- 1Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education, College of Animal Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2Qinghai Academy of Animal Science and Veterinary Medicine, Qinghai, China
- 3Sanjiaocheng Sheep Breeding Farm, Qinghai, China
- 4Animal Husbandry and Veterinary Station, Qinghai, China
Murine primary hair follicle induction is driven by the communication between the mesenchyme and epithelium and mostly governed by signaling pathways including wingless-related integration site (WNT), ectodysplasin A receptor (EDAR), bone morphogenetic protein (BMP), and fibroblast growth factor (FGF), as observed in genetically modified mouse models. Sheep skin may serve as a valuable system for hair research owing to the co-existence of sweat glands with wool follicles in trunk skin and asynchronized wool follicle growth pattern similar to that of human head hair follicles. However, the mechanisms underlying wool follicle development remain largely unknown. To understand how long non-coding RNAs (lncRNAs) and mRNAs function in primary wool follicle induction in carpet wool sheep, we conducted high-throughput RNA sequencing and revealed globally altered lncRNAs (36 upregulated and 26 downregulated), mRNAs (228 elevated and 225 decreased), and 80 differentially expressed novel transcripts. Several key signals in WNT (WNT2B and WNT16), BMP (BMP3, BMP4, and BMP7), EDAR (EDAR and EDARADD), and FGF (FGFR2 and FGF20) pathways, and a series of lncRNAs, including XLOC_539599, XLOC_556463, XLOC_015081, XLOC_1285606, XLOC_297809, and XLOC_764219, were shown to be potentially important for primary wool follicle induction. GO and KEGG analyses of differentially expressed mRNAs and potential targets of altered lncRNAs were both significantly enriched in morphogenesis biological processes and transforming growth factor-β, Hedgehog, and PI3K-Akt signaling, as well as focal adhesion and extracellular matrix-receptor interactions. The prediction of mRNA-mRNA and lncRNA-mRNA interaction networks further revealed transcripts potentially involved in primary wool follicle induction. The expression patterns of mRNAs and lncRNAs of interest were validated by qRT-PCR. The localization of XLOC_297809 and XLOC_764219 both in placodes and dermal condensations was detected by in situ hybridization, indicating important roles of lncRNAs in primary wool follicle induction and skin development. This is the first report elucidating the gene network of lncRNAs and mRNAs associated with primary wool follicle early development in carpet wool sheep and will shed new light on selective wool sheep breeding.
Introduction
Wool, the external portion of wool follicles, is a valuable natural fiber that differs greatly in diameter, crimp, and elasticity. The refinement of wool quality, especially fiber diameter, which is highly correlated with wool follicle development, could boost the economic performance of wool sheep husbandry (Rogers, 2006). Wool follicle development includes successive generation of primary wool follicles from approximately embryonic day 55 (E55) to E65, secondary wool follicles from approximately E75 to E85, and secondary-derived wool follicles from approximately E95 to E105 in sheep fetal skin (Rogers, 2006). The fleece of carpet or coarse wool sheep is dominated by long and thick medullated wool developed from primary wool follicles, which share anatomical structures with murine primary hair follicles during early skin morphogenesis (Paus et al., 1999; Rogers, 2006).
The development of murine pelage hair follicles has been carefully classified into nine specific stages (stages 0–8) based on previously established comprehensive criteria (Paus et al., 1999). Briefly, hair follicle development progresses through de novo formation of the hair germ (stages 0–1), down-growth and elongation of the hair peg (stages 2–3), advanced growth of the hair peg forming an inner root sheath, outer root sheath, sebocyte, and bulge (stages 4–6), and maturation of the hair follicle (stages 7–8) across the prenatal and neonatal periods (Paus et al., 1999). The first two stages (stages 0–1) are extremely interesting for investigation, as during the progression from stage 0 to stage 1, the embryonic skin displays simple but extensive morphological changes from thin skin with homogeneous epidermal layers at stage 0 to the occurrence of periodic hair follicles (hair placode with dermal condensation beneath) and the thickening of epidermal and dermal compartments at stage 1 (Paus et al., 1999). This periodic pattern formation in skin is the outcome of molecular and cellular crosstalk between the epithelium and mesenchyme represented by wingless-related integration site (WNT), ectodysplasin A receptor (EDAR), bone morphogenetic protein (BMP), and fibroblast growth factor (FGF) signaling pathways (Paus et al., 1999; Millar, 2002; Huh et al., 2013; Glover et al., 2017). The size of hair placode and number of dermal papilla cells are highly correlated with hair fiber thickness. Hence, the periodicity and diversity of pattern formation makes hair follicle a good model to investigate the mechanisms determining organ size and shape, tissue morphogenesis, homeostasis, and stem cell dynamics. Many signature genes have been identified and characterized in the different compartments formed during this process. Of those, one study isolated specific cell types from E14.5 dorsal skin from transgenic mouse lines and identified the corresponding signature genes by RNA-seq (Sennett et al., 2015). This large-scale data combined with numerous previous reports drew a comprehensive picture of the early molecular signaling networks engaged in murine primary hair follicle morphogenesis and skin development (Millar, 2002; Schmidt-Ullrich and Paus, 2005; Rishikaysh et al., 2014).
The general principles of hair follicle development, particularly for primary hair follicles, have been widely studied in mouse models (Nakamura et al., 2013). There are advantages to manipulating mouse models for skin and hair research because of the simplicity of the dorsal skin, which lacks sweat glands as well as the clear three-wave hair follicle development and synchronized postnatal hair growth cycle (Muller-Rover et al., 2001). However, understanding of the diversity of skin and hair in humans and other mammals is limited. For example, the mechanisms underlying murine hair cycling could not fully decipher the mystery of asynchronized human head hair growth. Therefore, wool follicle may serve as a distinctive system for hair and skin study owing to its unique features. Sheep wool follicles occur and grow independently from neighboring follicles, similarly to human head hair follicles (Rogers, 2006). Sheep adult trunk skin contains both wool follicles and sweat glands, thus sharing anatomical structures with human axillary skin (Saga, 2002; Rogers, 2006). These characteristics of skin anatomy and hair growth cycling pattern in human hair follicles and wool follicles are different from those of murine skin. Hence, the investigation of sheep skin development not only adds new knowledge to understand the enigma of hair development beyond established mouse and chicken models, but also potentially provides new ideas for addressing human skin health and skincare such as human head hair loss and axillary odor or hircismus.
Long non-coding RNAs (LncRNAs) are transcripts longer than 200 nucleotides that can regulate mRNA expression at both posttranscriptional and transcriptional levels (Kornienko et al., 2013). Though several lncRNAs relevant to skin biology have been reported, such as ANCR and SPRY4-IT1 (Wan and Wang, 2014), the detailed functional roles of lncRNAs in hair and wool follicle development are based on limited reports and, thus, largely unknown. A recent study detected 15 differentially expressed lncRNAs during secondary wool follicle induction in super-fine wool sheep, which possesses no primary wool follicles due to selective breeding (Yue et al., 2016). Another study identified lncRNAs which might contribute to the variable induction ability of dermal papilla cells under tissue culture conditions (Lin et al., 2014). No studies have examined prenatal primary wool follicle development. Therefore, we assessed this earlier time point in our current report.
In this study, we focused on elucidating the global gene expression changes during the first two stages (stages 0–1) of primary wool follicle formation. The aim of this study was to explore the dynamic gene regulation during primary wool follicle induction in carpet wool sheep and improve the understanding of hair follicle and skin development. This work will provide a global illustration of the lncRNA and mRNA interaction networks during primary wool follicle induction and potentially benefit wool quality control in the wool industry.
Materials and Methods
Experimental Animals
Tibetan carpet sheep fetuses intended to be discarded were scavenged and immediately placed in cold PBS at a local abattoir in Qinghai Province, China. The left-sided dorsal skins were excised, cut into strips, and fixed in 4% paraformaldehyde at 4°C for approximately one week prior to hematoxylin and eosin (H&E) staining. The right-sided dorsal skins were chopped into pieces and frozen in liquid nitrogen for RNA extraction. More than 60 individuals at unspecified embryonic stages were randomly collected and predicted to range approximately from E45 to E75 roughly based on the body size and length. The accurate developmental stages of wool follicles in sheep skin were determined by H&E staining in the next step. All experiments on animals were approved by the Standing Committee of Hubei People’s Congress and the ethics committee of Huazhong Agricultural University.
Morphological Identification of Primary Wool Follicle Induction in Carpet Wool Sheep
To identify the developmental stages of wool follicles, more than 60 (n ≥ 60) fixed sheep skin samples were dehydrated and processed in paraffin, according to the standard procedures. H&E staining was conducted using 5-μm skin sections. The stained skin sections were photographed individually and carefully grouped into different developmental stages based on previous reports (Paus et al., 1999). The induction stages of primary wool follicles were further investigated. Six individuals comparable to E55 and E65, corresponding to stages 0 and 1 of primary wool follicle morphogenesis were selected for RNA isolation and high throughput sequencing, respectively (n = 3).
RNA Extraction and cDNA Synthesis
Total RNA was extracted using TRIzol Reagent (Invitrogen, United StatesUSA) and processed for quality determination using a NanoDrop 2000 spectrophotometer (Thermo Scientific, United States). The cDNA was generated for qRT-PCR validation using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Japan).
Illumina Sequencing and Data Analysis
Six libraries of sheep skin samples were constructed and sequenced on the Illumina HiSeq 2000 (San Diego, CA, United States) with 150 base pair paired-end reads at Novogene (Beijing, China) following the manufacturer’s procedures. Briefly, total RNA was extracted using TRIzol reagent, and RNA integrity was assessed using the RNA Nano 6000 Assay Kit with the Bioanalyzer 2100 system (Agilent Technologies, United States). The sequencing libraries were prepared from total RNA devoid of ribosomal RNA using the NEBNext® UltraTM Directional RNA Library Prep Kit for Illumina® (NEB, United States). The products were purified with an AMPure XP system (Beckman Coulter, United States) and assessed on an Agilent Bioanalyzer 2100 system for quality control. After cluster generation, the libraries were sequenced. All reads were deposited in the NCBI Sequence Read Archive (SRA) database under accession number SRP126454. Clean reads were mapped to the sheep reference genome (version: Oarv3.1). Transcripts without coding potential, predicted by Coding-Non-Coding Index (CNCI) (v2) (Sun et al., 2013), Coding Potential Calculator (CPC) (0.9-r2) (Kong et al., 2007), Pfam-scan (v1.3) (Punta et al., 2012), and Phylogenetic codon substitution frequency (phyloCSF) (v20121028) (Lin et al., 2011), were candidate lncRNAs. The conservation of lncRNAs and mRNAs was analyzed using Phast (v1.3) software (Siepel et al., 2005). Coding genes 10 k/100 k upstream and downstream of lncRNAs were predicted as target genes in cis, while targets in trans were predicted via calculating the expressed correlation with lncRNAs. For biological replicates, lncRNAs and mRNAs with p-adjust <0.05 were regarded as differentially expressed between the two stages of primary wool follicle induction. GOseq R packages (Young et al., 2010) and KOBAS software (Mao et al., 2005) were used for Gene Ontology (GO) and KEGG analysis of the differentially expressed lncRNA targets and mRNAs, respectively.
