- 1College of Life Sciences, Nanjing Agricultural University, Nanjing, China
- 2Center for Soybean Research of the Partner State Key Laboratory of Agrobiotechnology and School of Life Sciences, The Chinese University of Hong Kong, Hong Kong, China
The family of chloride channel proteins that mediate Cl- transportation play vital roles in plant nutrient supply, cellular action potential and turgor pressure adjustment, stomatal movement, hormone signal recognition and transduction, Cl- homeostasis, and abiotic and biotic stress tolerance. The anionic toxicity, mainly caused by chloride ions (Cl-), on plants under salt stress remains poorly understood. In this work, we investigated the function of soybean Cl-/H+ antiporter GmCLC1 under salt stress in transgenic Arabidopsis thaliana, soybean, and yeast. We found that GmCLC1 enhanced salt tolerance in transgenic A. thaliana by reducing the Cl- accumulation in shoots and hence released the negative impact of salt stress on plant growth. Overexpression of GmCLC1 in the hairy roots of soybean sequestered more Cl- in their roots and transferred less Cl- to their shoots, leading to lower relative electrolyte leakage values in the roots and leaves. When either the soybean GmCLC1 or the yeast chloride transporter gene, GEF1, was transformed into the yeast gef1 mutant, and then treated with different chloride salts (MnCl2, KCl, NaCl), enhanced survival rate was observed. The result indicates that GmCLC1 and GEF1 exerted similar effects on alleviating the stress of diverse chloride salts on the yeast gef1 mutant. Together, this work suggests a protective function of GmCLC1 under Cl- stress.
Introduction
Ionic stress, osmotic stress, nutritional imbalance, and oxidative damage are the main causes for salt injury of plants, among which ionic stress is the primary factor (Xiao et al., 2013; Adem et al., 2014). Sodium chloride is the most common salt in environment. In salinized conditions, salt changed the water potential around the root first causing the osmotic stress, then inducing ionic stress by the accumulation of Na+ and Cl- (Deinlein et al., 2014; Gupta and Huang, 2014; Nguyen et al., 2015; Nie et al., 2015). Plants counter these stresses through strategies such as extracellular exclusion of excess ions across the plasma membrane or intracellular vacuolar compartmentalization to reduce the effective Na+ and Cl- levels inside the cell, especially in the aerial parts (Zhang et al., 2011; Hasegawa, 2013). In general, crops such as cotton, rice, and barley are more sensitive to Na+ than Cl- (Qu et al., 2009). So far, the researches on plant physiological and molecular mechanisms of salt tolerance have mostly focused on cation (Na+) poisoning and adaptations, such as the NSCCs (non-selective cation channels) and HKTs (High-affinity K+ Transporter), both controlling Na+ import into the cell, together with Na+/H+ antiporters, such as AtSOS1 (Arabidopsis Salt Overly Sensitive 1) located in the plasma membrane for excluding Na+ from cells, and AtNHX1 (Arabidopsis vacuolar Na+/H+ antiporter) located in the tonoplast for compartmentalizing Na+ mainly in the vacuoles (Adams and Shin, 2014; Deinlein et al., 2014; Nguyen et al., 2015). However, salt injuries to plants resulting from anions (mainly Cl-) have long been under-investigated.
Cl- is the main form of anions in plant cells besides nitrate (). As one of the essential micronutrient elements for higher plant growth and development, Cl- is involved in photosynthesis, stomatal movement, cellular osmotic pressure maintenance, charge balance, and disease resistance (Jossier et al., 2010; Tealle and Tyerman, 2010; Guo et al., 2014). Excessive amount of Cl- lead to various adverse effects, such as negatively impacting the absorption of macronutrient elements (nitrogen [N], phosphorus [P], and potassium [K]), leaf water potential, causing stomatal closure and the accumulation of reactive oxygen species (ROS) in chloroplasts, which severely affected crop quality and yield (Yu and Liu, 2004; Nguyen et al., 2015). Some studies reported that certain crops or woody species such as tobacco, tomato, barley, grapevine, citrus, Glycine max, Lotus, and poplar were more sensitive to Cl- than to Na+ under salt stress (Moya et al., 2003; Luo et al., 2005; Chen and Yu, 2007; Brumós et al., 2010; Tregeagle et al., 2010). The genetic differences in the control of Cl- transport from roots to shoots or the ability to maintain a low shoot Cl- level is the key determinant of Cl-/salt tolerance (Zhang et al., 2011; Henderson et al., 2014). The anion channels or transporters, widely distributed in all types of organelle membranes (such as plasma membrane, tonoplast, endoplasmic reticulum, mitochondria, and chloroplasts, etc.), predominantly mediate Cl- and flux across the membrane, playing a vital function in plant nutrition absorption and transportation, adjustment of cellular action potential and turgor pressure, stomatal movement, hormone signal recognition and transduction, Cl- homeostasis under abiotic (salt, heavy metal, low temperature, etc.) or biotic stress conditions (Jossier et al., 2010; Barbier-Brygo et al., 2011; Wei et al., 2013; Guo et al., 2014; Nguyen et al., 2015). Among them, the CLCs (Chloride channel proteins) family mainly mediate Cl- transport and have attracted the most research interests (Zifarelli and Pusch, 2010). Currently, plant CLCs have been identified in Arabidopsis, tobacco, rice, potato, corn, spinach, citrus, salt cress, soybean, and maize (Wei et al., 2013; Zhou et al., 2013; Li et al., 2014; Wang et al., 2015). In response to Cl- toxicity under salt stress, the NaCl-treated plants utilize the anion transporters such as CLCs to adjust and reduce Cl- accumulation in the cell cytoplasm. In soybean, Li et al. (2006) reported that the protein encoded by the salt- and polyethylene glycol-inducible GmCLC1 gene (GenBank accession: AY972079, or Phytozome database: Glyma05g14760) localizes on tonoplast and can transport and sequester Cl- into the vacuoles of plant cells. Wong et al. (2013) further found that the transmembrane Cl- transfer activity of the GmCLC1 protein depends on the cytoplasmic pH value, suggesting that it is most likely a kind of Cl-/H+ antiporter that participates in the maintenance of intracellular Cl- homeostasis and regulates Cl-/salt tolerance.
In the current study, we further investigated the functions of how GmCLC1 regulates Cl- transportation in salt-stressed plants and yeast. We found that overexpressing GmCLC1 in Arabidopsis thaliana, soybean hairy roots and composite plants, as well as yeast mutant could enhance salt tolerance by regulation Cl- homeostasis.
Materials and Methods
Plant Materials, Bacteria and Yeast Strains, and Plasmids
Plant seeds including wild-type (WT) A. thaliana (Columbia ecotype glabrous1), G. max (L.) Merr. cultivars Jackson (salt-sensitive), Lee68 (salt-tolerant) and N23674 (salt-tolerant), Escherichia coli DH5α, Agrobacterium tumefaciens strain GV3101, Agrobacterium rhizogenes strain K599, binary vector for plant transformation pCAMBIA1300, the yeast Δgef1 mutant (derived from Saccharomyces cerevisiae BY4741) and yeast expression plasmid pYES2 were used in this study.
GmCLC1 Gene Cloning and Vectors Construction
The seeds of G. max N23674 cultivar were surface-sterilized with 1 g dm-3 HgCl2 for 5 min, then fully rinsed in distilled water, soaked in distilled water for 6 h, and finally germinated at 25°C in the dark. The germinated seeds were grown on vermiculite irrigated with 1/2 Hoagland solution in a greenhouse with temperature at 25 ± 2°C and humidity ranging from 60 to 70%. Total RNA was extracted from 10-days-old seedlings using the Trizol reagent (Invitrogen, USA). First-strand cDNAs were synthesized with 2 μg total RNAs using a PrimeScriptTM II 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian) according to the manufacturer’s protocol. The full-length coding region of GmCLC1 was amplified from the cDNA using the following PCR protocol: 94°C 3 min; 30 cycles of 94°C 30 s, 55°C 30 s, and 72°C 2 min 30 s; and 72°C 5 min in a 25 μL reaction mixture [2 μL of first strand cDNA, 0.15 mM MgCl2, 0.2 mM dNTPs, 0.4 μM of each primer, 0.25 U Taq DNA polymerase (TaKaRa, Dalian) and 10× PCR buffer]. Primers used: 5′-ATGGGTGAGGAATCCAGTTT-3′ and 5′-CTTCCTCTTTGATTTTGCCAG-3′. The PCR products were then cloned into pMD19-T vector (TaKaRa, Dalian) for sequencing.
