- National Institute of Plant Genome Research, New Delhi, India
Drought is one of the leading factors responsible for the reduction in crop yield worldwide. Due to climate change, in future, more areas are going to be affected by drought and for prolonged periods. Therefore, understanding the mechanisms underlying the drought response is one of the major scientific concerns for improving crop yield. Plants deploy diverse strategies and mechanisms to respond and tolerate drought stress. Expression of numerous genes is modulated in different plants under drought stress that help them to optimize their growth and development. Plant hormone abscisic acid (ABA) plays a major role in plant response and tolerance by regulating the expression of many genes under drought stress. Transcription factors being the major regulator of gene expression play a crucial role in stress response. ABA regulates the expression of most of the target genes through ABA-responsive element (ABRE) binding protein/ABRE binding factor (AREB/ABF) transcription factors. Genes regulated by AREB/ABFs constitute a regulon termed as AREB/ABF regulon. In addition to this, drought responsive genes are also regulated by ABA-independent mechanisms. In ABA-independent regulation, dehydration-responsive element binding protein (DREB), NAM, ATAF, and CUC regulons play an important role by regulating many drought-responsive genes. Apart from these major regulons, MYB/MYC, WRKY, and nuclear factor-Y (NF-Y) transcription factors are also involved in drought response and tolerance. Our understanding about transcriptional regulation of drought is still evolving. Recent reports have suggested the existence of crosstalk between different transcription factors operating under drought stress. In this article, we have reviewed various regulons working under drought stress and their crosstalk with each other.
Introduction
Plants being sessile organisms frequently encounter a wide range of unfavorable conditions during their life cycle. These conditions have deleterious effects on their physiology leading to reduced growth and development. Such adverse conditions along with some other factors play a crucial role in determining the yield and geographical distribution of plants. These different unfavorable conditions are generally termed as stress.
Plants face both abiotic as well as biotic stresses during their life cycle. Various climatic factors such as extreme temperature, drought, salinity, and chemical contamination of soil fall in the category of abiotic stresses. However, stresses caused by various pathogens and other biological agents are grouped into biotic stress. Both kinds of stresses have detrimental effects on plants. Especially, abiotic stresses alter various cellular processes such as photosynthesis, growth, carbon partitioning, carbohydrate and lipid metabolism, protein synthesis, gene expression, and osmotic homeostasis. Thus, to survive under stress conditions, plants have evolved a wide range of mechanisms to avoid or tolerate these stresses.
Drought is an imperative factor limiting the crop productivity across the globe (Bray et al., 2000). It can be characterized by below normal precipitation for a certain period of months to year and drying winds leading to reduced soil water available to plants. In addition to this, it is generally accompanied with high temperature. In recent years, frequency of drought stress has increased due to irregular rain fall. Almost every year drought occurs in some part of the earth reducing the crop yield. The condition is going to be worse in coming years due to global warming responsible for increasing desertification. On the other hand, world population is anticipated to reach 9 billion by 2050 (http://www.fao.org/wsfs/world-summit/en). Taking into consideration the increase in population, it is important to increase crop yield. Therefore, it is important to understand the mechanism of drought stress tolerance in plants in order to improve crop productivity under stress conditions.
Understanding the mechanism underlying drought stress tolerance has been an active area of research. Till date, drought stress responses have been studied in various plants; including crops, vegetables, trees as well as horticulture plants. Plants respond to environmental stresses at various levels such as cellular responses, metabolic changes, molecular adaptations as well as epigenetic regulation (Krasensky and Jonak, 2012). Although, plant response to drought has been analyzed at all these levels but in past few years focus has shifted toward the molecular mechanism. Drought stress affects the expression of many genes. Most of the molecular studies have been done using Arabidopsis thaliana as a model plant (Ingram and Bartels, 1996; Shinozaki and Yamaguchi-Shinozaki, 2000). Genome sequence of Arabidopsis has provided valuable information pertaining genes, gene families, cis elements and other related factors; resulting in rapid progress regarding molecular responses of plants to drought (Hirayama and Shinozaki, 2010). Later on, in addition to genomics, incorporation of advanced omics approaches such as transcriptomics, proteomics and metabolomics have increased our knowledge in this area (Hirayama and Shinozaki, 2010).
During drought and other osmotic stresses, the phytohormone ABA (abscisic acid) plays a pivotal role in plant adaptation. Effect of ABA on plant response to stress has been extensively researched. ABA is accumulated under drought stress condition due to induction of ABA biosynthetic genes (Iuchi et al., 2001). ABA regulates the expression of many genes leading to some important physiological as well as biochemical changes that help plant to survive under stress (Umezawa et al., 2010). Molecular and genomic analyses have revealed the existence of ABA-independent signal transduction pathway in conjunction to the ABA-dependent signal transduction pathway during drought stress (Yamaguchi-Shinozaki and Shinozaki, 2006).
Transcriptional Regulatory Network
Plants respond to various environmental stresses including drought through changes ranging from physiological to molecular level. These changes help plants to optimize their growth and stress resistance. Drought stress changes the expression of many genes that are thought to play an important role in stress response and tolerance. Many of these genes have been identified and characterized (Yamaguchi-Shinozaki and Shinozaki, 2006; Todaka et al., 2015). Microarray analyses by various groups have revealed thousands of genes that are upregulated and downregulated in response to drought stress. A significant number of drought-inducible genes are also induced by high salinity, suggesting a cross-talk between drought and salt stress. Comparatively lesser number of drought-inducible genes are induced by cold stress (Yamaguchi-Shinozaki and Shinozaki, 2006). There is a very small overlap of only 27 genes that were found to be commonly induced in microarray studies (Bray, 2004). This lack of commonality may be attributed to the fact that different sets of probes were used during these microarray experiments and variations in conditions of plant growth and stress. Recently, 17 microarray experiments of Arabidopsis, rice, wheat, and barley were compared using a novel Cross-Species meta-Analysis of progressive Drought stress at the reproductive stage (CSA:Drought); and 225 differentially expressed genes were identified that were shared across studies and taxa (Shaar-Moshe et al., 2015).
Stress inducible genes in Arabidopsis can be classified into two categories: functional and regulatory genes (Yamaguchi-Shinozaki and Shinozaki, 2006). Genes encoding proteins required for cellular stress tolerance fall into the former category, for example, LEA (late embryogenesis abundant) proteins, molecular chaperones, reactive oxygen species detoxifying enzymes, and sugars or proline biosynthetic enzymes. Whereas, genes encoding proteins that are involved in signal transduction and gene expression come under the latter category, such as protein kinases, components of ABA signaling, enzymes for lipid signaling, and various transcription factors (Yamaguchi-Shinozaki and Shinozaki, 2006).
As stated above, plant hormone ABA plays an important role in response to water deficit including regulation of transcriptional network (Yamaguchi-Shinozaki and Shinozaki, 2006). A large number of genes that are induced by water deficit are also highly induced by exogenous application of ABA. Conversely, there are several genes that are induced by water deficit but are not affected by exogenous ABA. These findings suggested that transcriptional response to water deficit is regulated by both ABA-dependent and ABA-independent signal transduction pathways (Yamaguchi-Shinozaki and Shinozaki, 2006).