Interaction Network Construction
Protein-protein interaction analysis of differentially expressed genes was based on the commonly used STRING database. Due to the absence of sheep in this database and limited studies on skin and hair development of the relative species Bos taurus, we used the mouse genome for the interaction network construction. Briefly, the sequences of differentially expressed lncRNA targets and mRNAs were blasted (blastx) to the Mus musculus genome to predict protein-protein interactions using the STRING database1. Then, the mRNA-mRNA and lncRNA–mRNA interaction networks were visualized in Cytoscape (Shannon et al., 2003).
Quantitative Real-Time PCR (qRT-PCR) Validation
Several differentially expressed mRNAs and lncRNAs involved in primary wool follicle induction were selected and confirmed by qRT-PCR with GAPDH used as an internal reference. The primers for qRT-PCR are listed in Table 1. qRT-PCR was carried out with a Roche LightCycler® 96 using iTaqTM Universal SYBR® Green Supermix (Bio-Rad, United States). The amplification procedures were 95°C for 5 min initially, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. Quantification of mRNAs and lncRNAs was performed using the standard curve method with average cycle thresholds (Ct). The qRT-PCR data were generated from six independent samples per stage, including the three original samples subjected to RNA sequencing and three additional samples, and statistically analyzed using Student’s t-test (n = 6).
In Situ Hybridization
Antisense riboprobes were generated and labeled with Digoxigenin (Roche, United States) by in vitro transcription using linearized plasmids as templates, and then applied to 10-μm sheep skin sections for in situ hybridization as described previously with minor modifications (Lee et al., 2013). Briefly, the skin sections were dewaxed and rehydrated to process with proteinase K treatment. After a 15 min fixation in 4% paraformaldehyde and pre-hybridization, the sections were hybridized overnight at 58°C with riboprobes in the hybridization solution. The sections were rinsed in gradient SSC as descried previously (Lee et al., 2013). Then the sections were washed with TBST buffer for 10 min and incubated with 10% BSA, followed by incubation with Anti-Digoxigenin-AP Fab fragments (Roche, United States, 1:1000) overnight. The sections were washed with TBST buffer and incubated with the BCIP/NBT (Sigma, United States) solution to detect hybridized signal. The hybridized signals were photographed by microscopy (Olympus BX53, Tokyo, Japan). Corresponding sense riboprobes were generated and applied to the skin sections for in situ hybridization as negative controls.
Statistical Analyses
All data are presented as the mean ± SEM. All statistical tests were performed using GraphPad Prism version 6 software (United States). The Student’s t-test was used to assess the differences between two groups. Statistically significant differences were determined at a p-value < 0.05.
Results
Morphological Characterization of Primary Wool Follicle Induction in Carpet Wool Sheep Skin
The dorsal skin samples obtained from Tibetan carpet wool sheep fetuses (n ≥ 60) were randomly collected and grouped to identify the developmental stages of primary wool follicles by H&E staining (Figures 1A,B). The development of skin and wool follicles was detected across early and late stages, based on a series of H&E stained skin sections. The specific induction stages, stage 0 and stage 1 of the primary wool follicle development, which correspond to approximately embryonic day E55 and E65, respectively, were deliberately picked for further classification. At stage 0, the epidermis of the sheep fetal skin was observed thin and uniform, whereas at stage 1, it thickened dramatically with regularly spaced wool placodes attached with initiated dermal condensations. The dermis of the sheep skin was also thickened compared to that at stage 0. These characteristics were utilized to carefully classify wool follicles in sheep skin into developmental stage 0 and stage 1. (Figures 1A,B). More than six individuals in each group were further examined.
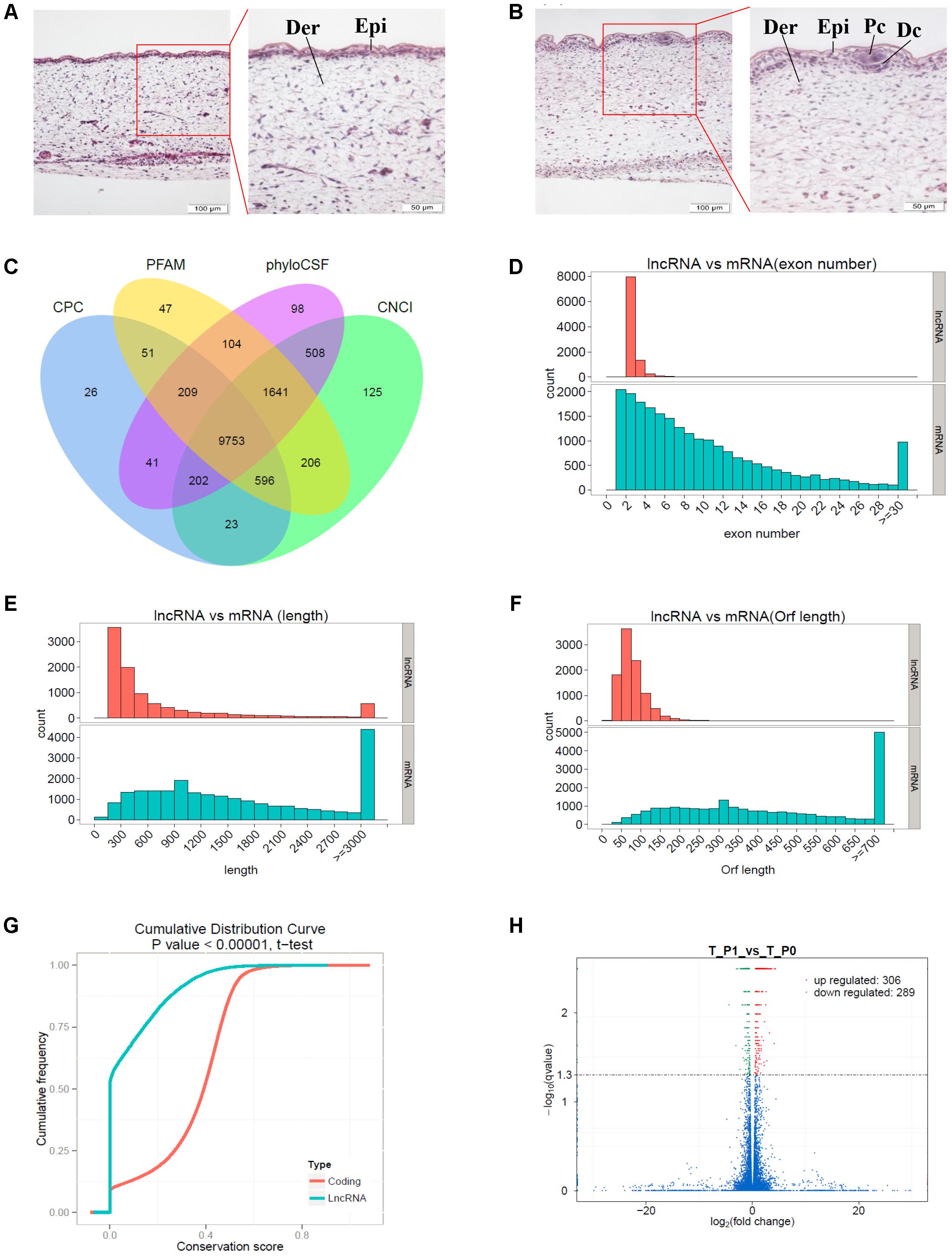
FIGURE 1. Identification of long non-coding RNAs (lncRNAs) and mRNAs involved in primary wool follicle induction stages in carpet wool sheep fetal skins. (A,B) The morphology of skin and wool follicles at stages 0 and 1 of primary wool follicle development were observed by hematoxylin and eosin (H&E) staining (Left) are 200×, scale bars represent 100 μm. (Right) are 400×, scale bars represent 50 μm, which is the magnification of the segment in the red frame). Epi, epidermis, Der, dermis, Pc, placode, Dc, dermal condensation. At stage 1 (B), the hair placode and dermal condensation were clearly detected compared to the homogeneous epidermal layers at stage (A). (C) Coding potentiality filter using Coding Potential Calculator, Pfam, Phylogenetic codon substitution frequency, and Coding-Non-Coding Index. (D) Exon number distribution of lncRNAs and mRNAs. (E) Transcript lengths distribution of lncRNAs and mRNAs. (F) Open reading frame length distribution of lncRNAs and mRNAs. (G) Conservation of the sequence in lncRNAs and mRNAs was analyzed using Phast (v1.3) software. (H) Differentially expressed transcripts in sheep skin between the two stages of primary wool follicle development. Red, green, and blue dots in the graph represent transcripts that were significantly upregulated, significantly downregulated, and not significantly changed between these two stages, respectively.
Identification of LncRNAs and mRNAs in Carpet Wool Sheep Fetal Skin
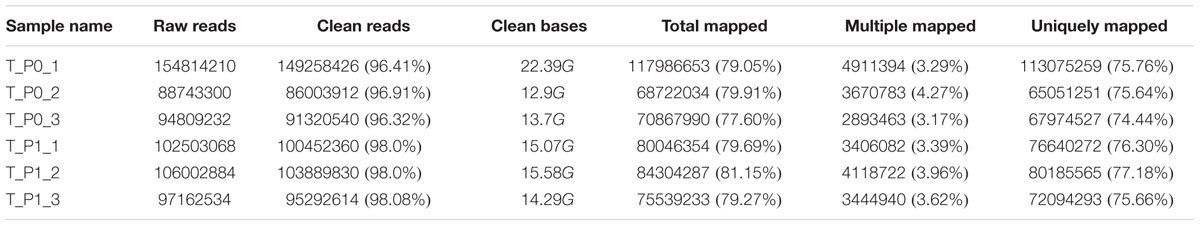
Illumina sequencing of six cDNA libraries derived from carpet wool sheep whole skin yielded a total of 338,366,742 and 305,668,486 raw reads for stage 0 and stage 1 samples, respectively. Clean reads devoid of adapter and low-quality sequences occupied more than 96% of the raw data (Table 2). Nearly 81.15% of the clean reads were mapped to the sheep reference genome, of which more than 74.44% were uniquely mapped.