Subsequently, the open reading frame of GmCLC1 was amplified from cDNA by PCR (95°C 3 min, 28 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 2.5 min) with KOD-Plus (TOYOBO, Japan), and ligated into the plasmid pCAMBIA1300 or pYES2 to obtain the recombinant plasmid pCAMBIA1300-GmCLC1 (primers used: 5′-GGGGTACCATGGGTGAGGAATCCAGTTT-3′ and 5′-GGACTAGTCTTCCTCTTTGATTTTGCCAG-3′) or pYES2-GmCLC1 (primers used: 5′-CCGGAATTCATGGGTGAGGAATCCAG-3′ and 5′-CCGCTCGAGTCACTTCCTCTTTGATTTTG-3′). After sequence verification, the recombinant plasmid pCAMBIA1300-GmCLC1 was transformed into A. tumefaciens GV3101 or A. rhizogenes K599, respectively.
Seed Germination and Root Elongation Experiments with WT and GmCLC1-Transgenic A. thaliana
The binary vector pCambia1300-GmCLC1 was transformed into WT Arabidopsis plants using the floral dip method mediated by A. tumefaciens (Clough and Bent, 1998). Transformants were selected by germination on Murashige and Skoog (MS) agar medium supplemented with 40 mg L-1 hygromycin B. Homozygous lines (L1–L5) of T2 plants were identified by PCR. The seeds of A. thaliana WT and a homozygous GmCLC1-transgenic line (L1) were sterilized and sown evenly on MS agar medium (pH = 5.8; Nie et al., 2015) containing 0, 150, and 200 mM NaCl, respectively. The agar plates were put in an illuminated growth chamber under a 14 h light/10 h dark cycle at 20 ± 2°C, with 60–70% relative humidity. The seed germination percentage (%) was recorded after 7 days. For measuring the root elongation of A. thaliana under salt stress, seeds of WT and GmCLC1-transgenic plants were cultured on MS agar medium (pH = 5.8) without NaCl for 5 days. Then seedlings with similar root lengths were selected and transferred onto MS agar medium (pH = 5.8) containing 0, 150, and 200 mM NaCl. After placing the agar plates vertically for 5 days, the seedlings of WT and GmCLC1-transgenics were photographed and the root lengths were measured.
Determination of Leaf Relative Water Content (RWC) and Chlorophyll Fluorescence (Fv/Fm), Root and Leaf Relative Electrolyte Leakage (REL) of WT and GmCLC1-Transgenic A. thaliana
Seeds of A. thaliana WT and GmCLC1-transgenic line (L1) were sterilized and sown on MS agar medium (pH = 5.8), and then placed in a growth chamber under a 14 h light/10 h dark cycle at 20 ± 2°C, with 60–70% relative humidity. After 8 days, the seedlings were then transferred to pots containing a sterilized peat moss and vermiculite mixture and grown for 5 days. Then the seedlings were treated with increasing concentrations of NaCl (50 mM NaCl for 2 days, 100 mM NaCl for the next 2 days, 150 mM NaCl solution for another 7 days). Finally, leaf relative water content (RWC) was measured according to the method described by Hu et al. (2016). Leaf chlorophyll fluorescence (Fv/Fm) was measured at room temperature with a plant efficiency analyzer (Handy PEA Fluorometer, Hansatech Instruments, UK; (Tian et al., 2014). Root or leaf relative electrolyte leakage (REL) was assayed using the method described by Hu et al. (2016) with a digital conductivity meter (DDS-307, Shanghai, China).
Analyses of Cl- Contents in Roots and Shoots of WT and GmCLC1-Transgenic A. thaliana Seedlings
Seeds of WT and GmCLC1-transgenic A. thaliana were sterilized and sown in pots containing a sterilized peat moss and vermiculite mixture and grown in a growth chamber under a 14 h light/10 h dark cycle at 20 ± 2°C, with 60–70% relative humidity. After 30 days, plants were treated with 1/2 X Hoagland solution containing 150 mM NaCl for 0, 4, 12, 24 h, 2, 4, and 6 days, respectively. Then the roots and shoots of Arabidopsis seedlings were sampled and Cl- contents were measured with the method described (Zhou and Yu, 2009).
Assays of Maximum Lengths and Fresh Weights of the Hairy Roots of Transformed Soybean Cotyledons, and REL and Cl- Contents of Soybean Hairy Roots Composite Plants
Hairy root transformation was performed according to Ali et al. (2012). A. rhizogenes strain K599 containing the recombinant binary vector pCAMBIA1300-GmCLC1 was grown in yeast extract peptone (YEP) medium containing 50 mg/L ampicillin (Amp) and 200 μM acetosyringone at 28°C for 16 h. Then the bacterial culture was centrifuged, and the pellet was resuspended gently in 10 mM MgSO4 solution followed by two washings, and adjusted to OD600 ≈ 0.5.
The cotyledons of soybean (Jackson and Lee68 cultivars) were scored with a scalpel and the wounds were infected with the pCAMBIA-containing A. rhizogenes infection solution for 1 h in dark at room temperature, and then transferred to moist filter paper and incubated in the dark for 5 days (at 25 ± 2°C). After that, the infected cotyledons were transferred to a growth chamber under a 12 h light/12 h dark cycle at 25 ± 2°C. New hair roots that sprout from the infected cotyledons that were free of Agrobacterium as screened by PCR and at similar lengths were selected and transferred into 1/2 X Hoagland solution (pH = 6.5) containing 0, 100, and 150 mM NaCl, respectively Hairy roots infected with A. rhizogenes strain K599 containing the empty vector pCAMBIA1300 served as the control. After 5 days treatment, hairy root growth (maximum root length and root fresh weight) was photographed and measured (Qi et al., 2014).
For whole-plant transformation, surface-sterilized soybean seeds (Jackson and Lee68 cultivars) were sown in pots containing a sterilized peat moss and vermiculite mixture. When the first pair of true leaves had fully expanded, the cotyledon nodes of the soybean seedlings were infected with the A. rhizogenes strain K599 infection solution for 1 h at room temperature by injection. Then the wounds were covered with moist vermiculite and incubated for 5 days at 28°C under a 12 h light/12 h dark cycle. After 7 days, hairy root lines that were free of Agrobacterium as screened by PCR were selected and the original roots removed from the seedlings. These seedlings were then cultured in 1/2 X Hoagland solution. Seedlings infected with A. rhizogenes strain K599 containing the empty vector pCAMBIA1300 served as the negative control. After 10 days, Jackson and Lee68 seedlings with similar-length hairy roots were selected and treated with 1/2 X Hoagland solution containing 120 mM NaCl for 3 and 5 days, respectively, and seedlings grown in Hoagland solution without NaCl served as the untreated control. All the solutions listed above were replaced every 3 days throughout the experiments. REL and Cl- contents in roots, stems, first true leaves, and the first and second trifoliates were measured accordingly.