Transcription factors are regulatory proteins that can modulate expression of a specific set of genes through binding to their promoter. They play an important role in converting the stress-induced signals to cellular responses. A single transcription factor can modulate the expression of a number of genes. A collection of genes under regulation of the same regulatory protein is called regulons. Plants activate many regulons under drought and other stresses to optimize plant growth, some of them have been well determined in Arabidopsis (Nakashima et al., 2009). Both ABA-dependent and ABA-independent pathways regulate the transcriptional response by affecting one or more regulons active under drought stress (Nakashima et al., 2009). In the following section, we will discuss in brief about water deficit induced regulons and pathways affecting them.
AREB/ABF Regulon
Abscisic acid-responsive element binding protein (AREB) /ABF (ABRE binding factor) regulon function in ABA-dependent regulation of gene expression under drought stress (Nakashima et al., 2009; Yoshida et al., 2015), (Figure 1). Many genes that are affected by water deficit also respond to the exogenous application of ABA (Nakashima et al., 2009). Promoter analysis revealed that most of these ABA-responsive genes are regulated by ABRE (ABA-responsive element) in their promoter region. ABRE is a conserved, 8 bp long cis element (PyACGTGG/TC) with a core ACGT sequence (Nakashima et al., 2009; Fujita et al., 2011). A single copy of an ABRE is not sufficient to induce ABA-responsive gene expression. To function as an active cis-acting element, ABRE requires in proximity other copies of ABRE or another specific cis-acting element, which is termed as the coupling element. Certain sequences have been shown to function as coupling elements such as, CE1 (coupling element1) and CE3 (coupling element3); and DRE (dehydration-responsive element)/CRT (C-repeat) cis element (major cis element in ABA-independent pathway; Shen et al., 1996; Narusaka et al., 2003). CEs are usually GC-rich sequence and they are similar to ABRE (Yamaguchi-Shinozaki and Shinozaki, 2006).
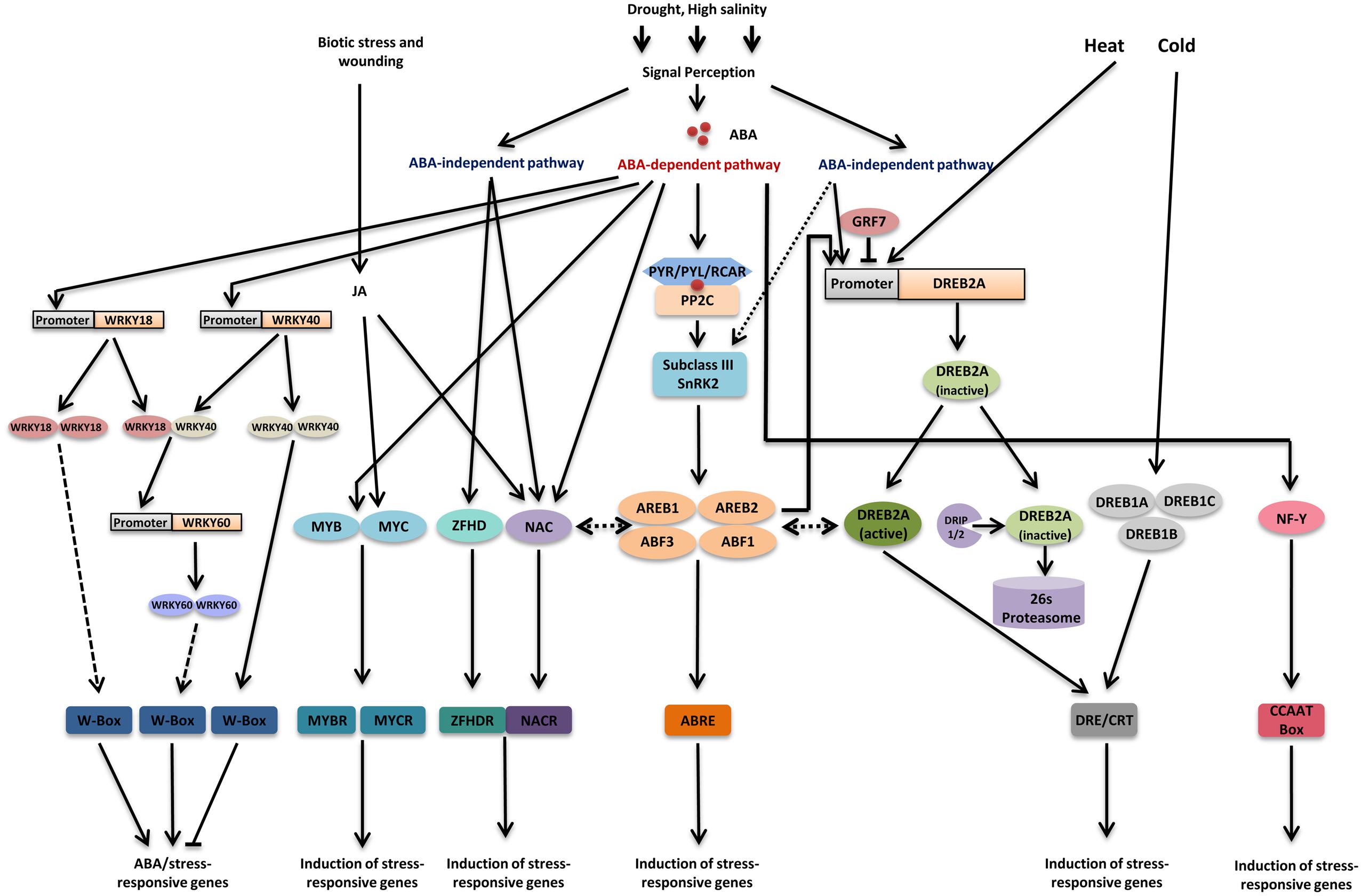
FIGURE 1. Major transcriptional regulatory networks of transcription factors involved in drought stress. Drought signal perception leads to activation of both abscisic acid (ABA)-dependent and ABA-independent pathways. In ABA-dependent pathway, accumulation of ABA leads to activation of sub class III SnRK2s through PYR/PYL/RCAR-PP2C receptor complex. Four transcription factors ABA-responsive element (ABRE) binding protein 1 (AREB1), AREB2, ABRE binding factor 3 (ABF3), and ABF1 are mainly phosphorylated through sub class III SnRK2s under drought stress and regulate most of the downstream genes by binding to the ABRE cis-element present in their promoter region. In addition to this, ABA also modulates the activity of MYB/MYCs, NACs, WRKYs, and NF-Y transcription factors. MYC2 and NAC proteins are also involved in JA signaling. ABA signal perception leads to induction of WRKY18 and WRKY40 and their product could bind to W-box present in WRKY60 and thereby induce it. DREB2A plays a pivotal role in ABA-independent gene expression regulation under drought stress. DREB2A expression is regulated by GRF7 under unstressed condition. Additionally, DRIPs regulate levels of DREB2A protein under unstressed condition. DREB2A also participate in gene expression regulation under heat stress. Three DREB1 proteins, DREB1A, DREB1B, and DREB1C are the key factors regulating gene expression under cold stress. Some NAC transcription factors also regulate gene expression in ABA-independent manner. Recently, it has been reported that AREB/ABFs can induce DREB2A and AREB/ABFs interact with DREB2A, based on these observations, ABA-dependent and ABA-independent pathways are thought to crosstalk under drought stress. Apart from this, some recent reports suggest a possible interaction between NACs and AREB/ABFs. Transcription factors and DNA-binding proteins are shown in colored ellipses. Dashed lines indicate possible although unconfirmed routes.
