- Department of Integrated Plant Protection, Agrotest Fyto Ltd, Kroměříž, Czechia
The primary genepool of barley comprises two subspecies – wild barley (Hordeum vulgare subsp. spontaneum) and cultivated barley H. vulgare. subsp. vulgare. The former originated 5.5 million years ago in southwest Asia and is the immediate ancestor of cultivated barley, which arose around 10,000 years ago. In this study, the specific resistance of a set of 146 wild barley accessions, maintained by the International Center for Agriculture Research in the Dry Areas (ICARDA), to 32 isolates of barley powdery mildew caused by Blumeria graminis f. sp. hordei was evaluated. The set comprised 146 heterogeneous accessions of a previously tested collection. Seed was obtained by single seed descent and each accession was usually represented by five single plant progenies. In total, 687 plant progenies were tested. There were 211 phenotypes of resistance among the accessions, 87 of which were found in single plants, while 202 plants contained the eight most common phenotypes. The most frequent phenotype was found in 56 plants that were susceptible to all pathogen isolates, whereas the second most frequent phenotype, which occurred in 46 plants, was resistant to all isolates. The broad resistance diversity that was revealed is of practical importance and is an aid to determining the extent and role of resistance in natural ecosystems.
Introduction
The grass family, Poaceae, originated during the Upper Cretaceous period and the Hordeum and Triticum genera diverged about 13 million years ago followed by the evolution of Hordeum one million years later (Nevo, 2013). More than 30 species of barley (Hordeum) are known, nearly all of which are included in the tertiary genepool of H. vulgare L. The secondary genepool comprises H. bulbosum L., which shares the basic genome with H. vulgare. Although there are interspecific crossability problems, especially regarding the tertiary genepool (Bothmer et al., 2003), discrete introgressions from H. bulbosum into the H. vulgare genome have been successful resulting in the transfer of disease resistance and other useful agronomic traits (Pickering et al., 2006; Shtaya et al., 2007; Wendler et al., 2015).
The primary genepool of barley is composed of two subspecies namely, wild barley [H. vulgare subsp. spontaneum (C. Koch) Tell.] and cultivated barley (H. vulgare subsp. vulgare) (hereafter denoted by Hvs and Hvv, respectively). H. vulgare originated 5.5 million years ago in southwest Asia (Dai et al., 2012). Hvv first arose from Hvs by independent polyphyletic domestication in three centres (Middle-East, Central Asia, and Tibet) around 10,000 years ago (Nevo, 2013). Hvs is two-rowed and diploid (2n = 2x = 14) and differs from Hvv in several important traits including brittle rachis and a tough (non-brittle) awn. There are no crossability barriers between the two subspecies and gene transfer is easily accomplished. The centre of distribution for Hvs is located mainly in southwest Asia, particularly in the Middle-East where vertical zonality and the diversity of environmental and climatic factors led to ecological and morphological differentiation (Bothmer et al., 2003).
Powdery mildew caused by Blumeria graminis (D.C.) Golovin ex Speer f. sp. hordei Em. Marchal (Bgh) is a world-wide disease of barley (Jørgensen, 1994; Dreiseitl, 2011a) infecting both Hvv and Hvs. Many host resistances to Bgh have been found in Hvs from the Middle-East (Fischbeck et al., 1976; Dreiseitl and Bockelman, 2003). Therefore, the region can be considered as a “specific” centre of diversity of barley resistance to Bgh. Regular occurrence of the pathogen (Dinoor and Eshed, 1990) and the long-term influence of evolutionary forces (McDonald and Linde, 2002) in the Middle-Eastern population of Bgh is indicated by the presence of many corresponding virulences (Eyal et al., 1973; Dreiseitl et al., 2006). Thus, the Middle-Eastern population of Bgh on Hvs is also the natural centre of diversity of this pathogen (Dreiseitl, 2014).
Hvs is still abundant in regions where it occurs naturally and because of its great diversity of useful traits for crop breeding there are also collections in gene banks. The resistance of Hvs against powdery mildew is usually done with a set of Bgh isolates. Isolate pathogenicity is determined using host genotypes that can differentiate the pathogen isolates. In both cases, the results are collated as response type arrays (RTAs) consisting of response types (RT = phenotype of plant × isolate interaction). The objective of this study was to determine the presence of specific resistances to Bgh in a set of heterogeneous Hvs accessions held in the International Center for Agriculture Research in the Dry Areas (ICARDA).