Tolerance Tests Using the Δgef1 Yeast Mutant
The yeast expression vector pYES2-GmCLC1 was transformed into Δgef1 mutant yeast (S. cerevisiae) using the PEG/LiAc procedure and transformants were screened by PCR (Gietz, 2014). Ten-fold serial dilutions (starting at OD550 ≈ 0.5) of each yeast culture were plated on agar supplemented with YPD medium (1% yeast extract/2% peptone/2% dextrose), YPG medium (1% yeast extract/2% peptone/2% galactose), or YPG medium supplemented with 1 M NaCl, 1 M KCl, or 3 mM MnCl2. Plates were then incubated at 30°C and photographs were taken after 60 h. For Cl- content measurements, the above-mentioned yeast cells were grown in the liquid YPG medium plus 1.0 M NaCl, and collected during the exponential growth phase (OD550 ≈ 0.2), and their Cl- contents were assayed by AgNO3 titration as described by Zhou and Yu (2009).
Statistical Analyses
Data were expressed as mean ± SD of three independent experiments and were analyzed using one-way analysis of variance by SPSS 19.0, and pairwise comparisons were performed using Duncan’s test.
Results
GmCLC1 Alleviates NaCl Stress on Transgenic A. thaliana by Reducing Cl- Accumulation in Leaves
The cDNA of GmCLC1 was cloned from G. max cultivar N23674. The encoded protein product has the same sequence as the annotated product of Glyma05g14760. We have constructed five independent transgenic lines expressing GmCLC1. All transgenic lines exhibited NaCl tolerance (Supplementary Figure S1) and hence we selected one typical line for detailed analysis. The A. thaliana plant was successfully transformed with GmCLC1 as shown with genotyping by PCR (Figure 1A). When grown on plate containing 150 or 200 mM NaCl, the seed germination rates of both WT and GmCLC1-transgenic line (L1) declined as the NaCl concentration increased. However, under 150 or 200 mM NaCl treatments, the transgenic line exhibited a higher germination rate than WT (Figure 1B). Also, under 200 mM NaCl treatment, the seedling growth of WT and GmCLC1-transgenic line in both roots and shoot were inhibited compared to the untreated control (Figure 1C). However, the root elongation of the GmCLC1-transgenic line was significantly higher than those of WT under either 150 or 200 mM NaCl stress (p < 0.01; Figure 1D). This indicates that ectopic expression of GmCLC1 in A. thaliana could enhance seed germination and seedling growth under salt stress.
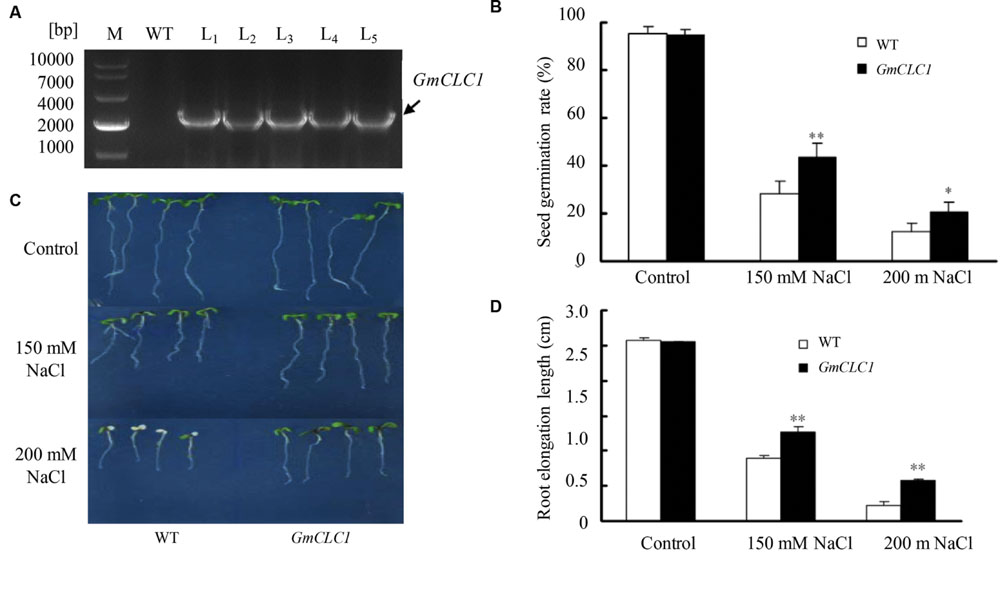
FIGURE 1. Growth characteristics of GmCLC1-transgenic Arabidopsis thaliana. (A) Genotyping by PCR of GmCLC1-transgenic A. thaliana lines (L1–L5) to confirm success in transformation. All five lines contain the transgene. Comparison in (B) seed germination rate, (C) seedlings growth, and (D) root lengths between A. thaliana wild-type (WT) and transgenic plants (GmCLC1; using L1 as the representative) sown on MS agar medium containing different concentrations of NaCl. (B) Seeds of WT and GmCLC1 (30 seeds per plate each with three replicates) were surface-sterilized and stratified at 4°C for 2–4 days and then sown on MS medium supplemented with 150 or 200 mM NaCl. After 7 days of incubation, the seed germination rate (%) was measured. (C,D) 5-days-old WT and GmCLC1 with nearly equal root lengths were transferred to MS agar medium supplemented with 0, 150, or 200 mM NaCl. After 5 days of vertical incubation, the seedlings’ growth phenotypes were photographed and the root lengths measured. M: DNA markers. ∗/∗∗: The difference between WT and GmCLC1 was significant at p < 0.05 and p < 0.01 respectively, using Duncan’s test. Each bar represents the mean and SD of for (C) 30 seeds per plate each with three replicates; for (D) more than 10 plants for each treatment.
When WT and GmCLC1-transgenic seedlings were continuously exposed to NaCl stress with increasing concentrations (50 mM NaCl solution for 2 days, and 100 mM NaCl solution for next 2 days, followed by 150 mM NaCl solution for 7 days), the transgenic plants appeared to be healthier than WT (Figure 2A), and RWC and Fv/Fm were also significantly higher than those of WT (p < 0.01; Figures 2B,C), while the REL values were significantly lower than those of WT (p < 0.01 for root, p < 0.05 for leaf; Figure 2D). This further shows that the GmCLC1-transgenic A. thaliana plants suffered less leaf water loss, exhibited more stable photosynthetic capacity and had reduced salt injuries compared to untransformed WT when under salt stress.
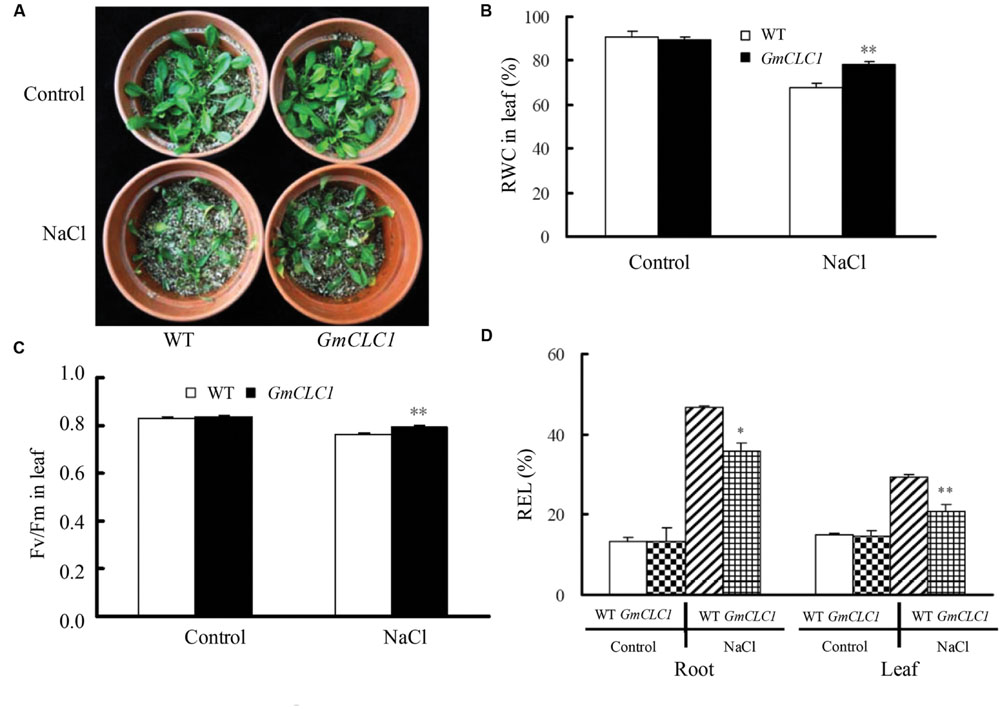
FIGURE 2. Effects of NaCl treatment on (A) growth, (B) leaf relative water content (RWC), (C) chlorophyll fluorescence (Fv/Fm), and (D) relative electrolyte leakage (REL) in roots and leaves of A. thaliana WT and transgenic GmCLC1 seedlings. Seeds of Arabidopsis WT and transgenic GmCLC1 plants were surface-sterilized and kept at 4°C for 2–4 days and then sown on MS medium. After 8 days of incubation, the seedlings were transferred into plastic pots filled with a sterilized peat moss and vermiculite mixture, and fertilized with ½-strength Hoagland nutrient solution for 5 days in the greenhouse. The nutrient solution was continuously replaced with half-strength Hoagland solution containing 50 mM NaCl for 2 days, followed by 100 mM NaCl for 2 days, and finally 150 mM NaCl for 7 days. No NaCl was added to the nutrient solution for the control treatment. ∗/∗∗: The difference between WT and GmCLC1 was significant at p < 0.05/0.01, respectively, using Duncan’s test. Each bar represents the mean and SD of at least three replicates.