Abscisic acid-responsive element binding protein or ABF transcription factors were found to bind to ABRE element in yeast one-hybrid screening. These are major transcription factors that bind to ABRE and regulate ABA-responsive gene expression (Choi et al., 2000; Uno et al., 2000), (Figure 1). AREB/ABF is a subfamily of the basic leucine zipper (bZIP) family that consists of 9 members in Arabidopsis. All AREB/ABF transcription factors contain four conserved domains in addition to bZIP domain (Fujita et al., 2011, 2013). In Arabidopsis, AREB1/ABF2, AREB2/ABF4, ABF1, and ABF3 are expressed mainly in the vegetative tissues in response to ABA and osmotic stress (Fujita et al., 2011). In contrast, some other members are expressed during seed maturation such as Arabidopsis ABI5, AREB3, DPBF2, and EEL (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Bensmihen et al., 2002). Transgenic plants overexpressing AREB1/ABF2, AREB2/ABF4, or ABF3 exhibit enhanced drought tolerance and increased ABA sensitivity (Kang et al., 2002; Fujita et al., 2005). Triple AREB/ABF mutant areb1 areb2 abf3 exhibits reduced tolerance to drought and decreased sensitivity to exogenous ABA compared to that of wild-type, single mutants, or double mutants (Yoshida et al., 2010). Transcriptome analysis of the triple mutant under osmotic stress conditions showed reduced levels of many osmotic stress-inducible genes (Yoshida et al., 2010). Recently, ABF1 has also been reported to play an important role in ABA-mediated gene expression under drought stress. Although, it is expressed in lesser quantity as compared to those three AREB/ABFs, the areb1 areb2 abf3 abf1 quadruple mutant plants show an increase in drought sensitivity and decreased ABA sensitivity in comparison to areb1 areb2 abf3 mutant. In quadruple mutant many dehydration-inducible genes including LEA protein genes and transcription factors show reduced expression (Yoshida et al., 2015). Both triple mutants, as well as quadruple mutants, exhibited decreased inflorescence heights as compared to wild-type; as well as delayed bolting. Except for this, all of them showed phenotypes similar to wild-type (Yoshida et al., 2010, 2015). Thus, these four AREB/ABFs have been shown to be central transcription factors that cooperatively function in ABA-dependent transcriptional activation through their ABREs under these abiotic stress conditions (Figure 1).
Abscisic acid-responsive element binding protein/ABF trans cription factors are fully activated only after phosphory lation of their conserved regions (Fujita et al., 2011). The phosphorylation is catalyzed by serine/threonine kinase SnRK2s (SNF1-related protein kinase) that are induced by ABA. The importance of phosphorylation was suggested by the observation that overexpression of AREB1/ABF2 activated downstream gene expression only when phosphorylated active form of this gene was used (Furihata et al., 2006). Thus, under water deficit, cellular ABA concentration is increased that is recognized by the ABA receptors PYR/PYL/RCARs (pyrabactin resistance/pyrabactin resistance1- like/regulatory component of ABA receptors) leading to the inhibition of phosphatase activity of PP2C (protein phosphatase 2C). PP2C is a negative regulator of ABA signaling which dephosphorylates and thereby inactivates subclass III SnRK2s. Subsequently, released subclass III SnRK2s get accumulated in the cell and phosphorylate AREB/ABF thereby inducing expression of AREB/ABF regulon genes (reviewed in Cutler et al., 2010; Umezawa et al., 2010), (Figure 1). There are 10 members of SnRK2 family in Arabidopsis that are divided into three groups including three members in subclass III SnRK2s, SRK2D/SnRK2.2, SRK2E/OST1/SnRK2.6, and SRK2I/SnRK2.3 (Umezawa et al., 2010). Out of these 10 SnRK2s, nine are activated by osmotic stress. However, subclass III SnRK2s are also strongly induced by ABA and mediate most of the ABA responses (Yoshida et al., 2014). Subclass III SnRK2s have been shown to phosphorylate AREB/ABFs in vitro as well as co-localize and interact with them in plant cell nuclei (Furihata et al., 2006; Fujii et al., 2007; Fujita et al., 2009; Yoshida et al., 2010). Studies involving triple mutant srk2d/e/i showed that expression of most of the AREB1/ABF2, AREB2/ABF4, and ABF3 regulated genes are highly reduced and ABA-dependent phosphorylation of AREB/ABFs is completely abolished (Fujii and Zhu, 2009; Fujita et al., 2009). All these results indicate that subclass III SnRK2s regulate ABA-responsive gene expression under drought stress by phosphorylating AREB/ABFs.
DREB1/CBF and DREB2 Regulons
Dehydration-responsive element binding protein 1 (DREB1)/CBF (C-repeat binding factor) and DREB2 regulons function in ABA-independent regulation of gene expression under drought stress (Nakashima et al., 2009), (Figure 1). In Arabidopsis, the RD29A/COR78/LTI78 gene is induced by drought and cold. This gene has been found to be ABA-inducible but at the same time it can be induced by drought and cold stress in mutants defective in ABA biosynthesis and signaling (Yamaguchi-Shinozaki and Shinozaki, 2006). The analysis of its promoter, together with expression studies suggested that the dehydration inducibility of this gene is regulated by both ABA-independent and ABA-dependent pathways through different cis-acting elements (Yamaguchi-Shinozaki and Shinozaki, 2006).
The promoter of RD29A gene contains DRE/CRT cis element in addition to ABRE (Yamaguchi-Shinozaki and Shinozaki, 1994, 2005). DRE element is responsible for ABA-independent induction of many genes in response to osmotic and cold stress in many plants including Arabidopsis (Yamaguchi-Shinozaki and Shinozaki, 2005). DRE is a conserved 9 bp long (TACCGACAT) cis element. Unlike ABRE, a single copy of DRE is sufficient to induce genes under osmotic and cold stress (Yamaguchi-Shinozaki and Shinozaki, 1994). Similar cis-acting elements, named C-repeat (CRT) and low-temperature responsive element (LTRE), were identified in low-temperature-inducible genes. These sequences share a common core sequence, A/GCCGAC, which is referred to as the DRE/CRT core sequence (Baker et al., 1994; Jiang et al., 1996; Thomashow, 1999). DRE/CRTs are found in promoters of many stress-inducible genes.
Dehydration-responsive element binding protein 1/CBF and DREB2 transcription factors recognize DRE/CRT and activate downstream genes (Figure 1). Both DREB1/CBF and DREB2 belong to the plant-specific AP2 (APETALA2)/ ERF (ethylene-responsive element-binding factor) family having AP2/ERF DNA-binding motif. There are 145 members in AP2/ERF transcription factor family in Arabidopsis (Sakuma et al., 2002). DREB transcription factors constitute a subfamily of AP2/ERF family. Arabidopsis has six and eight DREB1/CBF-type and DREB2-type genes, respectively. Among them, three DREB1/CBF-type transcription factors, DREB1A/CBF3, DREB1B/CBF1, and DREB1C/CBF2 are rapidly induced by low temperature and act as major transcription factors that activate gene transcription through DRE/CRT in response to cold stress (Nakashima et al., 2009), (Figure 1). Transgenic plants overexpressing DREB1 show enhanced tolerance to cold and accumulate osmoprotectants such as proline and various sugars (Gilmour et al., 2000). These transgenic plants also exhibit “dwarf” phenotype and pronounced prostrate growth habits. They have shorter petiole in comparison to wild-type plants with leaves having bluish-green tint. Additionally, plants overexpressing DREB1 show delayed bolting and flowering as well as lower yield in comparison to wild-type plants (Gilmour et al., 2000).