A total of 9,753 lncRNAs were identified after coding potential filter using CNCI, CPC, Pfam-scan, and phyloCSF software (Figure 1C). In addition, 22,800 mRNAs and 3,975 novel transcripts were identified. The exon number, transcript length, and ORF length of lncRNAs and mRNAs were calculated and graphed, as shown in Figures 1D–F. A majority of lncRNAs comprised two or three exons, whereas mRNAs contained a broad range of exon numbers from two to thirty (Figure 1D). The transcript length (Figure 1E) and ORF length (Figure 1F) of lncRNAs were significantly shorter than those of mRNAs. Moreover, most lncRNAs were less conserved than mRNAs (Figure 1G).
Functional Annotation of Differentially Expressed LncRNAs and mRNAs in Primary Wool Follicle Induction
During the early development of wool sheep fetus, anatomical changes were observed in trunk skin between stages 0 and 1 of primary wool follicle development (Figures 1A,B). The most prominent structures were the appearance of the follicle placode and dermal condensation (Figures 1A,B). RNA sequencing analyses detected 595 differentially expressed transcripts between these two stages (Figure 1H). Of these, there were 36 upregulated and 26 downregulated lncRNAs, together with 228 upregulated and 225 downregulated mRNAs (Supplementary Table S1). Additionally, 80 differentially expressed novel transcripts were obtained.
These differentially expressed transcripts were divided into four classes based on the structure changes of sheep embryonic skin during primary wool follicle induction (Figures 1A,B). The established hair follicle marker genes, including placode markers (EDAR, FOXI3, FGF20, SHH, and SOSTDC1), dermal condensation markers (TRPS1, CXCR4, BMP3, BMP4, BMP7, and TBX18), epidermal markers (KRT1, KRT10, DSC1, DMKN, and SLC7A11), and fibroblast markers (DPP4 and LOX) showed elevated expression at stage 1 when primary wool follicles were initiated (Sennett et al., 2015; Rezza et al., 2016). Further analyses revealed that the key signaling pathways regulating murine primary hair follicle induction, including WNT, BMP, EDAR, and FGF, were detected in our data. For example, several genes in the WNT signaling pathway (WNT16, LEF1, and WIF1) were significantly upregulated at stage 1. Three BMPs, including BMP3, BMP4, and BMP7, showed elevated expression in stage 1 samples. SOSTDC1, a modulator of both the WNT and BMP pathways, displayed increased expression at stage 1 in sheep skin. In addition, several components of the EDAR signaling pathway (EDAR, EDARADD, and the downstream effector FOXI3) were highly enriched at wool follicle induction stage 1.
Long non-coding RNAs may regulate gene expression by binding and modulating the activity and stability of the target genes. A total of 7,677 targets of 62 differentially expressed lncRNAs were predicted and classified based on the cis and trans principles (Supplementary Table S2). GO terms of these lncRNA potential targets were highly enriched for several developmental processes, such as system, organ, anatomical structure, multicellular organismal, and cellular development (Figure 2A and Supplementary Table S3). The predicted lncRNA targets partially overlapped with the differentially expressed mRNAs, which were highly enriched for morphogenesis biological processes, such as system development, anatomical structure development and morphogenesis, and organ development and morphogenesis (Figure 2B and Supplementary Table S4). Of these, 11, 8, and 19 overlapped target genes were identified and predicted to function in epidermal development and cell differentiation, hair follicle development, and skin development and morphogenesis, respectively (Figure 2C and Table 3). GO analyses revealed signaling pathways important in hair follicle development, including WNT, BMP, and FGF, which shared 16, 7, and 5 genes with lncRNA targets, respectively (Figure 2D and Table 3).
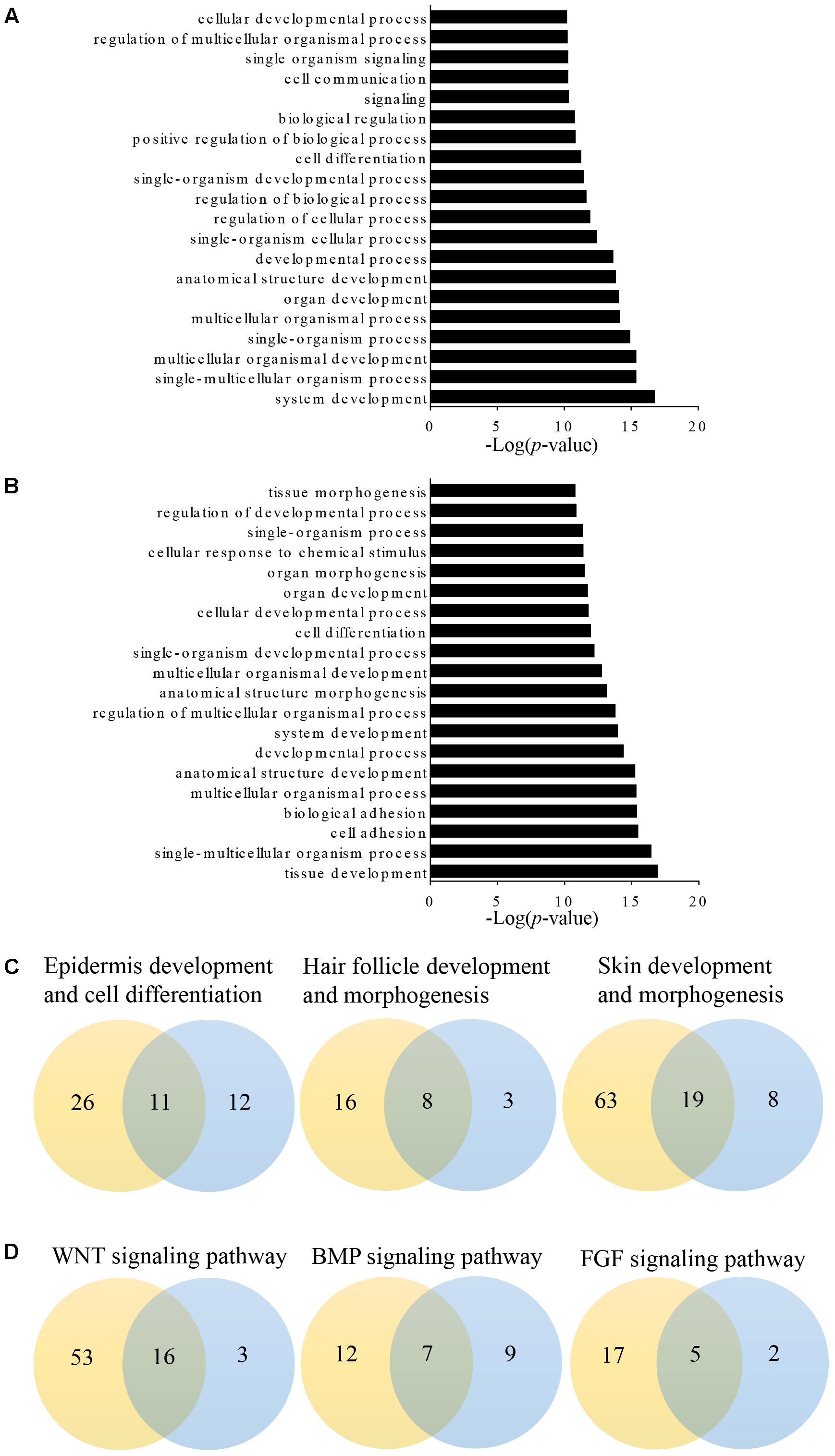
FIGURE 2. Gene Ontology (GO) analysis of differentially expressed lncRNAs targets and mRNAs in primary wool follicle induction. (A,B) The top 20 enrichment biological processes for differentially expressed lncRNA targets and mRNAs are listed. (C) One-to-one pairs of differentially expressed lncRNA targets and mRNAs involved in wool follicle and skin development. (D) One-to-one pairs of differentially expressed lncRNA targets and mRNAs involved in key signaling pathways in wool follicle development. The orange and blue circles represent the number of differentially expressed lncRNA targets and mRNAs, respectively. Their intersections represent the number of differentially expressed targets of differentially expressed lncRNAs.
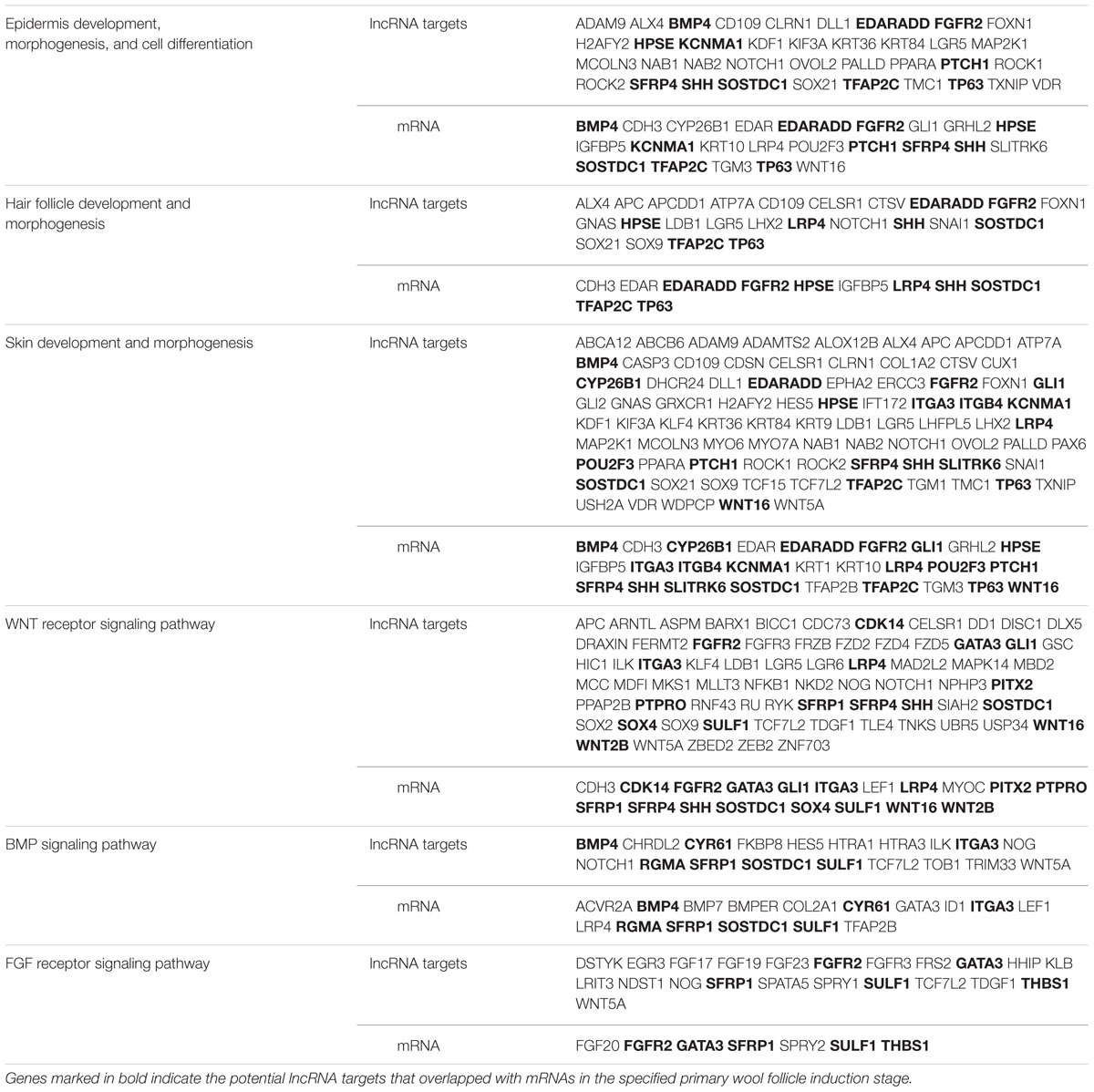
TABLE 3. Summary of differentially expressed long non-coding RNA (lncRNA) targets and mRNAs involved in primary wool follicle induction and skin development.