During the 6-days treatment with 150 mM NaCl, the Cl- contents in the roots of both WT and GmCLC1-transgenic seedlings increased significantly within the first 4 h and then leveled off (Figure 3A). However, the Cl- contents in the shoots of both WT and transgenic seedlings increased steadily throughout the duration of the experiment, with the increase in the shoots of WT being significantly higher than in the transgenic plants (p < 0.01; Figure 3B), indicating that GmCLC1 reduces salt stress partly by reducing the Cl- accumulation in shoots.
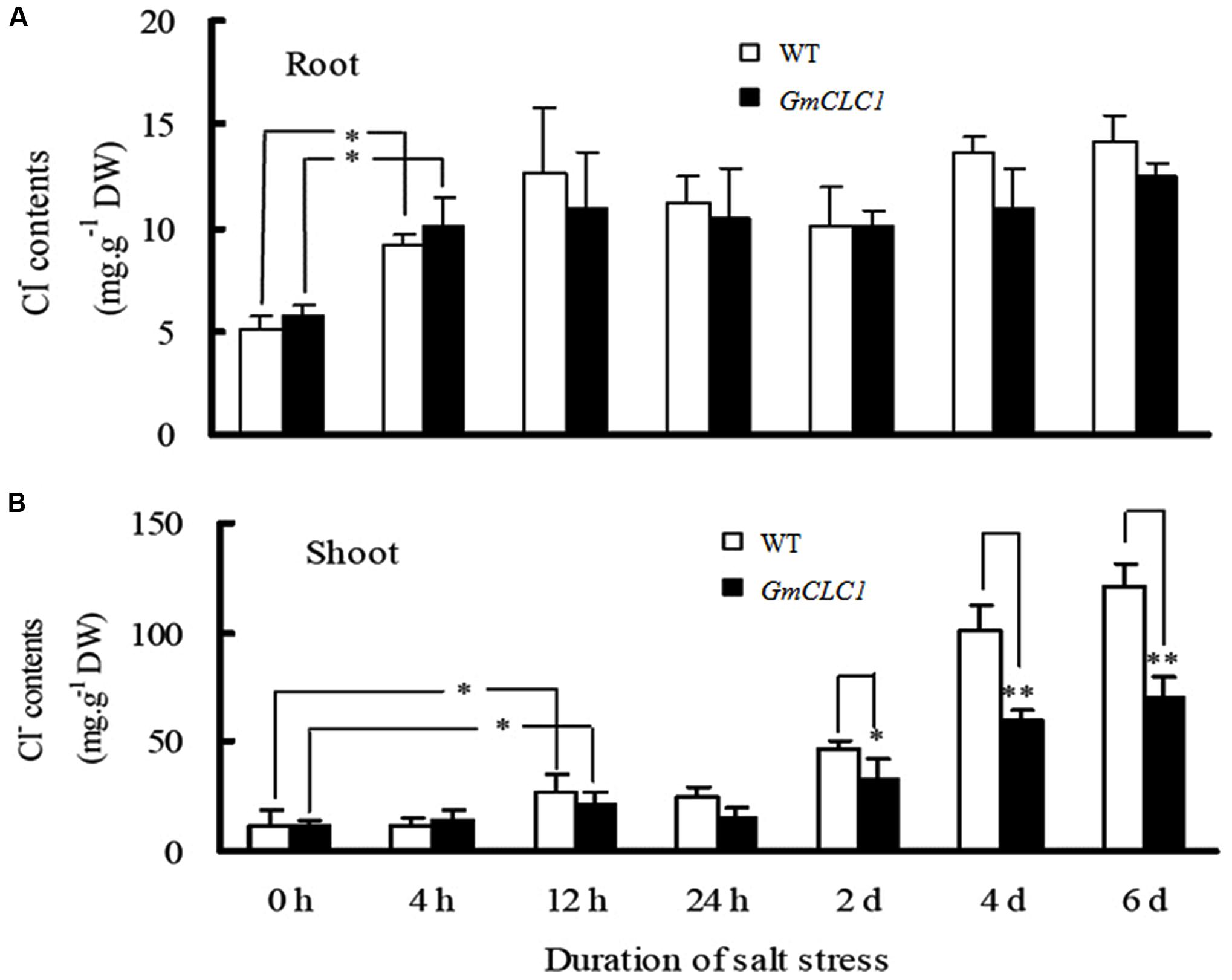
FIGURE 3. Dynamic changes in the chloride (Cl-) contents in (A) roots and (B) shoots of A. thaliana WT and transgenic GmCLC1 seedlings under 150 mM NaCl stress for 6 days. Seeds of WT and GmCLC1 transgenic A. thaliana were grown on pots containing the sterilized peat moss and vermiculite mixture and grown. After 30 days, plants were treated with 1/2 Hoagland solution containing 150 mM NaCl for 0, 4, 12, 24 h, 2, 4, and 6 days, respectively, then the roots (A) and shoot (B) of Arabidopsis seedlings were sampled and Cl- contents were measured. ∗/∗∗: The difference between WT and the GmCLC1 transgenic line was significant at p < 0.05/0.01, using Duncan’s test. Each bar represents the mean and SD of at least three replicates.
GmCLC1 Alleviates NaCl Stress on Transgenic Soybean Hairy Root Growth and Composite Plants
The transformation of soybean cotyledon hairy roots with GmCLC1 was shown to be successful through genotyping by PCR (Figure 4A). Under normal growing condition without additional NaCl, both empty vector- and GmCLC1-transformed hairy roots grew well, with no obvious difference (Figure 4B). However, under 100 mM NaCl stress, the growth of the hairy roots of both GmCLC1-transformed and empty vector-transformed lines were significantly inhibited, and the numbers of root branching were also reduced as compared to the untreated control. The maximum root lengths and fresh weights of empty vector-transformed hairy roots decreased more significantly than the GmCLC1-transformed ones (p < 0.01). At the NaCl concentration of 150 mM, the growth of the hairy roots of both GmCLC1- and empty vector-transformed cotyledons were inhibited, but there was no significant difference between the two (Figures 4C,D).