In contrast to DREB1, two DREB2-type transcription factors, DREB2A and DREB2B, are highly induced in response to osmotic stress conditions and are considered to be involved in DRE-mediated gene transcription in response to water deficit (Liu et al., 1998), (Figure 1). Later, it was found that DREB2A plays a pivotal role in gene expression regulation under salt stress whereas DREB2B regulates gene expression in response to drought stress (Nakashima et al., 2009). However, weak induction of several DREB1 genes such as DREB1D/CBF4, DREB1E/DDF2, and DREB1F/DDF1 under dehydration stress suggests that DREB1/CBF and DREB2 regulons interact with each other (Haake et al., 2002; Sakuma et al., 2002; Magome et al., 2004).
Although DREB2A and DREB1A were isolated together (Liu et al., 1998), later, it was discovered that both of them have slight difference in their downstream genes (Maruyama et al., 2009). Microarray analysis has suggested that products of most of the genes downstream to DREAB1A and DREB2A have similar putative functions, but carbohydrate metabolism genes have different expression pattern in DREB1A and DREB2A transgenic plants (Maruyama et al., 2009). Plants overexpressing DREB1A exhibit changes in expression of genes responsible for starch degradation, sucrose metabolism and sugar alcohol synthesis similar to that observed in dehydration and cold stress. These changes lead to accumulation of many kinds of sugar and sugar alcohols in plants that might be responsible for enhanced dehydration and cold stress tolerance. In contrast plants overexpressing DREB2A-CA (constitutively active form of DREB2A) do not exhibit the increase in these metabolites level (Maruyama et al., 2009). The reason for this might be the difference in their DNA-binding specificity (Nakashima et al., 2009). DREB1A has a high affinity to A/GCCGACNT sequences, whereas DREB2A preferentially binds ACCGAC motifs (Sakuma et al., 2006a) resulting in slight variation in target genes.
Although DREB2A regulates the expression of many genes involved in stress response and tolerance, it causes growth retardation and reduced reproduction rate in plants, therefore, its expression is tightly regulated (Yoshida et al., 2014). DREB2A expression is negatively regulated by GRF7 (growth-regulating factor7). GRF proteins are a family of putative transcription factors that consist of nine members in Arabidopsis (Kim et al., 2003). Among these nine members, GRF7 inhibits expression of DREB2A under normal conditions by binding to its short promoter region (Figure 1). Both knockdown and knockout mutants of GRF7 exhibit enhanced DREB2A expression under non-stressed condition (Kim et al., 2012a). These plants also exhibit enhanced salinity tolerance and retarded growth. Microarray analysis revealed that a large number of osmotic stress-responsive genes were upregulated in grf7 knockout mutants under non-stressed condition. These shreds of evidence suggest that GRF7 regulates a large number of osmotic stress responsive genes by regulating DREB2A expression (Kim et al., 2012a).
In addition to transcriptional regulation, DREB2A is also regulated at post-transcriptional level. Transgenic plant overexpressing DREB1/CBF under Arabidopsis stress-responsive RD29A promoter showed strong tolerance to stresses mainly against cold (Kasuga et al., 1999). However, transgenic plant overexpressing DREB2A did not exhibit any significant phenotypic change (Liu et al., 1998). Domain analysis revealed that central region of DREB2A protein has a negative regulatory domain (NRD), and removal of the NRD from DREB2A converts the protein into its constitutively active form (DREB2A-CA). The DREB2A-CA proteins are more stable in the nucleus than the wild-type protein. In contrast to wild-type DREB2A, overexpression of DREB2A-CA shows enhanced drought tolerance as well as a slight improvement in cold stress tolerance and upregulation of many stress-inducible genes (Sakuma et al., 2006a). Transgenic plants overexpressing DREB2A-CA exhibited growth retardation in comparison to wild-type as well as rounded, slightly darker leaves with short petiole. Additionally, extent of retardation and these mentioned phenotypes was in correlation to the expression of transgene (Sakuma et al., 2006a). All these reports suggest that stability control is a posttranslational regulatory mechanism of DREB2A.
Furthermore, DRIP1 (DREB2A-interacting protein 1), a ubiquitin E3 ligase is thought to degrade the leaky expression under normal conditions (Qin et al., 2008). DRIP1 harbors a C3HC4-type RING domain and has been found to interact with DREB2A in yeast two-hybrid screening. Transgenic Arabidopsis plants overexpressing DRIP1 exhibit delayed expression of DREB2A-regulated drought responsive genes, however, double knockout mutants of DRIP1 and its homolog DRIP2 exhibit increased expression of these genes. DRIP1 and DRIP2 function as E3 ubiquitin ligase and target DREB2A to 26S proteasome proteolysis and thereby negatively regulate the expression of drought-responsive genes (Qin et al., 2008), (Figure 1). Recently, it has been shown that stress signal play important role in stabilization of DREB2A (Morimoto et al., 2013). In drip1drip2 mutant, DREB2A protein levels were induced rapidly under heat stress suggesting their involvement in DREB2A degradation but the protein levels reduced in the mutant by prolonged heat stress (Morimoto et al., 2013). It has also been shown that stabilization of DREB2A is important but not sufficient for induction of downstream genes (Morimoto et al., 2013). All these results suggest that apart from DRIPs some other factors might be involved in degradation and activation of DREB2A.
Recently, transcription factor ERF53 (ethylene response factor53) and two homologous C3HC4-type RING E3 ligases, RGLG2 (RING domain ligase2) and RGLG1 has been identified that also function similar to DREB2A and DRIPs and regulate drought stress-responsive genes (Cheng et al., 2012). ERF53 is an AP2/ERF transcription factor that belongs to the non-DREB2 subfamily. AtERF53 expression increase significantly under drought and high salinity but it exhibit mild induction to exogenous ABA (Cheng et al., 2012; Hsieh et al., 2013). Overexpression of AtERF53 induces unstable drought tolerance. AtERF53 has been found to interact with RGLG2 and RGLG1, both of which act as E3 ubiquitin ligase and target it to proteasomal degradation. AtERF53-GFP fusion protein accumulates more stably in double mutant rglg1 rglg2 leading to enhanced drought stress tolerance. All these reports suggest that AtERF53 and RGLGs function in combination to regulate osmotic stress-responsive genes (Cheng et al., 2012; Hsieh et al., 2013).
Dehydration-responsive element binding protein 2A regulon also operates under heat shock stress (Figure 1). Overexpression studies of DREB2A-CA showed improved thermotolerance by inducing expression of heat shock related genes (Sakuma et al., 2006b; Mizoi et al., 2012). Similarly, transgenic plants overexpressing DREB2C showed increased expression of heat shock stress-inducible genes, leading to thermotolerance (Lim et al., 2007). Therefore, it is clear that DRE/CRT is involved in gene expression not only during dehydration but also under conditions of low and high temperature.
