCR-16-0613
101. Raja R, Kuziora M, Brohawn P, Higgs BW, Gupta A, Dennis PA, et al. Early reduction in ctDNA predicts survival in lung and bladder cancer patients treated with durvalumab. Clin Cancer Res. (2018). doi: 10.1158/1078-0432.CCR-18-0386. [Epub ahead of print].
Keywords: esophageal cancer (EC), circulating tumor DNA (ctDNA), targeted therapy (TT), immunotherapy, personalized medicine
Citation: Kosovec JE, Zaidi AH, Pounardjian TS and Jobe BA (2018) The Potential Clinical Utility of Circulating Tumor DNA in Esophageal Adenocarcinoma: From Early Detection to Therapy. Front. Oncol. 8:610. doi: 10.3389/fonc.2018.00610
Received: 04 September 2018; Accepted: 28 November 2018;
Published: 11 December 2018.
Edited by:
Pashtoon Murtaza Kasi, Mayo Clinic, United StatesReviewed by:
Paolo Giorgi Rossi, Azienda Sanitaria Unità Locale di Reggio Emilia, ItalyJohan Nicolay Wiig, Oslo University Hospital, Norway
Copyright © 2018 Kosovec, Zaidi, Pounardjian and Jobe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali H. Zaidi, ali.zaidi@ahn.org
 Juliann E. Kosovec
Juliann E. Kosovec Ali H. Zaidi
Ali H. Zaidi Tamar S. Pounardjian
Tamar S. Pounardjian