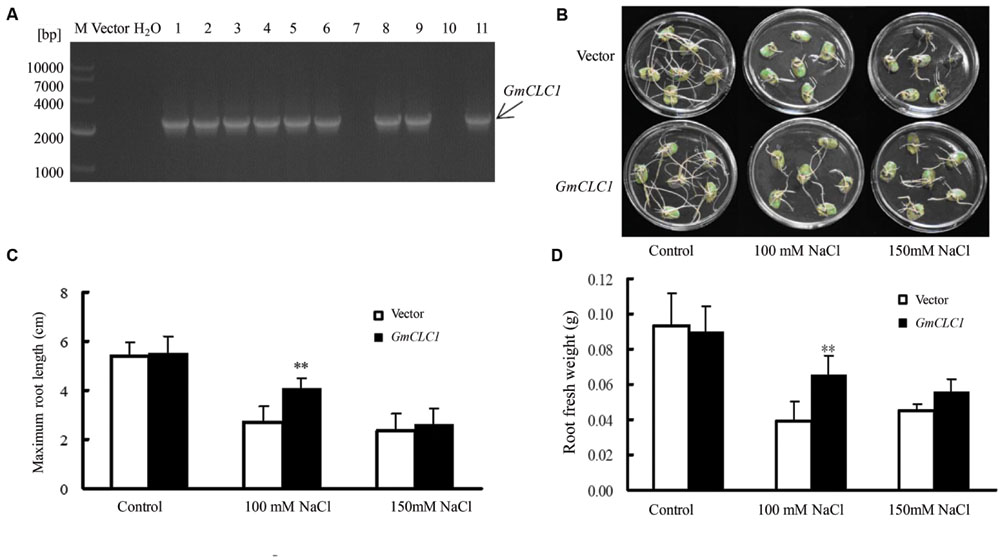
FIGURE 4. Growth characteristics of transgenic soybean (hair root GmCLC1) under NaCl treatment. (A) PCR verification of the presence of the transgene, GmCLC1, in the transgenic soybean hairy roots (GmCLC1) that had undergone Agrobacterium rhizogenes-mediated cotyledon transformation. Nine out of the 11 lines were successfully transformed. M: DNA marker; Vector: empty vector, as negative control; ddH2O: distilled deionized water, as negative control. Comparisons of (B) hairy root phenotype, (C) maximum root length, and (D) root fresh weight of empty vector-transformed control and GmCLC1 grown in with 1/2 Hoagland solution containing 0, 100, and 150 mM NaCl for 5 days. Each bar represents the mean and SD of three replicates each with six soybean cotyledons. ∗∗: The difference between was significant at p < 0.01, using Duncan’s test.
Building on the results we obtained using the GmCLC1-transgenic soybean cotyledon hairy root system, we further tested the response of whole GmCLC1-transgenic soybean hairy root composite plants to salt stress. First, the presence of the transgene in the soybean plants was confirmed by PCR (Figure 5A). Without any additional NaCl in the culture medium, there was no obvious phenotypic difference between the GmCLC1-transgenic soybean (including both Jackson and Lee68 cultivars) hairy root composite plants and the empty vector-transformed plants (Figure 5B). There was also no significant difference in the REL values of roots, first true leaves, the first and second trifoliate leaves among all the genotypes when grown in NaCl-free medium (Figure 5C).
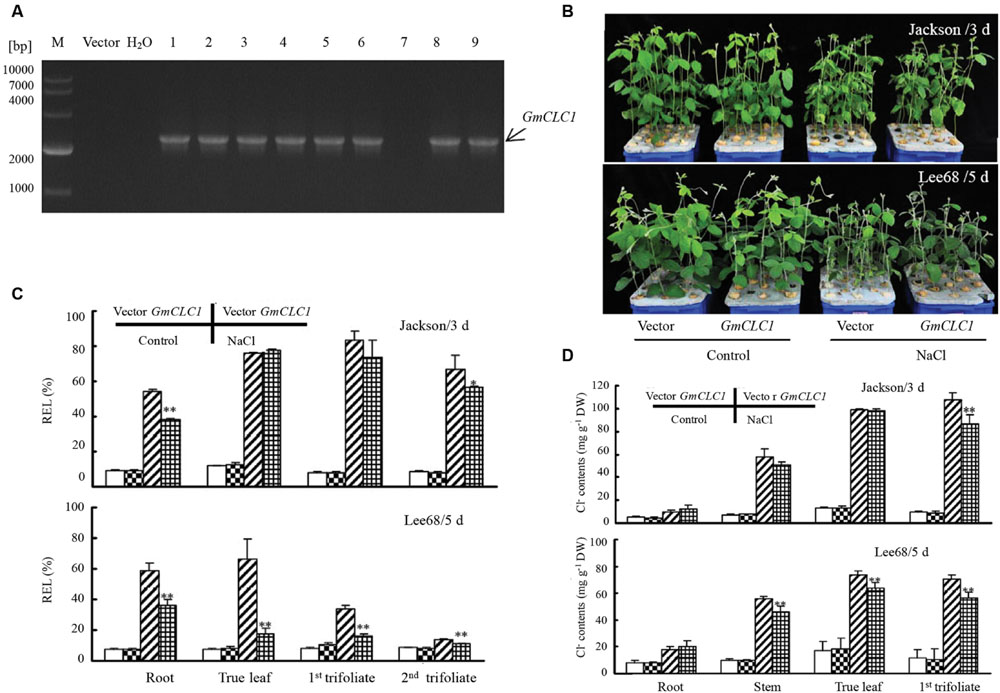
FIGURE 5. Growth phenotype and physiological responses of soybean hairy root composite plants (GmCLC1) under NaCl treatment. (A) PCR verification of the presence of the transgene, GmCLC1, in the soybean hairy root composite plants (lines 1–6 and 8–9). M: DNA markers; Vector: Empty vector-transformed plants (negative control); ddH2O: distilled deionized water (negative control). (B) Phenotypes of empty vector-transformed and GmCLC1-transgenic plants. Surface-sterilized soybean seeds were sown in container till the first pair of unifoliate leaves was fully expanded, the cotyledon node site of soybean seedlings were infected with the A. rhizogenesin K599 for 1 h at room temperature. The wound was then covered with moist vermiculite and co-cultivated with A. rhizogenesin K599 for 5 days at 25 ± 2°C under a 12/12 h light/dark cycle. After 7 days, hairy root lines were identified by PCR for free of bacterium. The original roots were removed and the seedlings with positive hairy roots were cultured in 1/2 Hoagland solution. After 10 days, seedlings were treated with 1/2 Hoagland solutions containing 120 mM NaCl for 3 or 5 days, respectively, and solutions without NaCl as the untreated control. (C) REL and (D) Cl- content of GmCLC1-transgenic soybean plants versus empty vector-transformed controls (Vector) in different parts of the plant. Control: no NaCl was given to the growth medium; DW: dry weight; ∗/∗∗: The difference between was significant at p < 0.05/0.01, using Duncan’s test. Each bar represents the mean and SD of three replicates.
When Jackson or Lee68 GmCLC1-transgenic soybean hairy root composite plants and the corresponding empty vector-transformed plants were exposed to 150 mM NaCl solution, the relatively salt-sensitive Jackson cultivar (both vector-only and GmCLC1) displayed obvious salt injury symptoms (with severely withered leaves) on the 3rd day, while the salt-tolerant Lee68 cultivar (both vector-only and GmCLC1) showed only mildly withered leaves up to the 5th day (Figure 5B). Furthermore, the hairy root composite plants of both cultivars that were transformed with GmCLC1 showed better salt adaptation than their empty vector-transformed counterparts, especially for the more salt-tolerant Lee68 cultivar (Figure 5B). When examining the REL values in separate parts of the composite soybean plants, we found the REL values of roots, first true leaves, the first and second trifoliate leaves of GmCLC1 were all significantly lower than their empty vector-transformed counterparts from the Lee68 cultivar (p < 0.01). The REL values of the second trifoliates of the GmCLC1-transgenic Lee68 cultivar, especially, were comparable to those of the water control. The REL values of the roots and second trifoliate leaves of GmCLC1 of the Jackson cultivar were also significantly lower than their empty vector-transformed counterparts (p < 0.01 and 0.05, respectively; Figure 5C).