NAC Regulon
NAM, ATAF, and CUC (NAC) are plant-specific transcription factors that constitute one of the largest families of plant transcription factors. There are more than a hundred members in Arabidopsis and rice that have been classified in 10 groups based on their phylogenetic relationship (Jensen et al., 2010). NAC transcription factors have a highly conserved N-terminal DNA-binding domain and variable C-terminal region this C-terminal region is thought to play a crucial role in determination of their target genes (Nuruzzaman et al., 2013). NAC transcription factors are involved in various developmental processes from shoot meristem development to auxin signaling (Olsen et al., 2005). There are reports demonstrating their involvement in abiotic and biotic stress signaling (reviewed in Olsen et al., 2005; Nakashima et al., 2012; Nuruzzaman et al., 2013). NAC transcription factors involved in stress response and tolerance have been classified in stress-responsive NAC (SNAC) group.
The ERD1 (early response to dehydration1) gene encodes a ClpA homolog of Arabidopsis (Nakashima et al., 1997). ERD1 was found to be induced by osmotic stress such as dehydration and salt stress as well as during senescence; however, exogenous ABA application could not strongly induce it (Nakashima et al., 1997). Two different cis-acting elements, a MYC-like sequence (CATGTG) and a 14-bp rps1 site 1-like sequence are necessary for dehydration induced expression of ERD1 (Simpson et al., 2003). Three NAC transcription factors; ANAC019, ANAC055, and ANAC072/RD26 were reported to bind MYC-like sequence present in the promoter of ERD1 (Tran et al., 2004). These three proteins are included in the SNAC group of NAC transcription factors (Nakashima et al., 2012). Detailed DNA binding assay of these NAC transcription factors determined NACRS (NAC recognition sequence) ANNNNNTCNNNNNNNACACGCATGT, containing CATGT and harboring CACG as the core DNA-binding site (Tran et al., 2004), (Figure 1). These NAC genes were found to be expressed within 1–2 h of ABA treatment suggesting that they are induced through ABA-independent pathway under drought stress (Tran et al., 2004). Transgenic plants overexpressing ANAC019 and ANAC072 showed phenotype and the time course of growth similar to that of vector control (Tran et al., 2004). In contrast, plants overexpressing ANAC055 exhibited growth rate similar to that of vector control until they reached rosette stage; after this point, plants in which the expression of transgene was at the medium level, showed a little delay in bolting as compared to vector control whereas; plants in which the transgene was overexpressed at high level remained at rosette stage for an additional few days before first bolting (Tran et al., 2004). Transgenic plant overexpressing ANAC019, ANAC055, or ANAC072/RD26 induced expression of many stress-inducible genes but failed to up-regulate ERD1 (Fujita et al., 2004; Tran et al., 2004).
Subsequently, a zinc-finger homeodomain (ZFHD) transcription factor, ZFHD1, was identified as a transcriptional activator that recognizes the 14-bp rps1 site1-like sequence (CACTAAATTGTCAC) and this sequence was termed as ZFHDR (zinc finger homeodomain recognition sequence; Tran et al., 2007), (Figure 1). Expression of the ERD1 gene was induced only when both the NAC and ZFHD proteins were overproduced simultaneously in a transgenic plant. Thus, these transcription factors cooperatively activate the transcription of the ERD1 gene (Tran et al., 2007). Studies suggest that in addition to the role of ZFHD1 in cooperation of NAC transcription factor it can also function as transcriptional activator alone (Tran et al., 2007).
Overexpression of several stress-responsive NAC factors in Arabidopsis and rice has imparted drought tolerance in transgenic plants. For example, transgenic plants overexpressing ANAC072/RD26 exhibited enhanced drought tolerance as well as increased sensitivity to ABA (Fujita et al., 2004; Tran et al., 2004). Microarray analysis of ANAC072/RD26 overexpressing plants showed upregulation of stress-inducible genes and ABA-responsive genes suggesting that RD26 is involved in regulation of drought-responsive genes in ABA-dependent manner (Fujita et al., 2004). Similarly, overexpression of another NAC factor ATAF1 resulted in improved drought tolerance (Wu et al., 2009). Recently, NAC genes such as ANAC096 and ANAC016 are associated with drought response and tolerance (Xu et al., 2013; Sakuraba et al., 2015). In case of rice, overexpression of NAC genes such as SNAC1, OsNAC6/SNAC2, OsNAC5, and OsNAC10 improved drought tolerance (reviewed in Nakashima et al., 2014). NTLs (NAC with transmembrane motif 1-like) such as NTL4 and NTL6 are also involved in drought stress. Transgenic plants overexpressing NTL6 and ntl4 null mutants exhibit drought tolerance, suggesting that these two work antagonistic to each other during drought stress (Lee and Park, 2012; Kim et al., 2012b).
Several stress-inducible NAC genes are also induced by jasmonates and/or during senescence in Arabidopsis and rice (Figure 1). Thus, these stress-responsive NAC transcription factors not only function in the transcriptional response to abiotic stress conditions, including water stress, but are likely involved in the cross talk between abiotic and biotic stress responses (Nakashima et al., 2012).
Other Transcriptional Pathways Involved in Water Stress Responses
In addition to the above-mentioned pathways of cis-acting elements and transcription factors, many other transcriptional pathways function in water stress responses. The Arabidopsis RD22 gene is inducible by drought stress in ABA-dependent manner (Yamaguchi-Shinozaki and Shinozaki, 1993). Although it is induced by ABA, it does not have ABRE cis-element in its promoter region; in spite of that its expression is regulated by two cis-acting elements, MYC and MYB recognition elements (Abe et al., 1997). A MYC like transcription factor, MYC2, and a MYB transcription factor, MYB2 bind to these cis-acting elements and cooperatively activate the transcription of this gene (Abe et al., 1997, 2003), (Figure 1). Transgenic plants overexpressing AtMYC2 and AtMYB2 exhibit higher ABA sensitivity as well as osmotic tolerance (Abe et al., 2003). Transgenic plants overexpressing AtMYC2 show morphology similar to wild-type. However, overexpression of AtMYB2 causes growth retardation in transgenic plants growing on soil. Similarly, overexpression of both transcription factors causes severe growth retardation in plants growing on soil (Abe et al., 2003). Although, AtMYC2 overexpressing plants have the morphology similar to wild-type, they have characteristically irregular shaped leaf epidermal cells. In contrast, plants overexpressing AtMYB2 and both transcription factors have leaf epidermal as well as parenchymal cells similar to wild-type but smaller in shape (Abe et al., 2003). Microarray analysis has suggested that their target genes include many ABA-inducible genes. Conversely, a mutation in MYC2 decreased the expression of target genes, including RD22 (Abe et al., 2003). All these results suggest that in addition to ABRE mediated gene regulation, MYB and MYC transcription factors regulate gene expression in response to ABA under drought stress (Figure 1).
MYB transcription factors are one of the largest transcription factor families in plants that have characteristic MYB domain in their DNA binding region (Lindemose et al., 2013). Analysis of transcriptome data present in GENEVESTIGATOR database showed that 51% of Arabidopsis MYB genes are upregulated and 41% are downregulated under drought stress (Baldoni et al., 2015). Furthermore, many MYB genes have been found to be involved in drought stress responses (reviewed in Baldoni et al., 2015).