KEGG analyses revealed that these differentially expressed lncRNA target genes related to hair follicle and skin development were enriched in 32 significant terms, such as WNT, TGF-β, Hedgehog, and PI3K-Akt signaling pathways, focal adhesion, ECM-receptor interaction, and signaling pathways regulating pluripotency of stem cells (Figure 3A and Supplementary Table S5). KEGG analyses of differentially expressed mRNAs were also enriched in four of the five pathways mentioned above except for the WNT signaling pathway (Figure 3B and Supplementary Table S6).
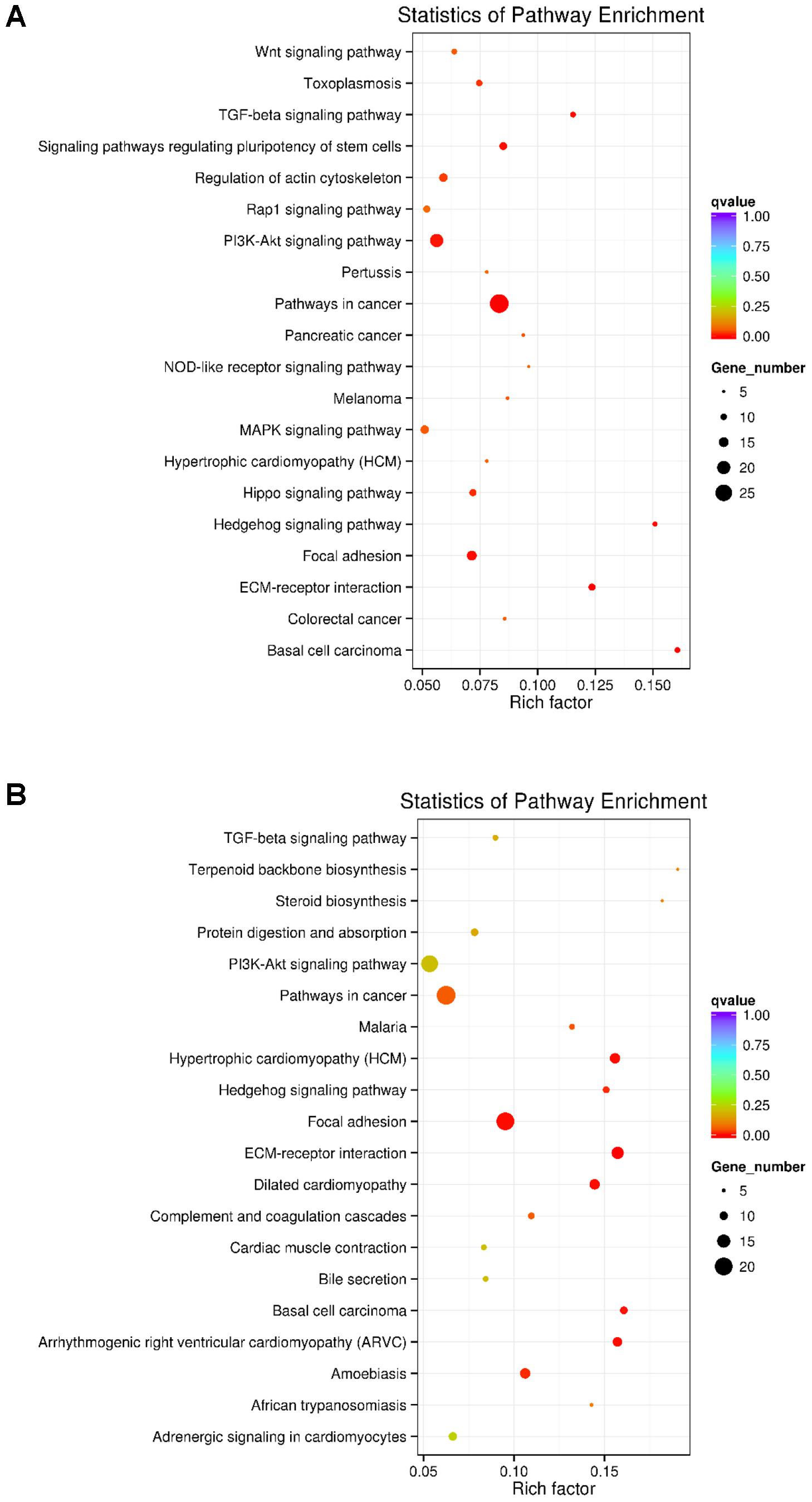
FIGURE 3. KEGG analysis of differentially expressed lncRNAs targets and mRNAs in primary wool follicle induction. (A,B) The top 20 KEGG enrichment pathways for differentially expressed lncRNA targets and mRNAs are presented. The longitudinal and horizontal axis represents the enrichment pathways and rich factor of these pathways, respectively. Spot size represents the number of differentially expressed genes enriched in each pathway, and the color of the spot represents the q-value of each pathway.
mRNA–mRNA and LncRNA–mRNA Interaction Network in Primary Wool Follicle Induction
A number of transcripts related to hair and skin development were applied to construct mRNA–mRNA and lncRNA–mRNA interaction networks. Of these, differentially expressed mRNAs were grouped and found to regulate the proliferation and differentiation of epidermal keratinocytes (KRT1, KRT4, KRT10, KRT15, and TP63), dermal-epidermal junction and basement membrane (COL17A1, LAMA1, LAMC2, LAMC3, ITGA3, ITGA8, and ITGB4), and dermal compartment (COL5A3 and COL2A1) (Pulkkinen et al., 1994; Kivirikko et al., 1995; McGrath et al., 1995; Mills et al., 1999; Imamura et al., 2000; Natsuga et al., 2011; Has et al., 2012; Talbot et al., 2016). Transcripts regulating hair placode and dermal condensation were also enriched and represented by the WNT (WNT2B, WNT16, SFRP1, SFRP4, SERPINF1, LEF1, and WIF1), EDAR (EDAR, EDARADD, and TRAF5), BMP (BMP4, BMP7, BMPER, ACVR2A, RGMA, and SOSTDC1), SHH (SHH, PTCH1, and GLI1), and FGF (FGFR2 and FGF20) signaling pathways (Figure 4A).
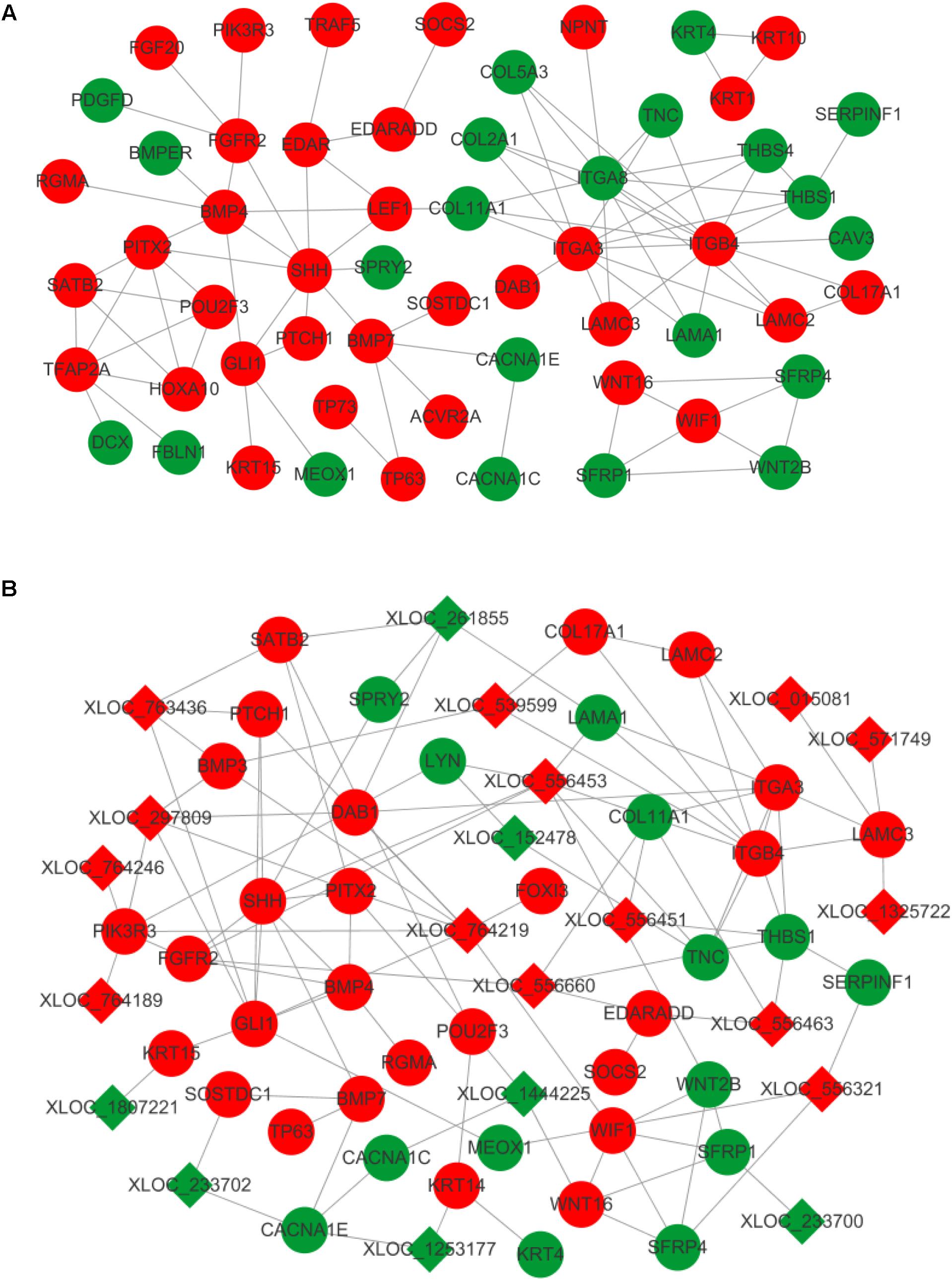
FIGURE 4. Interaction network in primary wool follicle induction. (A) mRNA–mRNA interaction network involved in hair follicle development was constructed and is presented. (B) lncRNA–mRNA interaction network related to hair follicle development is displayed. Red and green represent upregulated and downregulated, respectively. Circles and squares represent mRNAs and lncRNAs, respectively.