In addition, under salt stress, the Cl- contents in roots, stems, first true leaves and the first trifoliate leaves of both Jackson and Lee68 hairy root composite plants (including both GmCLC1 and vector-only) were dramatically increased compared to the water control. It is clear that the GmCLC1-transgenic plants of the Lee68 cultivar, when compared to their empty vector-transformed counterparts, had significantly lower Cl- accumulation in their stems, first true leaves and the first trifoliate leaves (p < 0.01), and GmCLC1-transgenic plants of the Jackson cultivar also had significantly lower Cl- accumulation in the first trifoliates (p < 0.01; Figure 5D). This indicates that the ectopic expression of GmCLC1 in Jackson or Lee68 hairy root composite plants can reduce the Cl- transportation and accumulation in the aerial parts of the plant, especially for the salt-tolerant Lee68 cultivar.
GmCLC1 Enhances Survival of the Chloride-Channel-Deficient Yeast Mutant Δgef1 under MnCl2, NaCl, or KCl
All the yeast strains, including the mutant Δgef1 (a GEF1-deficient mutant), and Δgef1 transformed with empty vector, GEF1, and GmCLC1, grew well on both YPD and YPG media under normal growing conditions. When cultured on YPG medium supplemented with 3 mM MnCl2, the mutant Δgef1 and Δgef1/Vector were unable to grow, but the mutants transformed with yeast GEF1 or soybean GmCLC1 could. Similarly, mutants transformed with yeast GEF1 or soybean GmCLC1 grew better on YPG medium supplemented with 1.0 M NaCl or 1.0 M KCl (Figure 6A). The Cl- contents in the cells of Δgef1/GEF1 and Δgef1/GmCLC1 grown in liquid YPG medium plus 1.0 M NaCl were significantly higher than in the control cells without intracellular vesicle-localized GEF1 transporter activity (Δgef1 and Δgef1/Vector; p < 0.01; Figure 6B). Thus GmCLC1 may have similar functions much like the yeast GEF1.
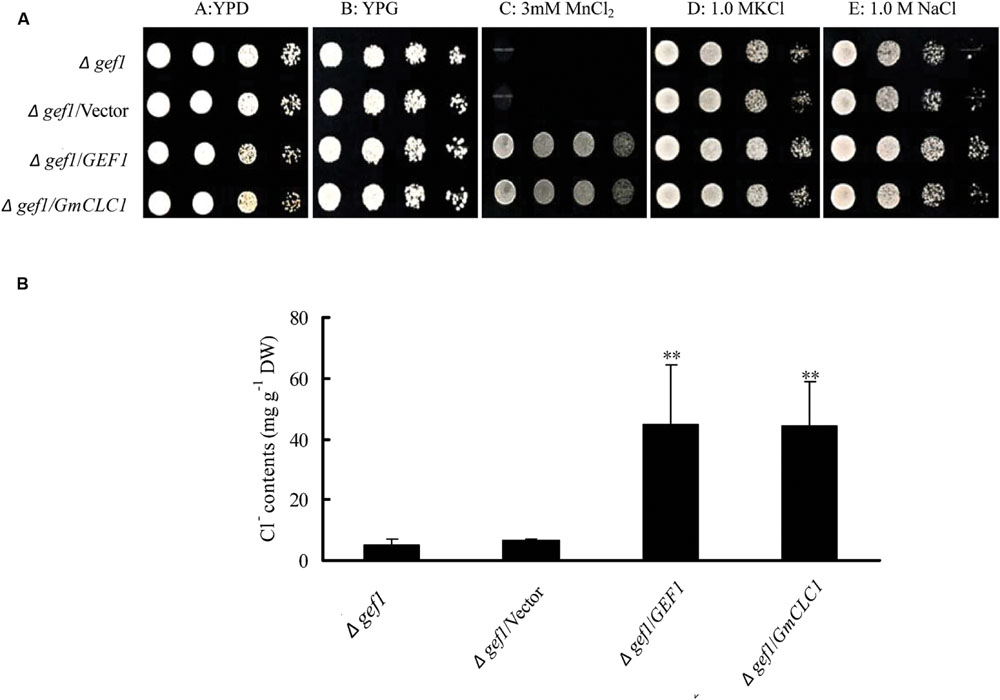
FIGURE 6. Effects of expressing GmCLC1 on the survival of the Δgef1 yeast mutant under MnCl2, NaCl, or KCl. (A) GmCLC1 could enhance the survival of the chloride channel-deficient Δgef1 yeast mutant in the same way as GEF1, when the yeast was grown in media supplemented with MnCl2, KCl, and NaCl. The Δgef1 mutant was also transformed with an empty vector as a negative control. The expression vector pYES2 containing GEF1 or GmCLC1 was introduced into the Δgef1 mutant. Ten-fold serial dilutions (starting at OD550 ≈ 0.5) of each sample were plated on media of YPD, YPG, or YPG supplemented with 3 mM MnCl2, 1.0 M KCl, or 1.0 M NaCl. Plates were incubated at 30°C and photographs were taken after 60 h. (B) The expression of GmCLC1 in the gef1 yeast mutant increased Cl- accumulation. Cells of the above-mentioned Δgef1 mutant, Δgef1/Vector, Δgef1/GEF1, or Δgef1/GmCLC1 were grown in liquid YPG medium with 1.0 M NaCl. When cultures reached OD550 ≈ 0.2, cells were collected by filtration, and their Cl- contents were determined. DW, dry weigh. Data shown are the means and standard deviations of ion contents of three independent cultures of each strain. ∗∗: The difference between was significant at p < 0.01, using Duncan’s test.
Discussion
Ionic toxicity is the main cause of salt injury for plants or crops, and Cl- is the main culprit. The predominant strategies for plants to reduce the effects of salt stress are via active Cl- efflux or vacuolar Cl- partitioning to reestablish intracellular Cl- homeostasis (Tealle and Tyerman, 2010; Wei et al., 2013; Wong et al., 2013), especially for chloride-intolerant plants such as the cultivated soybean, citrus, grape, potato, tobacco, and so on (Moya et al., 2003; Henderson et al., 2014).
In the Arabidopsis genome, a total of seven genes (AtCLCa-g) encoding putative CLC proteins have been identified. For example, AtCLCc is mainly expressed in stomatal guard cells, and the AtCLCc protein located in the tonoplast displays transmembrane Cl--transporting activity, which not only aids in the regulation of stomatal movements, but also in enhancing salt tolerance (Lv et al., 2009; Jossier et al., 2010). Nguyen et al. (2015) reported that a knock-out mutant AtCLCg, a member of the Arabidopsis CLC family localized in the vacuolar membrane, showed a decrease in biomass and an accumulation of chloride in shoots when grown under NaCl stress. AtCLCg was expressed in mesophyll cells, hydathodes and phloem while AtCLCc (with 62% similarity to AtCLCg protein) was expressed in stomata. A atclcc/atclccg double mutant was not more sensitive to NaCl than the single mutants, which demonstrated that AtCLCc and AtCLCg formed part of a regulatory network controlling chloride sensitivity and were both important for chloride tolerance but not redundant.
The soybean genome has eight CLC genes in total: Glyma01g44950, Glyma05g14760 (GmCLC1), Glyma09g28620, Glyma11g00690, Glyma13g23080, Glyma16g06190, Glyma16g33351, and Glyma19g25680 located on chromosomes 1, 5, 9, 11, 13, 16, and 19, respectively (Li et al., 2014).
We previously reported that GmCLC1 encodes a putative Cl-/H+ antiporter (Wong et al., 2013) that is localized on tonoplast (Li et al., 2006). Protective functions of GmCLC1 were only shown using transgenic tobacco cells (Li et al., 2006). In this study, we conducted functional tests in planta.