MYC transcription factors belong to bHLH (basic-helix-loop-helix) transcription factor family of plants that have a characteristic bHLH domain (Kazan and Manners, 2013). Guard cell transcriptome analysis showed that ABA-responsive genes having MYC-binding motifs in the promoter region are present in large number in these cells (Wang et al., 2011). MYC2 protein has emerged as a master player in jasmonic acid signaling as well as cross-talk between jasmonic acid and ABA signaling (Kazan and Manners, 2013).
The involvement of WRKY transcription factors in drought stress has been reported recently. WRKY transcription factors constitute a family that has one or two WRKY domains which is involved in DNA binding. WRKY transcription factors bind to a conserved sequence named as W box and regulate gene expression (Rushton et al., 2010), (Figure 1). WRKY transcription factors are involved in various plant processes including biotic stress responses (Ulker and Somssich, 2004; Rushton et al., 2010). Recently, they have been reported to be involved in abiotic stress responses (Rushton et al., 2010; Banerjee and Roychoudhury, 2015). Various WRKY transcription factors have been found to be involved in ABA signaling. WRKY18 and WRKY60 act as positive regulators of ABA signaling during seed germination, and stress response while WRKY40 has the opposite effect on ABA signaling. WRKY18 and WRKY60 act as weak transcriptional activator whereas WRKY40 binds to the promoters of multiple stress-inducible transcription factor genes, including DREB1A/CBF3, DREB2A, and MYB2, and represses their expression (Chen et al., 2010; Shang et al., 2010). ABA signal perception leads to induction of WRKY18 and WRKY40 and their product could bind to W-box present in WRKY60 promoter and thereby induce it (Chen et al., 2010), (Figure 1). In another report, WRKY gene, WRKY63/ABO3 (ABA Overly Sensitive3) has been demonstrated to be involved in drought responses. abo3 mutant exhibits hypersensitive response for ABA in the seedling stage as well as reduced drought tolerance. WRKY63 has been shown to bind to the promoter of AREB1/ABF2 and thereby positively regulating its expression (Ren et al., 2010). Apart from these WRKY genes, many other have been reported to be involved in drought and salt stress responses (Bakshi and Oelmüller, 2014; Banerjee and Roychoudhury, 2015)
In addition to these, NF-Y (nuclear factor-Y) transcription factors also take part in drought stress response and tolerance mechanisms. NF-Y transcription factors are heterotrimeric proteins with three distinct subunits NF-YA, NF-YB, and NF-YC and bind to CCAAT box in the promoter region of target genes (Figure 1). NF-Y transcription factors are crucial factors in nodulation in nitrogen-fixing plants and nitrogen assimilation but there are several reports suggesting their role in stress responses especially in drought stress response and tolerance (Li et al., 2008; Petroni et al., 2012; Laloum et al., 2013; Xu et al., 2014; Quach et al., 2015). Under drought stress, AtNF-YA5 has been shown to be upregulated in ABA-dependent manner in leaf and roots of Arabidopsis plants (Li et al., 2008), (Figure 1). Transgenic plants overexpressing AtNF-YA5 exhibited improved drought resistance and reduced water loss; whereas, Atnf-ya5 mutant plants were found to be hypersensitive to drought (Li et al., 2008).
Interaction Between Different Transcription Factors Involved in Drought Stress Response
Evidence suggests that drought-responsive transcription factors work cooperatively to regulate gene expression. The subclass III SnRK2s play a very important role in this context. These subclass III SnRK2s are induced by both ABA and drought stress. In-gel kinase assay using ABA-insensitive and –deficient mutants has suggested that drought stress activate SnRK2s independent of ABA (Yoshida et al., 2006; Boudsocq et al., 2007), (Figure 1). Transcriptome analysis of srk2d/e/i triple mutants suggests that they modulate the expression of genes involved in ABA-dependent as well as ABA-independent pathways (Fujita et al., 2009). Recently, phosphoproteome study has shown that 5 min of ABA treatment results in phosphorylation of phosphopeptide corresponding subclass III SnRK2s, but osmotic stress fails to do the same. However, short osmotic stress led to phosphorylation of a phosphopeptide corresponding to subclass I SnRK2s, which are not activated by ABA, and phosphopeptides corresponding to MAP3K and MAP4K (E Stecker et al., 2014). Novel proteins are thought to be involved in osmotic stress-dependent activation of SnRK2s.
As mentioned earlier, the target site of DREB/CBF, i.e., DRE/CRT motif also function as a coupling element for ABRE and is present in many ABA-inducible drought responsive genes (Narusaka et al., 2003). AREB/ABF proteins have been shown to physically interact with DREB/CBFs including DREB1A, DREB2A, and DREB2C (Lee et al., 2010), (Figure 1). Recent reports have shown that ABRE sequence in the promoter region is required for induction of DREB2A under osmotic stress. Furthermore, transient-expression analyses coupled with ChIP (Chromatin Immunoprecipitation) assays has shown AREB/ABFs such as AREB1, AREB2, and ABF3 can bind to the promoter of DREB2A and thereby induce them in an ABRE-dependent manner (Kim et al., 2011), (Figure 1). In grf7 mutant expression of ABA-inducible genes and osmotic stress-responsive genes is upregulated (Kim et al., 2012a). All these reports suggest a complex interaction between the AREB and DREB regulons that need to be further studied in order to create a more comprehensive picture of transcriptional regulation under drought stress.
There are several reports indicating the interaction between AREB/ABFs and NACs. SNAC transcription factor ATAF1 has been reported to bind to the promoter of NCED3 and thereby regulating ABA hormone levels, giving rise to a probability those SNACs may be involved in regulation of ABA-dependent gene expression of AREB/ABF regulons (Jensen et al., 2013). Conversely, ABRE sequences have been reported in the promoter region of SNAC genes (Nakashima et al., 2012). ANAC096 directly interacts with ABF2 and ABF4 but not with ABF3. Evidence suggests that ANAC096 acts cooperatively with ABFs in activation of ABA-inducible drought responsive genes (Xu et al., 2013), (Figure 1). Sakuraba et al. (2015) found that ANAC016 negatively regulates drought stress tolerance. They also found that ANAC016 directly binds to the promoter of AREB1 and represses its expression. In addition to these two, five other drought-responsive NAC transcription factors (NAC019, NAP, NAC053/NTL4, NAC055, and NAC072) have been found to be associated with ABA signaling (Tran et al., 2004; Lee and Park, 2012; Zhang and Gan, 2012); suggesting they might act as additional tier in regulation of ABA-dependent drought responsive genes.
Conclusion
Plant response to drought is a complex process comprising many changes from morphological to molecular level. Under drought stress, many transcription factors operate both exclusively and cooperatively forming a web of interactions. In this review, we have summarized major transcription factors that play a pivotal role in drought stress response and tolerance.