The altered lncRNAs were also highly correlated with development of the epidermis, dermis, dermal condensation, and wool placode. The elevated expression of lncRNAs was predicted to target laminins (LAMC3), collagens (COL17A1), WNT signals (WIF1 and SFRP4), BMP signals (BMP3 and BMP4), SHH signals (SHH, PTCH1, and GLI1), and EDAR signals (EDARADD and FOXI3). This group of lncRNAs may regulate epidermis, dermis, and placode formation through their targets. The downregulated lncRNAs were predicted to interact with keratins (KRT14 and KRT15), BMP signals (SOSTDC1), WNT signals (WNT16 and SFRP1), and laminins (LAMA1), to mainly influence epidermal and wool placode development. These results demonstrate a complex regulatory relationship between lncRNAs and mRNAs in primary wool follicle induction (Figure 4B).
Validation of Potential mRNAs and LncRNAs Functioning in Primary Wool Follicle Induction by qRT-PCR and in Situ Hybridization
Twelve mRNAs highly related to wool placode and dermal condensation formation (EDAR, EDARADD, FOXI3, SHH, SOSTDC1, BMP3, BMP4, BMP7, WNT16, WIF1, LEF1, and SMOC1) and four lncRNAs (XLOC_539599, XLOC_556463, XLOC_015081, and XLOC_1285606) possibly essential for primary wool follicle induction were selected and validated by qRT-PCR at the first two induction stages in wool follicle development. The qRT-PCR results of these mRNAs and lncRNAs were consistent with those from the sequencing data (Figure 5).
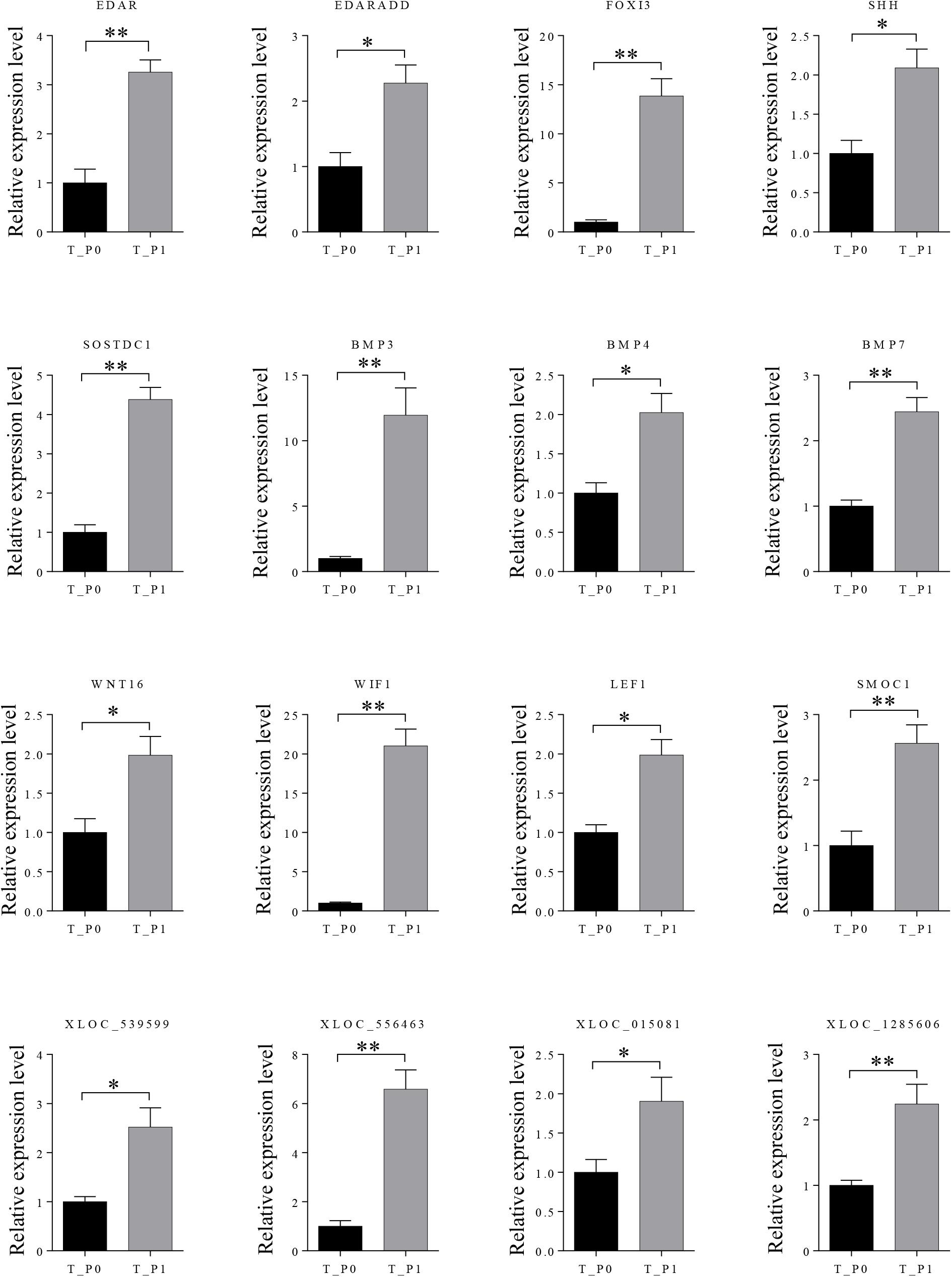
FIGURE 5. Expression levels of differently expressed mRNAs and lncRNAs involved in primary wool follicle induction were validated by qRT-PCR. Data are presented as mean ± SEM (n = 6). ∗p < 0.05, ∗∗p < 0.01 (Student’s t-test).
Two lncRNAs (XLOC_297809 and XLOC_764219), which potentially target BMP3 and FOXI3, were chosen to detect the expression patterns at stage 1 in primary wool follicle induction by in situ hybridization. Both lncRNAs were detected in wool placode and dermal condensation. XLOC_297809 was restricted in the placode and dermal condensation, whereas positive XLOC_764219 signals were strong in these two structures, but weaker in the epidermal layers (Figure 6). In negative controls hybridized with corresponding sense riboprobes, no hybridized signal was detected in the skin sections with either the XLOC_297809 or XLOC_764219 probe (Supplementary Figure S1).
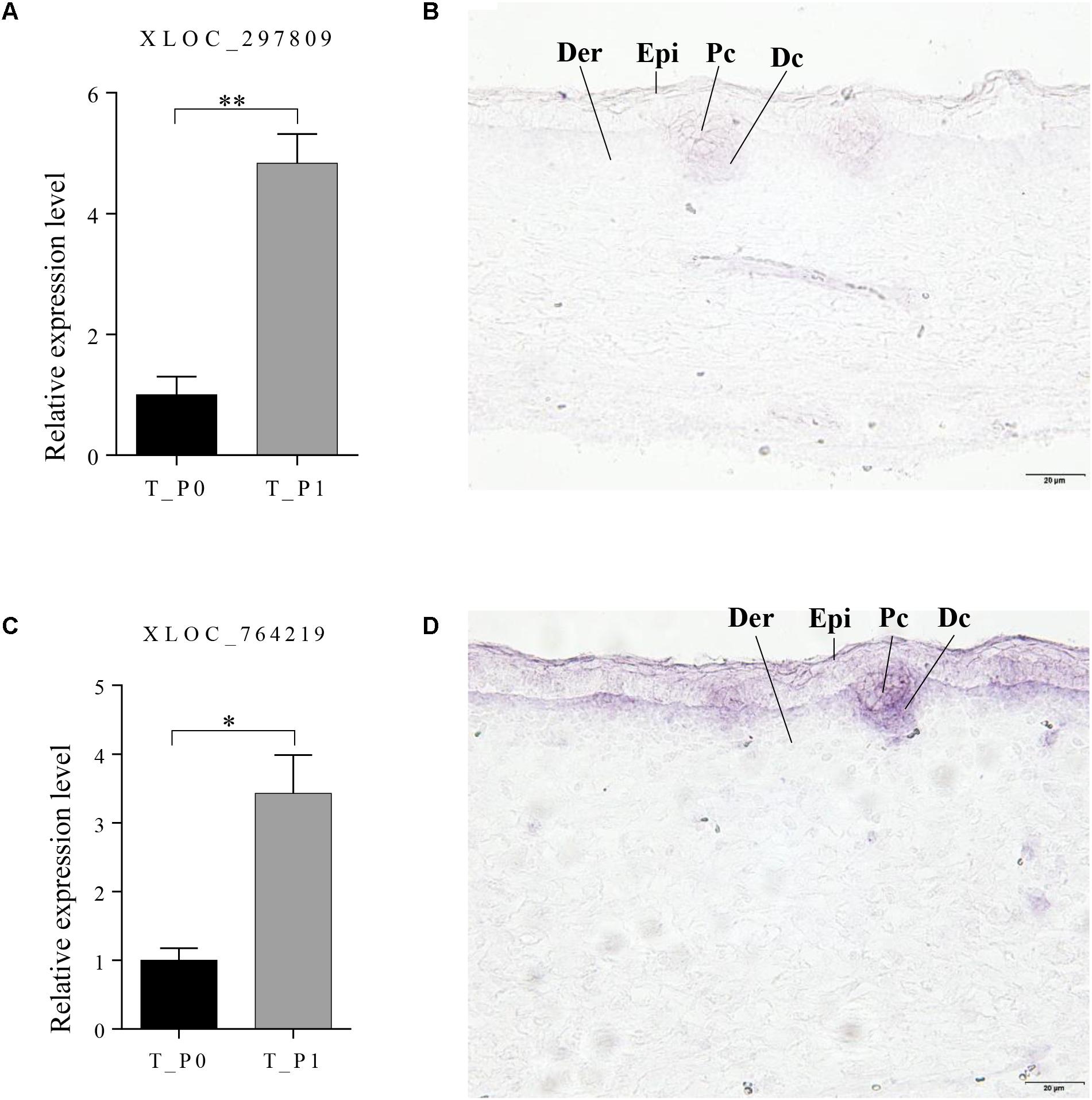
FIGURE 6. Expression pattern of XLOC_297809 and XLOC_764219 at stage 1 of primary wool follicle induction. (A,B) qRT-PCR and in situ hybridization of XLOC_297809. XLOC_297809 was expressed in placodes and dermal condensations. (C,D) qRT-PCR and in situ hybridization of XLOC_764219. XLOC_764219 was expressed in epidermis, placodes, and dermal condensations. Data are presented as the mean ± SEM (n = 6). ∗p < 0.05, ∗∗p < 0.01 (Student’s t-test). Skin sections at stage 1 of primary wool follicle development in sheep fetus were displayed at 400× (scale bars represent 20 μm). Epi, epidermis, Der, dermis, Pc, placode, Dc, dermal condensation.