Our results showed that the ectopic expression of GmCLC1 in Arabidopsis (Figure 1A) significantly enhanced the transgenic seeds germination rate and subsequent seedling growth (Figures 1B–D), and the transgenic plants were better able to adapt to NaCl stress (Figure 2A). Moreover, the alleviation of salt injuries to GmCLC1-transgenic Arabidopsis plants was correlated with reduction in Cl- transport and accumulation in shoots compared to WT plants (Figure 3B). Since GmCLC1 is tonoplast-localized, it is unlikely that this transporter will exclude Cl- to prevent accumulating of this ion. Therefore, the root Cl- concentration did not differ significantly by expressing GmCLC1 (Figure 3A). In contrast, since GmCLC1 may help to compartmentalize Cl- into vacuole, it could delay Cl- transporting from root to shoot and hence lead to a lower Cl- concentration in the aerial part, in both transgenic A. thaliana (Figure 3B) and composite soybean plants (Figure 5D).
In a previous study Ali et al. (2012), it was found that over-expression of nine TFs (such as GmWRKY, GmNAC2, GmbZIP110, and GmMYB92) in hairy roots could enhance the survival or tolerance to 200 mM NaCl stress in mosaic or composite soybean plants. We also demonstrated that cotyledon hairy roots could be used as a rapid gain-of-function test for ion transporters (Qi et al., 2014). In this study, we employed soybean cotyledon hairy roots transformed with GmCLC1 to investigate the role of this chloride transporter in enhancing salt tolerance (Figures 4B–D). We found that overexpression of GmCLC1 in Jackson or Lee68 hairy root composite plants increased the Cl- sequestering in roots, and, as a consequence, reduced Cl- transportation and accumulation in aerial parts, and alleviated their salt injuries as represented by the REL values in roots and various stages of leaf development. This was especially true for the salt-tolerant Lee68 cultivar (Figure 5). All these findings inferred that in GmCLC1 plays a vital role in enabling the soybean plant to adapt to chloride/salt stress.
To provide more understanding on how the GmCLC1 protein may function to control Cl- accumulation, we made use of a yeast system. The yeast GEF1 gene encodes a CLC-type chloride channel protein, which is co-localized with Ccc2 proteins to control Cu2+ in the intracellular vesicles, and drives Cl- and H+ transmembrane exchanges. Thus the yeast GEF1 protein is involved in the co-transport of Cu2+ or Fe3+, the regulation of cation homeostasis and the growth of yeast cells. In the Δgef1 loss-of-function yeast mutant, the Cl- transport into the intracellular vesicles (vacuole or Golgi apparatus) was blocked, resulting in hypersensitivity to extracellular cations such as Cu2+, Fe3+, and Mn2+, hygromycin B, tetramethyl ammonium chloride, and thus growth were hindered (Gaxiola et al., 1998; Lv et al., 2009; Sasvari et al., 2013). Among the seven Arabidopsis CLC paralogs (AtCLCa-g), over-expressions of AtCLCa, AtCLCd, and AtCLCf in the yeast gef1 mutant could complement the deficiency in GEF1 protein functions or growth phenotype (Lv et al., 2009; Barbier-Brygo et al., 2011). When the soybean GmCLC1 or the yeast GEF1 gene was transformed into the Δgef1 mutant and cultured in YPG media containing different chloride salts (MnCl2, KCl, NaCl), the growth of the transformed mutants was much enhanced compared to the original mutant. Therefore, GmCLC1 may help restore the Cl- transportation into intercellular vesicles.
Author Contributions
PW and LW conducted the experiments, collected and analyzed all data. BY and H-ML designed the experiments. BY, H-ML, and AL interpreted the data and wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 30871462) and the Transgenic Engineering Crops Breeding Special Funds of China (No. 2009ZX08004-008B) awarded to BY and grants from Hong Kong Research Grants Council (Collaborative Research Fund: CUHK3/CRF/11G; Area of Excellence: AoE/M-05/12) and Lo Kwee-Seong Biomedical Research Fund awarded to H-ML.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Dayong Zhang (Institute of Biotechnolgy, Jiangsu Academy of Agricultural Sciences, China) for kindly providing the A. rhizogenes strain K599 and related technical guidance on soybean transformation. We also thank Ms. Jee-Yan Chu for copy-editing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01082
FIGURE S1 | Growth characteristics of GmCLC1-transgenic Arabidopsis thaliana. (A) Growth of WT and transgenic Arabidopsis GmCLC1 (L1–L5) under salt stress. Comparison in (B) fresh weight (FW) and (C) shoot fresh weight (FW) of WT and GmCLC1-transgenic Arabidopsis plants (L1, L2, L5). Seeds of WT and Arabidopsis transgenic GmCLC1 plants were surface-sterilized and kept at 4°C for 2–4 days and then sown on MS medium. After 8 days of incubation, the seedlings were transferred into plastic pots filled with a sterilized peat moss and vermiculite mixture, and fertilized with 1/2-strength Hoagland nutrient solution for 5 days in the greenhouse. The nutrient solution was continuously replaced with half-strength Hoagland solution containing 50 mM NaCl for 2 days, followed by 100 mM NaCl for 2 days, and finally 150 mM NaCl for 6 days. No NaCl was added to the nutrient solution for the control treatment. ∗/∗∗: The difference between WT and GmCLC1 was significant at p < 0.05/0.01, respectively, using Duncan’s test. Each bar represents the mean and SD of at least three replicates.
References
Adams, E., and Shin, R. (2014). Transport, signaling, and homeostasis of potassium and sodium in plants. J. Integr. Plant Biol. 56, 231–249. doi: 10.1111/jipb.12159
Adem, G. D., Roy, S. J., Zhou, M. X., Bowman, J. P., and Shabala, S. (2014). Evaluating contribution of ionic, osmotic and oxidative stress components towards salinity tolerance in barley. BMC Plant Biol. 14:113. doi: 10.1186/1471-2229-14-113
Ali, Z., Zhang, D. Y., Xu, Z. L., Xu, L., Yi, J. X., He, X. L., et al. (2012). Uncovering the salt response of soybean by unraveling its wild and cultivated functional genomes using tag sequencing. PLoS ONE 7:e48819. doi: 10.1371/journal.pone.0048819
Barbier-Brygo, H., Angeli, A. D., Filleur, S., Frachisse, J. M., Gambale, F., Thomine, S., et al. (2011). Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu. Rev. Plant Biol. 62, 25–51. doi: 10.1146/annurev-arplant-042110-103741
Brumós, J., Talón, M., Bouhlal, R., and Colmenero-Flores, J. M. (2010). Cl- homeostasis in includer and excluder citrus rootstocks: transport mechanisms and identification of candidate genes. Plant Cell Environ. 33, 2012–2027. doi: 10.1111/j.1365-3040.2010.02202.x
Chen, X. Q., and Yu, B. J. (2007). Ionic effects of Na+ and Cl- on photosynthesis in Glycine max seedlings under isoosmotic salt stress. J. Plant Physiol. Mol. Biol. 33, 294–300.
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G. H., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Gaxiola, R. A., Yuan, D. S., Klausner, R. D., and Fink, G. R. (1998). The yeast CLC chloride channel functions in cation homeostasis. Proc. Natl. Acad. Sci. U.S.A. 95, 4046–4050. doi: 10.1073/pnas.95.7.4046
Gietz, R. D. (2014) “Yeast transformation by the LiAc/SS carrier DNA/PEG method,” in Yeast Protocols, Methods in Molecular Biology. Vol. 1163, ed. W. Xiao (New York, NY: Springer Science+Business Media), 33–44.