Drought activates many pathways in plants that have been broadly classified in two categories, i.e., ABA-dependent pathways and ABA-independent pathways. AREB/ABFs, DREBs and NACs are the vital transcription factors regulating a large fraction of drought inducible genes (Figure 1). Along with these, some other transcription factors such as MYB/MYC factors, WRKY and NF-Y have also been demonstrated to be involved in one or more drought responsive mechanisms (Abe et al., 1997, 2003; Yamaguchi-Shinozaki and Shinozaki, 2006; Baldoni et al., 2015). Four AREB/ABFs, AREB1/ABF2, AREB2/ABF4, ABF3, and ABF1 are the central players of ABA-mediated regulation of gene expression. These AREB/ABFs carry out ABA-regulated responses through binding to the ABRE cis-elements present in the promoter region of target genes (Yoshida et al., 2015). On the other hand, DREB1/CBF and DREB2 transcription factors regulate gene expression in ABA-independent manner. Three DREB1/CBFs, DREB1A/CBF3, DREB1B/CBF1, and DREB1C/CBF2 regulate cold-responsive gene expression whereas, DREB2A and DREB2B are mainly involved in regulation of osmotic stress-responsive gene expression (Nakashima et al., 2009). Additionally, heat stress-inducible genes are also regulated by DREB2A (Mizoi et al., 2012). However, under unstressed conditions, DREB2A levels are tightly regulated at the transcriptional and post-transcriptional levels through GRF7 and DRIPs, respectively (Yoshida et al., 2014). These DREBs bind to DRE/CRT element present in the promoters of genes acting downstream to them (Nakashima et al., 2009). Different NAC factors regulate drought-inducible gene expression by binding to NACRS cis elements (Nakashima et al., 2012, 2014).
Drought-responsive transcription factors interact with each other as well as components of other stress pathways resulting in overlap of target genes of these pathways. In this context, subclass III SnRK2s might act as a nodal point as these kinases can regulate the expression of AREB/ABFs as well as DREB2A (Yoshida et al., 2014). Furthermore, there are several pieces of evidence suggesting that AREB/ABF transcription factors interact with DREB and NAC transcription factors (Nakashima et al., 2014).
Transcription factors are potent candidates for engineering stress tolerant plants as a single transcription factor can modulate a large set of genes. Many of drought-responsive transcription factors have been used to improve drought tolerance in different crops such as rice, wheat, soybean, and maize (Nakashima et al., 2014; Krannich et al., 2015).
All these transcription factors along with other interacting partners constitute a complex network that has been extensively studied but not yet completely understood. In future, further studies about transcriptional regulatory network should provide a more comprehensive picture of the pathways as well as cross-talks. Identification of crucial factors of these pathways along with evolution of new technologies to produce genetically engineered plants should lead to development of plants with improved drought tolerance under field condition with minimal negative effects on crop yield and development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors are thankful to University Grants Commission for research fellowship to DS.
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868.
Baker, S. S., Wilhelm, K. S., and Thomashow, M. F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–13. doi: 10.1007/BF00029852
Bakshi, M., and Oelmüller, R. (2014). WRKY transcription factors: jack of many trades in plants. Plant Signal. Behav. 9:e27700 doi: 10.4161/psb.27700
Baldoni, E., Genga, A., and Cominelli, E. (2015). Plant MYB transcription factors: their role in drought response mechanisms. Int. J. Mol. Sci. 16, 15811–15851. doi: 10.3390/ijms160715811
Banerjee, A., and Roychoudhury, A. (2015). WRKY proteins: signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015:807560. doi: 10.1155/2015/807560
Bensmihen, S., Rippa, S., Lambert, G., Jublot, D., Pautot, V., Granier, F., et al. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14, 1391–1403. doi: 10.1105/tpc.000869
Boudsocq, M., Droillard, M. J., Barbier-Brygoo, H., and Laurière, C. (2007). Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol. 63, 491–503. doi: 10.1007/s11103-006-9103-1
Bray, E. A. (2004). Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J. Exp. Bot. 55, 2331–2341. doi: 10.1093/jxb/erh270
Bray, E. A., Bailey-Serres, J., and Weretilnyk, E. (2000). “Responses to abiotic stresses,” in Biochemistry and Molecular Biology of Plants, eds B. B. Buchanan, W. Gruissem, and R. L. Jones (Rockville, MD: American Society of Plant Physiologists), 1158–1203.
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., and Xu, X. (2010). Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10:281. doi: 10.1186/1471-2229-10-281
Cheng, M. C., Hsieh, E. J., Chen, J. H., Chen, H. Y., and Lin, T. P. (2012). Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 158, 363–375. doi: 10.1104/pp.111.189738
Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. doi: 10.1074/jbc.275.3.1723
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
E Stecker, K., Minkoff, B. B., and Sussman, M. R. (2014). Phosphoproteomic analyses reveal early signaling events in the osmotic stress response. Plant Physiol. 165, 1171–1187. doi: 10.1104/pp.114.238816
Finkelstein, R. R., and Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.1105/tpc.12.4.599
Fujii, H., Verslues, P. E., and Zhu, J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19, 485–494. doi: 10.1105/tpc.106.048538
Fujii, H., and Zhu, J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106. 8380–8385. doi: 10.1073/pnas.0903144106
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi: 10.1105/tpc.105.035659
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132. doi: 10.1093/pcp/pcp147
Fujita, Y., Yoshida, T., and Yamaguchi-Shinozaki, K. (2013). Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 147, 15–27. doi: 10.1111/j.1399-3054.2012.01635.x
Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103, 1988–1993. doi: 10.1073/pnas.0505667103
Gilmour, S. J., Sebolt, A. M., Salazar, M. P., Everard, J. D., and Thomashow, M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. doi: 10.1104/pp.124.4.1854
Haake, V., Cook, D., Riechmann, J. L., Pineda, O., Thomashow, M. F., and Zhang, J. Z. (2002). Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 130, 639–648. doi: 10.1104/pp.006478
Hirayama, T., and Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Hsieh, E. J., Cheng, M. C., and Lin, T. P. (2013). Functional characterization of an abiotic stress-inducible transcription factor AtERF53 in Arabidopsis thaliana. Plant Mol. Biol. 82, 223–237. doi: 10.1007/s11103-013-0054-z
Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403. doi: 10.1146/annurev.arplant.47.1.377
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., Kato, T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Jensen, M. K., Kjaersgaard, T., Nielsen, M. M., Galberg, P., Petersen, K., O’Shea, C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426, 183–196. doi: 10.1042/BJ20091234
Jensen, M. K., Lindemose, S., de Masi, F., Reimer, J. J., Nielsen, M., Perera, V., et al. (2013). ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open. Biol. 3, 21–7. doi: 10.1016/j.fob.2013.07.006
Jiang, C., Iu, B., and Singh, J. (1996). Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol. Biol. 30, 679–684. doi: 10.1007/BF00049344
Kang, J. Y., Choi, H. I., Im, M. Y., and Kim, S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. doi: 10.1105/tpc.010362
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Kazan, K., and Manners, J. M. (2013). MYC2: the master in action. Mol. Plant. 6, 686–703. doi: 10.1093/mp/sss128
Kim, J. H., Choi, D., and Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313X.2003.01862.x
Kim, J. S., Mizoi, J., Kidokoro, S., Maruyama, K., Nakajima, J., Nakashima, K., et al. (2012a). Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24, 3393–3405. doi: 10.1105/tpc.112.100933