Discussion
Carpet wool sheep are characterized by long and thick wool fiber which is developed from primary wool follicles. In sheep skin, the primordia of primary follicles develop in a trio pattern defined by one central and two lateral follicles and finally organize in a hexagonal pattern (Rogers, 2006). This occurs between E55 and E65 (Ruttle and Sorensen, 1965; du Cros et al., 1992; Wynn et al., 1995; Xavier et al., 2013), which is comparable to the developmental stages E13.5 to E14.5 of primary hair follicle in murine dorsal skin (Millar, 2002; Schmidt-Ullrich and Paus, 2005). During these particular primary follicle induction stages, murine and sheep skin share complicated anatomical changes including de novo formation of the wool placode and dermal condensation, as well as thickening of the epidermis and dermis, which are marked with different signature genes. The similar morphological changes and different duration of induction periods indicate that the mechanism regulating wool and hair primary follicle induction could be mostly conserved with subtle differences. It may also explain the differences observed in our data compared to mouse data. A previous RNA sequencing analysis identified 102, 395, and 254 signature genes specific for development of the placode, dermal condensation, and epidermis in E14.5 murine skin, respectively (Sennett et al., 2015). Of these, only 14, 29, and 19 genes were differentially expressed in the wool placode, dermal condensation, and epidermis in our data, respectively (Table 4). The remaining genes did not show significantly altered expression. Interestingly, a total of 27, 90, and 66 genes specific for these three structures, respectively, were not found in our data. This could be partially explained that the murine data showed slight differences among the different skin compartments at the hair placode stage, while our data generated from the whole skin compared gene regulation between two stages (Sennett et al., 2015). Another explanation might be due to some genes being restricted by expressing location without changing the expression levels. These enriched mRNAs in our data revealed that primary wool follicle induction shares a number of key genes driving morphological changes as those in murine primary hair follicles.

TABLE 4. List of significantly expressed signature genes in placode, dermal condensation, and epidermis.
The mRNA–mRNA interaction network showed two complex systems responsible for follicle placode and dermal condensation as well as epidermal differentiation and homeostasis, implying conserved signals functioned in both primary hair follicle and primary wool follicle induction. At the earliest stage of primary hair follicle morphogenesis, the communication between the epithelium and mesenchyme drives formation of the placode, dermal condensation niche, and thickened epidermis and dermis, which requires a cascade of gene networks including WNT, EDAR, BMP, and FGF signaling pathways (Paus et al., 1999; Millar, 2002; Huh et al., 2013; Glover et al., 2017).
Canonical WNT/β-catenin signaling is essential for hair follicle induction, as ectopically expressed WNT inhibitor Dickkopf 1 (Dkk1) or specifically knocked-out β-catenin in epidermis both resulted complete blockage of hair follicle induction (Huelsken et al., 2001; Andl et al., 2002). Two WNTs (WNT2B and WNT16) and other signaling molecules in WNT (SFRP1, SFRP4, LRP4, LEF1, and WIF1) were significantly enriched in our data, suggesting crucial roles of the WNT signaling pathway in primary wool follicle induction. Wnt2 was mainly expressed in mouse primary hair placode (Fu and Hsu, 2013). WNT2B, also known as WNT13, was significantly downregulated by approximately twofold at stage 1 in our data, which shares similar expression trend with WNT2 during the induction of secondary wool follicle development, as reported previously (Yue et al., 2016). The expression of WNT2B was stimulated in human keratinocytes following BMP2 application (Yang et al., 2006), indicating that WNT2B might act as a potential BMP target in skin and hair follicle development. In our data, WNT16 was upregulated by approximately twofold during primary wool follicle induction and may be regulated by XLOC_1444225. Wnt16 was mostly expressed in the inter-follicular epithelium, suggesting that it restricts the hair placode edge in the placode stage (Reddy et al., 2001). However, its actual function in early development has not been examined in detail, although it may function in hair follicle bulge stem cell regulation (Kandyba et al., 2013). WNT inhibitory factor 1 (Wif1) is a marker of dermal condensation that displays a spot-like expression pattern in the developing murine whisker follicles (Hsieh et al., 1999). WIF1, a potential target of XLOC_764219 in our data, was upregulated at the placode stage, indicating its importance in primary wool follicle dermal condensation formation. Unexpectedly, both WNT10A and WNT10B, which are important in early hair follicle formation (Reddy et al., 2001; Andl et al., 2002; Zhang et al., 2009), did not show altered expression in our sequencing data. Whether their expression location is changed rather than their expression levers requires further elucidation.
The activation of WNT/β-catenin signaling drives the homogenous epidermis to become competent and gain follicular or inter-follicular cellular fates forming rudimentary hair follicle placodes (Zhang et al., 2008). These follicle precursors need other signals to reinforce and shape the pre-pattern to finally establish the dense and clear-edged hair follicle placode. Our previous studies together with other reports confirmed that Edar functions as a key activator negotiating with the inhibitor, Bmp, to refine the pre-pattern into fine-patterned, well-organized hair follicle placode (Headon and Overbeek, 1999; Botchkarev et al., 2002; Mou et al., 2006). Extensive studies showed that the receptor Edar, not its ligand Eda, is a key stimulator for primary hair follicle induction, as overexpression of Edar in the epidermis could rescue primary hair follicle formation in a cross of Edar transgenic mice with Eda mutant mice (Mou et al., 2008). The elevated expression of EDAR and its intracellular adaptor EDARADD indicates their important roles in wool placode induction. EDA did not show convincing expression change supporting the notion that EDA is not a key signal during the induction stage. Foxi3, a potential target of XLOC_764219, is a downstream effector of Eda in hair placode formation. However, unlike other molecules involved in EDA/EDAR/NF-kB signaling, Foxi3 knockout mice have normal primary hair placodes (Shirokova et al., 2013, 2016). Mostly likely, EDA, and FOXI3 are not key signals that stimulate primary wool follicle induction. These results suggest that the EDAR signaling pathway is important for primary wool follicle induction. It would be interesting to explore sheep EDAR function in later developmental stages.
Edar and Bmps act as a stimulator and inhibitor, respectively, in primary hair follicle formation as proposed in our and other previous reports (Headon and Overbeek, 1999; Mou et al., 2006; Pummila et al., 2007). BMPs have been extensively studied in early and late development as well as during cycling growth of hair follicles (Botchkarev et al., 2002; Botchkarev and Sharov, 2004; Guha et al., 2004; Plikus et al., 2008). Application of BMP or its antagonist, NOGGIN in skin cultures abolished the occurrence of hair placodes or generated normal hair placodes with increased density, respectively (Mou et al., 2006). These results together with other genetically modified mouse models revealed the general repressive roles of BMP signals in hair follicle induction and development. The observation of significant upregulation of the dermal condensation markers, BMP3, BMP4, and BMP7 at stage 1 in our data, implied that BMPs also contribute to the formation of primary wool follicle dermal condensation (Takahashi and Ikeda, 1996). The similar expression patterns of EDAR, BMP4, and BMP7 in our data suggest that the balance of activator and inhibitor functioned in murine primary hair follicle induction may be conserved in primary wool follicle induction (Headon and Overbeek, 1999; Mou et al., 2006; Pummila et al., 2007).
Though the mechanisms underlying murine hair follicle induction have been well studied, lncRNAs functioned in hair follicle and skin development remain largely unclear. We identified 62 differentially expressed lncRNAs and predicted their potential targets which partially overlapped with differentially expressed mRNAs and were highly enriched in pathways including WNT, TGF-β, Hedgehog, PI3K-Akt, focal adhesion, and ECM-receptor interaction. These pathways were found to be important in hair and feather follicle morphogenesis and skin development (Jung et al., 1998; Noramly and Morgan, 1998; Huelsken et al., 2001; Andl et al., 2002; Pummila et al., 2007; Michon et al., 2008; Xu et al., 2013; Zhu et al., 2013; Pan et al., 2015). Of these predicted targets, EDARADD, FGFR2, HPSE, LRP4, SHH, SOSTDC1, TFAP2C, and TP63 were differentially expressed during induction of primary wool follicles and were found to play important roles in hair follicle development and morphogenesis (Table 3; Sutton and van Bokhoven, 1993; Li et al., 2001; Thesleff and Mikkola, 2002; Wang et al., 2008; Augusto et al., 2011; Cui et al., 2011; Narhi et al., 2012; Ahn et al., 2013; Mukhopadhyay et al., 2013).
In addition to potential marker genes of wool placode and dermal condensation discussed above, a cascade of genes related to epidermal differentiation and homeostasis were grouped, indicating the specification of granular and spinous layers of the epidermis in sheep skin. This is consistent with the anatomical changes between these two stages. When primary wool follicles formed at stage 1, the epidermis thickened dramatically from one or two homogenous layers to differentiated and stratified layers. TP63, a homologue of tumor suppressor P53, is a key regulator of epidermal development, as Tp63 deficient mice showed blocked epidermal proliferation and stratification and no hair follicle formation in early embryonic stages (Mills et al., 1999). Upregulation of TP63 in stage 1 suggests its important role in the embryonic epidermal development of sheep skin. Elevated expression of three type II keratins, KRT1, KRT4, and KRT10, were also highly correlated with epidermal differentiation and stratification at stage 1 of wool placode skin samples. Of these, KRT1 and KRT10 are two specific keratins enriched in differentiated and stratified spinous and granular layers of the skin epidermis. One type I keratin, KRT15, which was enriched in stage 1 samples and potentially regulated by XLOC_1807221, is mostly expressed in the hair placode and epidermis of murine E14.5 skin. Another keratin, KRT14, which marks the epidermal basal layer, showed elevated expression, possibly regulated by XLOC_1253177. A group of genes is involved in the adhesion of the basal epidermal sheet to the basement membrane including those encoding three laminins, three integrins, and one collagen (Col17a1) (McGrath et al., 1995). Two collagens, COL5A3 and COL2A1, showed decreased expression, indicating their potential roles in development of the fetal sheep skin dermis. This interaction network provides a complex picture of wool follicle initiation driven by crosstalk between mRNAs and lncRNAs and strongly supports the view that these differentially expressed lncRNAs and mRNAs play important roles in sheep primary wool follicle and skin development. Additional studies are needed to determine the precise function of this small group of lncRNAs and mRNAs in wool follicle induction.