Guo, W., Zuo, Z., Cheng, X., Sun, J., Li, H., Li, L., et al. (2014). The chloride channel family gene CLCd negatively regulates pathogen-associated molecular pattern (PAMP)-triggered immunity in Arabidopsis. J. Exp. Bot. 65, 1205–1215. doi: 10.1093/jxb/ert484
Gupta, B., and Huang, B. (2014). Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 1–18. doi: 10.1155/2014/701596
Hasegawa, P. M. (2013). Sodium (Na+) homeostasis and salt tolerance of plants. Environ. Exp. Bot. 92, 19–31. doi: 10.1016/j.envexpbot.2013.03.001
Henderson, S. W., Baumann, U., Blackmore, D. H., Walker, A. R., Walker, R. R., and Gilliham, M. (2014). Shoot chloride exclusion and salt tolerance in grapevine is associated with differential ion transporter expression in roots. BMC Plant Biol. 14:273. doi: 10.1186/s12870-014-0273-8
Hu, S. B., Zhou, Q., An, J., and Yu, B. J. (2016). Cloning of PIP genes in drought-tolerant vetiver grass and responses of transgenic VzPIP2;1 soybean plants to water stress. Biol. Plant. doi: 10.1007/s10535-016-0631-5
Jossier, M., Kroniewicz, L., Dalmas, F., Le Thiec, D., Ephritikhine, G., Thomine, S., et al. (2010). The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J. 64, 563–576. doi: 10.1111/j.1365-313X.2010.04352.x
Li, W., Wang, L. C., and Yu, B. J. (2014). Bioinformatics analysis of CLC homologous genes family in soybean genome. J. Nanjing Agric. Univ. 37, 35–43.
Li, W. Y. F., Wong, F. L., Tsai, S. N., Phang, T. H., Shao, G. H., and Lam, H. M. (2006). Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environ. 29, 1122–1137. doi: 10.1111/j.1365-3040.2005.01487.x
Luo, Q. Y., Yu, B. J., and Liu, Y. L. (2005). Differential sensitivity to chloride and sodium ions in seedlings of Glycine max and G. soja under NaCl stress. J. Plant Physiol. 162, 1003–1012. doi: 10.1016/j.jplph.2004.11.008
Lv, Q. D., Tang, R. J., Liu, H., Gao, X. S., Li, Y. Z., Zheng, H. Q., et al. (2009). Cloning and molecular analyses of the Arabidopsis thaliana chloride channel gene family. Plant Sci. 176, 650–661. doi: 10.1016/j.plantsci.2009.02.006
Moya, J. L., Gómez-Cadenas, A., Primo-Millo, E., and Talon, M. (2003). Chloride absorption in salt-sensitive Carrizo citrange and salt-tolerant Cleopatra mandarin citrus rootstocks is linked to water use. J. Exp. Bot. 54, 825–833. doi: 10.1093/jxb/erg064
Nguyen, C. T., Agorio, A., Jossier, M., Depré, S., Thomine, S., and Filleur, S. (2015). Characterization of the chloride channel-like, AtCLCg, involved in chloride tolerance in Arabidopsis thaliana. Plant Cell Physiol. 57, 764–775. doi: 10.1093/pcp/pcv169
Nie, W. X., Xu, L., and Yu, B. J. (2015). A putative soybean GmsSOS1 confers enhanced salt tolerance to transgenic Arabidopsis sos1-1 mutant. Protoplasma 252, 127–134. doi: 10.1007/s00709-014-0663-7
Qi, X. P., Li, M. W., Xie, M., Liu, X., Ni, M., Shao, G. H., et al. (2014). Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 5:4340. doi: 10.1038/ncomms5340
Qu, Y. N., Zhou, Q., and Yu, B. J. (2009). Effects of Zn2+ and niflumic acid on photosynthesis in Glycine soja and Glycine max seedlings under NaCl stress. Environ. Exp. Bot. 65, 304–309. doi: 10.1016/j.envexpbot.2008.11.005
Sasvari, Z., Kovalev, N., and Nagy, P. D. (2013). The GEF1 Proton-Chloride exchanger affects Tombusvirus replication via regulation of copper metabolism in yeast. J. Virol. 87, 1800–1810. doi: 10.1128/JVI.02003-12
Tealle, N. L., and Tyerman, S. D. (2010). Mechanisms of Cl- transport contributing to salt tolerance. Plant Cell Environ. 33, 566–589. doi: 10.1111/j.1365-3040.2009.02060.x
Tian, F., Jia, T. J., and Yu, B. J. (2014). Physiological regulation of seed soaking with soybean isoflavones on drought tolerance of Glycine max and Glycine soja. Plant Growth Regul. 74, 229–237. doi: 10.1007/s10725-014-9914-z
Tregeagle, J. M., Tisdall, J. M., Tester, M., and Walker, R. R. (2010). Cl–uptake, transport and accumulation in grapevine rootstocks of differing capacity for Cl—exclusion. Funct. Plant Biol. 37, 665–673. doi: 10.1071/FP09300
Wang, S., Su, S. Z., Wu, Y., Li, S. P., Shan, X. H., Liu, H. K., et al. (2015). Overexpression of maize chloride channel gene ZmCLC-d in Arabidopsis thaliana improved its stress resistance. Biol. Plant. 59, 55–64. doi: 10.1007/s10535-014-0468-8
Wei, Q. J., Liu, Y. Z., Zhou, G. F., Li, Q. H., Yang, C. Q., and Peng, S. A. (2013). Overexpression of CsCLCc, a chloride channel gene from Poncirus trifoliata, enhances salt tolerance in Arabidopsis. Plant Mol. Biol. Rep. 31, 1548–1557. doi: 10.1007/s11105-013-0592-1
Wong, T. H., Li, M. W., Yao, X. Q., and Lam, H. M. (2013). The GmCLC1 protein from soybean functions as a chloride ion transporter. J. Plant Physiol. 170, 101–104. doi: 10.1016/j.jplph.2012.08.003
Xiao, Y. H., Xi, H., Shen, Y. Z., and Huang, Z. J. (2013). A novel wheat alpha-amylase inhibitor gene, TaHPS, significantly improves the salt and drought tolerance of transgenic Arabidopsis. Physiol. Plant. 148, 273–283. doi: 10.1111/j.1399-3054.2012.01707.x
Yu, B. J., and Liu, Y. L. (2004). Chlorine, chloride channel and chlorine tolerance in plants. Chin. Bull. Bot. 21, 402–410.
Zhang, X. K., Zhou, Q. H., Cao, J. H., and Yu, B. J. (2011). Differential Cl-/Salt tolerance and NaCl-induced alternations of tissue and cellular ion fluxes in Glycine max, Glycine soja and their hybrid seedlings. J. Agron. Crop Sci. 197, 329–339. doi: 10.1111/j.1439-037X.2011.00467.x
Zhou, C., Wang, H. P., Zhu, J., and Liu, Z. X. (2013). Molecular cloning, subcellular localization and functional analysis of thclc-a from Thellungiella Halophila. Plant Mol. Biol. Rep. 31, 783–790. doi: 10.1007/s11105-012-0545-0
Zhou, Q., and Yu, B. J. (2009). Accumulation of inorganic and organic osmolytes and their role in osmotic adjustment in nacl-stressed vetiver grass seedlings. Russ. J. Plant Physiol. 56, 678–875. doi: 10.1134/S1021443709050148
Keywords: chloride transporter, Cl- toxicity, GmCLC1, salt stress, soybean root transformation, transgenic Arabidopsis
Citation: Wei P, Wang L, Liu A, Yu B and Lam H-M (2016) GmCLC1 Confers Enhanced Salt Tolerance through Regulating Chloride Accumulation in Soybean. Front. Plant Sci. 7:1082. doi: 10.3389/fpls.2016.01082
Received: 03 June 2016; Accepted: 08 July 2016;
Published: 25 July 2016.
Edited by:
Urs Feller, University of Bern, SwitzerlandReviewed by:
Amarendra Narayan Misra, Central University of Jharkhand, IndiaMichiel Van Eijk, KeyGene, Netherlands
Copyright © 2016 Wei, Wang, Liu, Yu and Lam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingjun Yu, bjyu@njau.edu.cn Hon-Ming Lam, honming@cuhk.edu.hk
 Peipei Wei
Peipei Wei Longchao Wang1
Longchao Wang1 Ailin Liu
Ailin Liu Bingjun Yu
Bingjun Yu Hon-Ming Lam
Hon-Ming Lam