Kim, M. J., Park, M. J., Seo, P. J., Song, J. S., Kim, H. J., and Park, C. M. (2012b). Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 448, 353–363. doi: 10.1042/BJ20120244
Kim, J. S., Mizoi, J., Yoshida, T., Fujita, Y., Nakajima, J., Ohori, T., et al. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell. Physiol. 52, 2136–2146. doi: 10.1093/pcp/pcr143
Krannich, C. T., Maletzki, L., Kurowsky, C., and Horn, R. (2015). Network candidate genes in breeding for drought tolerant crops. Int. J. Mol. Sci. 16, 16378–16400. doi: 10.3390/ijms160716378
Krasensky, J., and Jonak, C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. doi: 10.1093/jxb/err460
Laloum, T., De Mita, S., Gamas, P., Baudin, M., and Niebel, A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18, 157–166. doi: 10.1016/j.tplants.2012.07.004
Lee, S., and Park, C. M. (2012). Regulation of reactive oxygen species generation under drought conditions in Arabidopsis. Plant Signal. Behav. 7, 599–601. doi: 10.4161/psb.19940
Lee, S. J., Kang, J. Y., Park, H. J., Kim, M. D., Bae, M. S., Choi, H. I., et al. (2010). DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 153, 716–727. doi: 10.1104/pp.110.154617
Li, W. X., Oono, Y., Zhu, J., He, X. J., Wu, J. M., Iida, K., et al. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20, 2238–2251. doi: 10.1105/tpc.108.059444
Lim, C. J., Hwang, J. E., Chen, H., Hong, J. K., Yang, K. A., Choi, M. S., et al. (2007). Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem. Biophys. Res. Commun. 362, 431–436. doi: 10.1016/j.bbrc.2007.08.007
Lindemose, S., O’Shea, C., Jensen, M. K., and Skriver, K. (2013). Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 14, 5842–5878. doi: 10.3390/ijms14035842
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Lopez-Molina, L., and Chua, N. H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41, 541–547. doi: 10.1093/pcp/41.5.541
Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., and Oda, K. (2004). Dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37, 720–729. doi: 10.1111/j.1365-313X.2003.01998.x
Maruyama, K., Takeda, M., Kidokoro, S., Yamada, K., Sakuma, Y., Urano, K., et al. (2009). Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150, 1972–1980. doi: 10.1104/pp.109.135327
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Morimoto, K., Mizoi, J., Qin, F., Kim, J. S., Sato, H., Osakabe, Y., et al. (2013). Stabilization of Arabidopsis DREB2A is required but not sufficient for the induction of target genes under conditions of stress. PLoS ONE 8:e80457. doi: 10.1371/journal.pone.0080457
Nakashima, K., Ito, Y., and Yamaguchi-Shinozaki, K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Nakashima, K., Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1997). A nuclear gene, erd1, encoding a chloroplast-targeted Clp protease regulatory subunit homolog is not only induced by water stress but also developmentally up-regulated during senescence in Arabidopsis thaliana. Plant J. 12, 851–861. doi: 10.1046/j.1365-313X.1997.12040851.x
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170. doi: 10.3389/fpls.2014.00170
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148. doi: 10.1046/j.1365-313X.2003.01708.x
Nuruzzaman, M., Sharoni, A. M., and Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248. doi: 10.3389/fmicb.2013.00248
Olsen, A. N., Ernst, H. A., Leggio, L. L., and Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Petroni, K., Kumimoto, R. W., Gnesutta, N., Calvenzani, V., Fornari, M., Tonelli, C., et al. (2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24, 4777–4792. doi: 10.1105/tpc.112.105734
Qin, F., Sakuma, Y., Tran, L. S., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20, 1693–1707. doi: 10.1105/tpc.107.057380
Quach, T. N., Nguyen, H. T., Valliyodan, B., Joshi, T., Xu, D., and Nguyen, H. T. (2015). Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol. Genet. Genomics 290, 1095–1115. doi: 10.1007/s00438-014-0978-2
Ren, X., Chen, Z., Liu, Y., Zhang, H., Zhang, M., Liu, Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313X.2010.04248.x
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006a). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006b). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 18822–18827. doi: 10.1073/pnas.0605639103
Sakuraba, Y., Kim, Y. S., Han, S. H., Lee, B. D., and Paek, N. C. (2015). The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27, 1771–1787. doi: 10.1105/tpc.15.00222
Shaar-Moshe, L., Hübner, S., and Peleg, Z. (2015). Identification of conserved drought-adaptive genes using a cross-species meta-analysis approach. BMC Plant Biol. 15:111. doi: 10.1186/s12870-015-0493-6
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shen, Q., Zhang, P., and Ho, T. H. (1996). Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8, 1107–1119. doi: 10.1105/tpc.8.7.1107
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. doi: 10.1016/S1369-5266(00)00067-4
Simpson, S. D., Nakashima, K., Narusaka, Y., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 33, 259–270. doi: 10.1046/j.1365-313X.2003.01624.x
Thomashow, M. F. (1999). PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Todaka, D., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2015). Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 18:684. doi: 10.3389/fpls.2015.00084
Tran, L. S., Nakashima, K., Sakuma, Y., Osakabe, Y., Qin, F., Simpson, S. D., et al. (2007). Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 49, 46–63. doi: 10.1111/j.1365-313X.2006.02932.x
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Ulker, B., and Somssich, I. E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. doi: 10.1016/j.pbi.2004.07.012
Umezawa, T., Nakashima, K., Miyakawa, T., Kuromori, T., Tanokura, M., Shinozaki, K., et al. (2010). Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839. doi: 10.1093/pcp/pcq156
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisicacid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 97, 11632–11637. doi: 10.1073/pnas.190309197
Wang, R. S., Pandey, S., Li, S., Gookin, T. E., Zhao, Z., Albert, R., et al. (2011). Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12:216. doi: 10.1186/1471-2164-12-216
Wu, Y., Deng, Z., Lai, J., Zhang, Y., Yang, C., Yin, B., et al. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19, 1279–1290. doi: 10.1038/cr.2009.108
Xu, L., Lin, Z., Tao, Q., Liang, M., Zhao, G., Yin, X., et al. (2014). Multiple NUCLEAR FACTOR Y transcription factors respond to abiotic stress in Brassica napus L. PLoS ONE 30:e111354. doi: 10.1371/journal.pone.0111354
Xu, Z. Y., Kim, S. Y., Hyeon do, Y., Kim, D. H., Dong, T., Park, Y., et al. (2013). The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724. doi: 10.1105/tpc.113.119099
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993). The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol. Gen. Genet. 238, 17–25.
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yoshida, R., Umezawa, T., Mizoguchi, T., Takahashi, S., Takahashi, F., and Shinozaki, K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281, 5310–5318. doi: 10.1074/jbc.M509820200
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi: 10.1111/pce.12351
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Keywords: ABA, drought, regulons, cross-talk, transcription factors
Citation: Singh D and Laxmi A (2015) Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6:895. doi: 10.3389/fpls.2015.00895
Received: 15 August 2015; Accepted: 08 October 2015;
Published: 29 October 2015.
Edited by:
Maik Boehmer, Westfälische Wilhelms-Universität Münster, GermanyReviewed by:
Xinguang Zhu, Chinese Academy of Sciences, ChinaTae-Houn Kim, Duksung Women’s University, South Korea
Copyright © 2015 Singh and Laxmi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashverya Laxmi, ashverya_laxmi@nipgr.ac.in
 Dhriti Singh
Dhriti Singh Ashverya Laxmi
Ashverya Laxmi