This investigation delineated global gene expression changes in mRNAs and lncRNAs during primary wool follicle induction. The fundamental knowledge of early development of primary wool follicles established in this report not only improve the understanding of wool follicle and skin development and potential refinement of wool quality, but also facilitates use of wool follicles as an additional system to assist human skin health and care. The wool developed from primary wool follicles in coarse and fine wool sheep displays very large differences in fiber thickness. The former is long and thick, whereas the latter is short and thin. It would be interesting to further elucidate the mechanisms underlying the differences between coarse and fine wool and contribute to selective breeding of wool sheep, particularly for fiber thickness.
Author Contributions
CM designed the experiments and wrote the manuscript. YN wrote the manuscript, analyzed the data, and performed the qPCR and in situ validation. WC, XZ, XL, SL, and MY were involved in sample collection and staining. ZL, YH, HQ, QQ, QP, and DC were involved in sample collection and processing.
Funding
This work was funded by Special Funds for Basic Research Projects in Central Universities (2662015PY007) and grants from the National High Technology Research and Development Program (“863” Program) of China (2013AA102506).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ARAM and handling Editor declared their shared affiliation.
Acknowledgments
We thank Dr. Liqing Ma and Mr. Xiaoqiang Zhang, and all the facilities involved including Animal Husbandry and Veterinary Station, as well as the local abattoir for their support during this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00446/full#supplementary-material
FIGURE S1 | Negative controls for in situ hybridization are presented applying with sense riboprobes of either XLOC_297809 (A) or XLOC_764219 (B) at stage 1 of primary wool follicle induction. In the negative controls, no hybridized signal was detected in sense riboprobes of either XLOC_297809 or XLOC_764219. Skin sections at stage 1 of primary wool follicle development in sheep fetus are shown at 400× (scale bars represent 20 μm). Epi, epidermis, Der, dermis, Pc, placode, Dc, dermal condensation.
TABLE S1 | Differentially expressed transcripts between stage 0 and stage 1 of primary wool follicle induction in carpet wool sheep fetal skin.
TABLE S2 | Annotation of targeted genes of differentially expressed lncRNAs.
TABLE S3 | Significantly enriched GO terms of differentially expressed lncRNA target genes.
TABLE S4 | Significantly enriched GO terms of differentially expressed mRNAs.
TABLE S5 | Significantly enriched KEGG pathways of differentially expressed lncRNA target genes related to hair follicle and skin development.
TABLE S6 | Significantly enriched KEGG pathways of differentially expressed mRNAs.
Footnotes
References
Ahn, Y., Sims, C., Logue, J. M., Weatherbee, S. D., and Krumlauf, R. (2013). Lrp4 and Wise interplay controls the formation and patterning of mammary and other skin appendage placodes by modulating Wnt signaling. Development 140, 583–593. doi: 10.1242/dev.085118
Andl, T., Reddy, S. T., Gaddapara, T., and Millar, S. E. (2002). WNT signals are required for the initiation of hair follicle development. Dev. Cell 2, 643–653. doi: 10.1016/S1534-5807(02)00167-3
Augusto, T. M., Rosa-Ribeiro, R., and Carvalho, H. F. (2011). Neonatal exposure to high doses of 17beta-estradiol results in inhibition of heparanase-1 expression in the adult prostate. Histochem. Cell Biol. 136, 609–615. doi: 10.1007/s00418-011-0860-9
Botchkarev, V. A., Botchkareva, N. V., Sharov, A. A., Funa, K., Huber, O., and Gilchrest, B. A. (2002). Modulation of BMP signaling by noggin is required for induction of the secondary (nontylotrich) hair follicles. J. Invest. Dermatol. 118, 3–10. doi: 10.1046/j.1523-1747.2002.01645.x
Botchkarev, V. A., and Sharov, A. A. (2004). BMP signaling in the control of skin development and hair follicle growth. Differentiation 72, 512–526. doi: 10.1111/j.1432-0436.2004.07209005.x
Cui, C. Y., Kunisada, M., Childress, V., Michel, M., and Schlessinger, D. (2011). Shh is required for Tabby hair follicle development. Cell Cycle 10, 3379–3386. doi: 10.4161/cc.10.19.17669
du Cros, D. L., Isaacs, K., and Moore, G. P. (1992). Localization of epidermal growth factor immunoreactivity in sheep skin during wool follicle development. J. Invest. Dermatol. 98, 109–115.
Fu, J., and Hsu, W. (2013). Epidermal Wnt controls hair follicle induction by orchestrating dynamic signaling crosstalk between the epidermis and dermis. J. Invest. Dermatol. 133, 890–898. doi: 10.1038/jid.2012.407
Glover, J. D., Wells, K. L., Matthaus, F., Painter, K. J., Ho, W., Riddell, J., et al. (2017). Hierarchical patterning modes orchestrate hair follicle morphogenesis. PLoS Biol. 15:e2002117. doi: 10.1371/journal.pbio.2002117
Guha, U., Mecklenburg, L., Cowin, P., Kan, L., O’Guin, W. M., D’Vizio, D., et al. (2004). Bone morphogenetic protein signaling regulates postnatal hair follicle differentiation and cycling. Am. J. Pathol. 165, 729–740. doi: 10.1016/s0002-9440(10)63336-6
Has, C., Sparta, G., Kiritsi, D., Weibel, L., Moeller, A., Vega-Warner, V., et al. (2012). Integrin alpha3 mutations with kidney, lung, and skin disease. N. Engl. J. Med. 366, 1508–1514. doi: 10.1056/NEJMoa1110813
Headon, D. J., and Overbeek, P. A. (1999). Involvement of a novel Tnf receptor homologue in hair follicle induction. Nat. Genet. 22, 370–374. doi: 10.1038/11943
Hsieh, J. C., Kodjabachian, L., Rebbert, M. L., Rattner, A., Smallwood, P. M., Samos, C. H., et al. (1999). A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature 398, 431–436. doi: 10.1038/18899
Huelsken, J., Vogel, R., Erdmann, B., Cotsarelis, G., and Birchmeier, W. (2001). beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 105, 533–545. doi: 10.1016/S0092-8674(01)00336-1
Huh, S. H., Narhi, K., Lindfors, P. H., Haara, O., Yang, L., Ornitz, D. M., et al. (2013). Fgf20 governs formation of primary and secondary dermal condensations in developing hair follicles. Genes Dev. 27, 450–458. doi: 10.1101/gad.198945.112
Imamura, Y., Scott, I. C., and Greenspan, D. S. (2000). The pro-alpha3(V) collagen chain. Complete primary structure, expression domains in adult and developing tissues, and comparison to the structures and expression domains of the other types V and XI procollagen chains. J. Biol. Chem. 275, 8749–8759. doi: 10.1074/jbc.275.12.8749
Jung, H. S., Francis-West, P. H., Widelitz, R. B., Jiang, T. X., Ting-Berreth, S., Tickle, C., et al. (1998). Local inhibitory action of BMPs and their relationships with activators in feather formation: implications for periodic patterning. Dev. Biol. 196, 11–23. doi: 10.1006/dbio.1998.8850
Kandyba, E., Leung, Y., Chen, Y. B., Widelitz, R., Chuong, C. M., and Kobielak, K. (2013). Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proc. Natl. Acad. Sci. U.S.A. 110, 1351–1356. doi: 10.1073/pnas.1121312110
Kivirikko, S., McGrath, J. A., Baudoin, C., Aberdam, D., Ciatti, S., Dunnill, M. G., et al. (1995). A homozygous nonsense mutation in the alpha 3 chain gene of laminin 5 (LAMA3) in lethal (Herlitz) junctional epidermolysis bullosa. Hum. Mol. Genet. 4, 959–962. doi: 10.1093/hmg/4.5.959
Kong, L., Zhang, Y., Ye, Z. Q., Liu, X. Q., Zhao, S. Q., Wei, L., et al. (2007). CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345–W349. doi: 10.1093/nar/gkm391
Kornienko, A. E., Guenzl, P. M., Barlow, D. P., and Pauler, F. M. (2013). Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11:59. doi: 10.1186/1741-7007-11-59
Lee, D., Xiong, S., and Xiong, W. C. (2013). General introduction to in situ hybridization protocol using nonradioactively labeled probes to detect mRNAs on tissue sections. Methods Mol. Biol. 1018, 165–174. doi: 10.1007/978-1-62703-444-9_16
Li, C., Guo, H., Xu, X., Weinberg, W., and Deng, C. X. (2001). Fibroblast growth factor receptor 2 (Fgfr2) plays an important role in eyelid and skin formation and patterning. Dev. Dyn. 222, 471–483. doi: 10.1002/dvdy.1205
Lin, C. M., Liu, Y., Huang, K., Chen, X. C., Cai, B. Z., Li, H. H., et al. (2014). Long noncoding RNA expression in dermal papilla cells contributes to hairy gene regulation. Biochem. Biophys. Res. Commun. 453, 508–514. doi: 10.1016/j.bbrc.2014.09.119
Lin, M. F., Jungreis, I., and Kellis, M. (2011). PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275–i282. doi: 10.1093/bioinformatics/btr209
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
McGrath, J. A., Gatalica, B., Christiano, A. M., Li, K., Owaribe, K., McMillan, J. R., et al. (1995). Mutations in the 180-kD bullous pemphigoid antigen (BPAG2), a hemidesmosomal transmembrane collagen (COL17A1), in generalized atrophic benign epidermolysis bullosa. Nat. Genet. 11, 83–86. doi: 10.1038/ng0995-83
Michon, F., Forest, L., Collomb, E., Demongeot, J., and Dhouailly, D. (2008). BMP2 and BMP7 play antagonistic roles in feather induction. Development 135, 2797–2805. doi: 10.1242/dev.018341
Millar, S. E. (2002). Molecular mechanisms regulating hair follicle development. J Invest. Dermatol. 118, 216–225. doi: 10.1046/j.0022-202x.2001.01670.x
Mills, A. A., Zheng, B., Wang, X. J., Vogel, H., Roop, D. R., and Bradley, A. (1999). p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713. doi: 10.1038/19531
Mou, C., Jackson, B., Schneider, P., Overbeek, P. A., and Headon, D. J. (2006). Generation of the primary hair follicle pattern. Proc. Natl. Acad. Sci. U.S.A. 103, 9075–9080. doi: 10.1073/pnas.0600825103
Mou, C., Thomason, H. A., Willan, P. M., Clowes, C., Harris, W. E., Drew, C. F., et al. (2008). Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Hum. Mutat. 29, 1405–1411. doi: 10.1002/humu.20795
Mukhopadhyay, A., Krishnaswami, S. R., Cowing-Zitron, C., Hung, N. J., Reilly-Rhoten, H., Burns, J., et al. (2013). Negative regulation of Shh levels by Kras and Fgfr2 during hair follicle development. Dev. Biol. 373, 373–382. doi: 10.1016/j.ydbio.2012.10.024
Muller-Rover, S., Handjiski, B., van der Veen, C., Eichmuller, S., Foitzik, K., McKay, I. A., et al. (2001). A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol. 117, 3–15. doi: 10.1046/j.0022-202x.2001.01377.x
Nakamura, M., Schneider, M. R., Schmidt-Ullrich, R., and Paus, R. (2013). Mutant laboratory mice with abnormalities in hair follicle morphogenesis, cycling, and/or structure: an update. J. Dermatol. Sci. 69, 6–29. doi: 10.1016/j.jdermsci.2012.10.001
Narhi, K., Tummers, M., Ahtiainen, L., Itoh, N., Thesleff, I., and Mikkola, M. L. (2012). Sostdc1 defines the size and number of skin appendage placodes. Dev. Biol. 364, 149–161. doi: 10.1016/j.ydbio.2012.01.026
Natsuga, K., Nishie, W., Shinkuma, S., Nakamura, H., Arita, K., Yoneda, K., et al. (2011). A founder effect of c.1938delC in ITGB4 underlies junctional epidermolysis bullosa and its application for prenatal testing. Exp. Dermatol. 20, 74–76. doi: 10.1111/j.1600-0625.2010.01177.x
Noramly, S., and Morgan, B. A. (1998). BMPs mediate lateral inhibition at successive stages in feather tract development. Development 125, 3775–3787.
Pan, L., Liu, Y., Wei, Q., Xiao, C., Ji, Q., Bao, G., et al. (2015). Solexa-sequencing based transcriptome study of plaice skin phenotype in rex rabbits (Oryctolagus cuniculus). PLoS One 10:e0124583. doi: 10.1371/journal.pone.0124583
Paus, R., Muller-Rover, S., Van Der Veen, C., Maurer, M., Eichmuller, S., Ling, G., et al. (1999). A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J. Invest. Dermatol. 113, 523–532. doi: 10.1046/j.1523-1747.1999.00740.x
Plikus, M. V., Mayer, J. A., de la Cruz, D., Baker, R. E., Maini, P. K., Maxson, R., et al. (2008). Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 451, 340–344. doi: 10.1038/nature06457
Pulkkinen, L., Christiano, A. M., Airenne, T., Haakana, H., Tryggvason, K., and Uitto, J. (1994). Mutations in the gamma 2 chain gene (LAMC2) of kalinin/laminin 5 in the junctional forms of epidermolysis bullosa. Nat. Genet. 6, 293–297. doi: 10.1038/ng0394-293
Pummila, M., Fliniaux, I., Jaatinen, R., James, M. J., Laurikkala, J., Schneider, P., et al. (2007). Ectodysplasin has a dual role in ectodermal organogenesis: inhibition of Bmp activity and induction of Shh expression. Development 134, 117–125. doi: 10.1242/dev.02708
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2012). The Pfam protein families database. Nucleic Acids Res. 40, D290–D301. doi: 10.1093/nar/gkr1065
Reddy, S., Andl, T., Bagasra, A., Lu, M. M., Epstein, D. J., Morrisey, E. E., et al. (2001). Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of Sonic hedgehog in hair follicle morphogenesis. Mech. Dev. 107, 69–82. doi: 10.1016/S0925-4773(01)00452-X
Rezza, A., Wang, Z., Sennett, R., Qiao, W., Wang, D., Heitman, N., et al. (2016). Signaling networks among stem cell precursors, transit-amplifying progenitors, and their niche in developing hair follicles. Cell Rep. 14, 3001–3018. doi: 10.1016/j.celrep.2016.02.078
Rishikaysh, P., Dev, K., Diaz, D., Qureshi, W. M., Filip, S., and Mokry, J. (2014). Signaling involved in hair follicle morphogenesis and development. Int. J. Mol. Sci. 15, 1647–1670. doi: 10.3390/ijms15011647
Rogers, G. E. (2006). Biology of the wool follicle: an excursion into a unique tissue interaction system waiting to be re-discovered. Exp. Dermatol. 15, 931–949. doi: 10.1111/j.1600-0625.2006.00512.x
Ruttle, J. L., and Sorensen, A. M. (1965). Prenatal development of the wool follicles in rambouillet sheep1. J. Anim. Sci. 24, 69–75. doi: 10.2527/jas1965.24169x
Saga, K. (2002). Structure and function of human sweat glands studied with histochemistry and cytochemistry. Prog. Histochem. Cytochem. 37, 323–386. doi: 10.1016/S0079-6336(02)80005-5
Schmidt-Ullrich, R., and Paus, R. (2005). Molecular principles of hair follicle induction and morphogenesis. Bioessays 27, 247–261. doi: 10.1002/bies.20184
Sennett, R., Wang, Z., Rezza, A., Grisanti, L., Roitershtein, N., Sicchio, C., et al. (2015). An integrated transcriptome atlas of embryonic hair follicle progenitors, their niche, and the developing skin. Dev. Cell 34, 577–591. doi: 10.1016/j.devcel.2015.06.023
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shirokova, V., Biggs, L. C., Jussila, M., Ohyama, T., Groves, A. K., and Mikkola, M. L. (2016). Foxi3 deficiency compromises hair follicle stem cell specification and activation. Stem Cells 34, 1896–1908. doi: 10.1002/stem.2363
Shirokova, V., Jussila, M., Hytonen, M. K., Perala, N., Drogemuller, C., Leeb, T., et al. (2013). Expression of Foxi3 is regulated by ectodysplasin in skin appendage placodes. Dev. Dyn. 242, 593–603. doi: 10.1002/dvdy.23952
Siepel, A., Bejerano, G., Pedersen, J. S., Hinrichs, A. S., Hou, M., Rosenbloom, K., et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050. doi: 10.1101/gr.3715005
Sun, L., Luo, H., Bu, D., Zhao, G., Yu, K., Zhang, C., et al. (2013). Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 41:e166. doi: 10.1093/nar/gkt646
Sutton, V. R., and van Bokhoven, H. (1993). “TP63-related disorders,” in GeneReviews(R), eds M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. H. Bean, H. C. Mefford, et al. (Seattle, WA: University of Washington).
Takahashi, H., and Ikeda, T. (1996). Transcripts for two members of the transforming growth factor-beta superfamily BMP-3 and BMP-7 are expressed in developing rat embryos. Dev. Dyn. 207, 439–449. doi: 10.1002/(SICI)1097-0177(199612)207:4<439::AID-AJA8>3.0.CO;2-I
Talbot, J. C., Nichols, J. T., Yan, Y. L., Leonard, I. F., BreMiller, R. A., Amacher, S. L., et al. (2016). Pharyngeal morphogenesis requires fras1-itga8-dependent epithelial-mesenchymal interaction. Dev. Biol. 416, 136–148. doi: 10.1016/j.ydbio.2016.05.035
Thesleff, I., and Mikkola, M. L. (2002). Death receptor signaling giving life to ectodermal organs. Sci STKE 2002:pe22. doi: 10.1126/stke.2002.131.pe22
Wan, D. C., and Wang, K. C. (2014). Long noncoding RNA: significance and potential in skin biology. Cold Spring Harb. Perspect. Med. 4:a015404. doi: 10.1101/cshperspect.a015404
Wang, X., Pasolli, H. A., Williams, T., and Fuchs, E. (2008). AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J. Cell Biol. 183, 37–48. doi: 10.1083/jcb.200804030
Wynn, P. C., Brown, G., and Moore, G. P. (1995). Characterization and distribution of epidermal growth factor receptors in the skin and wool follicles of the sheep fetus during development. Domest. Anim. Endocrinol. 12, 269–281. doi: 10.1016/0739-7240(95)00024-9
Xavier, S. P., Gordon-Thomson, C., Wynn, P. C., McCullagh, P., Thomson, P. C., Tomkins, L., et al. (2013). Evidence that Notch and Delta expressions have a role in dermal condensate aggregation during wool follicle initiation. Exp. Dermatol. 22, 659–662. doi: 10.1111/exd.12217
Xu, T., Guo, X., Wang, H., Hao, F., Du, X., Gao, X., et al. (2013). Differential gene expression analysis between anagen and telogen of Capra hircus skin based on the de novo assembled transcriptome sequence. Gene 520, 30–38. doi: 10.1016/j.gene.2013.01.068
Yang, L., Yamasaki, K., Shirakata, Y., Dai, X., Tokumaru, S., Yahata, Y., et al. (2006). Bone morphogenetic protein-2 modulates Wnt and frizzled expression and enhances the canonical pathway of Wnt signaling in normal keratinocytes. J. Dermatol. Sci. 42, 111–119. doi: 10.1016/j.jdermsci.2005.12.011
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Yue, Y., Guo, T., Yuan, C., Liu, J., Guo, J., Feng, R., et al. (2016). Integrated analysis of the roles of long noncoding RNA and coding RNA expression in sheep (Ovis aries) skin during initiation of secondary hair follicle. PLoS One 11:e0156890. doi: 10.1371/journal.pone.0156890
Zhang, Y., Andl, T., Yang, S. H., Teta, M., Liu, F., Seykora, J. T., et al. (2008). Activation of beta-catenin signaling programs embryonic epidermis to hair follicle fate. Development 135, 2161–2172. doi: 10.1242/dev.017459
Zhang, Y., Tomann, P., Andl, T., Gallant, N. M., Huelsken, J., Jerchow, B., et al. (2009). Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Dev. Cell 17, 49–61. doi: 10.1016/j.devcel.2009.05.011
Keywords: wool follicle, induction, expression pattern, skin development, long non-coding RNA
Citation: Nie Y, Li S, Zheng X, Chen W, Li X, Liu Z, Hu Y, Qiao H, Qi Q, Pei Q, Cai D, Yu M and Mou C (2018) Transcriptome Reveals Long Non-coding RNAs and mRNAs Involved in Primary Wool Follicle Induction in Carpet Sheep Fetal Skin. Front. Physiol. 9:446. doi: 10.3389/fphys.2018.00446
Received: 24 October 2017; Accepted: 10 April 2018;
Published: 15 May 2018.
Edited by:
Timothy J. Moss, The Ritchie Centre, AustraliaReviewed by:
Annie Rene Alison McDougall, Hudson Institute of Medical Research, AustraliaAdam John Watkins, University of Nottingham, United Kingdom
Copyright © 2018 Nie, Li, Zheng, Chen, Li, Liu, Hu, Qiao, Qi, Pei, Cai, Yu and Mou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunyan Mou, chunyanmou@mail.hzau.edu.cn
 Yangfan Nie1
Yangfan Nie1 Chunyan Mou
Chunyan Mou