Materials and Methods
Plant Material and Pathogen Isolates
Resistance of a set of 146 accessions of Hvs maintained by ICARDA was studied (Table 1). This set comprised accessions from 363 previously tested accessions whose heterogeneity prevented a definitive assessment of RTAs (Dreiseitl, unpublished). Seed was obtained using single seed descent and progenies from five single plants were used for screening 128 accessions. The remaining 18 accessions had fewer plants available for testing. In total, 687 plant progenies were tested at the Agrotest Fyto Ltd.
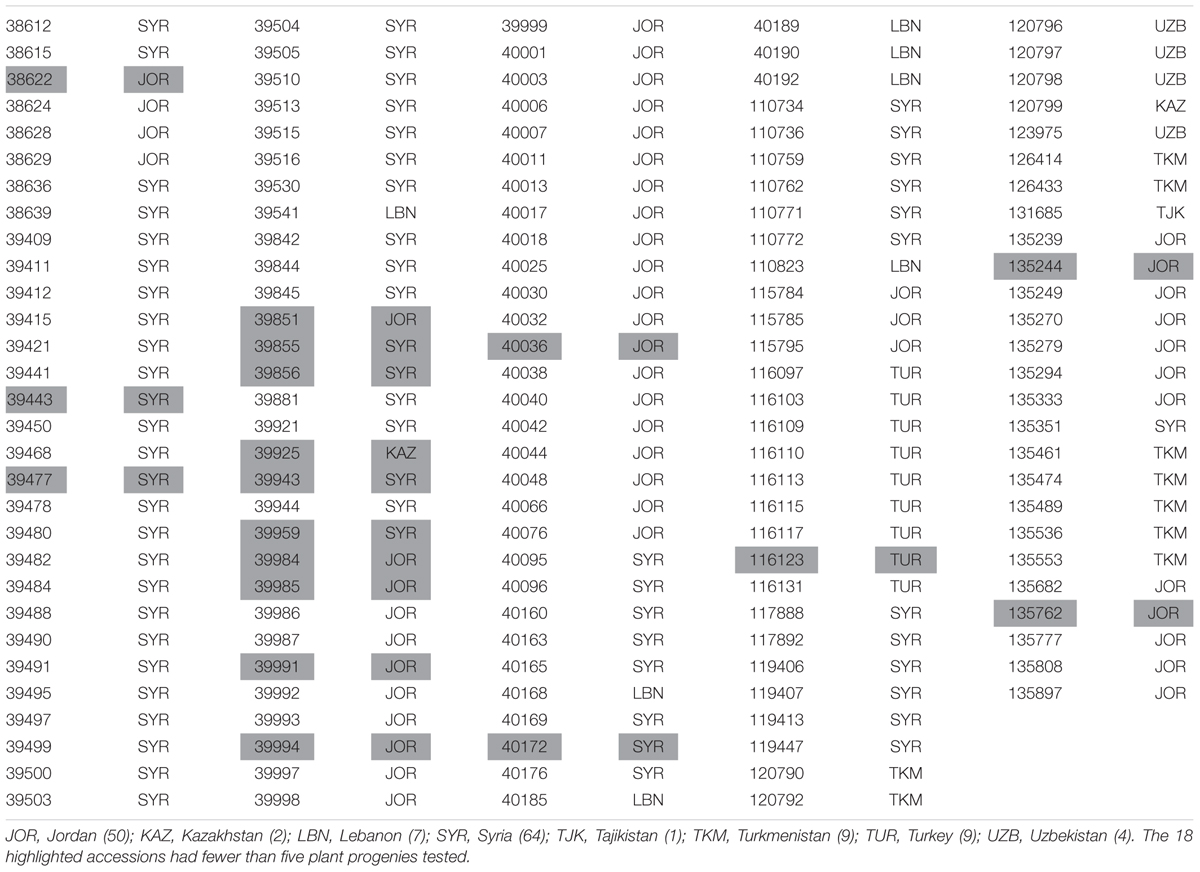
TABLE 1. Origin of 146 Hordeum vulgare subsp. spontaneum accessions maintained by the International Center for Agriculture Research in the Dry Areas (ICARDA) tested with 32 Blumeria graminis f. sp. hordei isolates.
Thirty-two reference isolates of Bgh from the pathogen gene bank at Agrotest Fyto Ltd. representing the world diversity of the pathogen were used for resistance tests (Table 2). Five of them were collected on Hvs naturally occurring in Israel. Before inoculation, each isolate was purified, verified as the correct pathogenicity phenotype on standard barley lines (Kølster et al., 1986) and multiplied on leaf segments of susceptible B-3213.
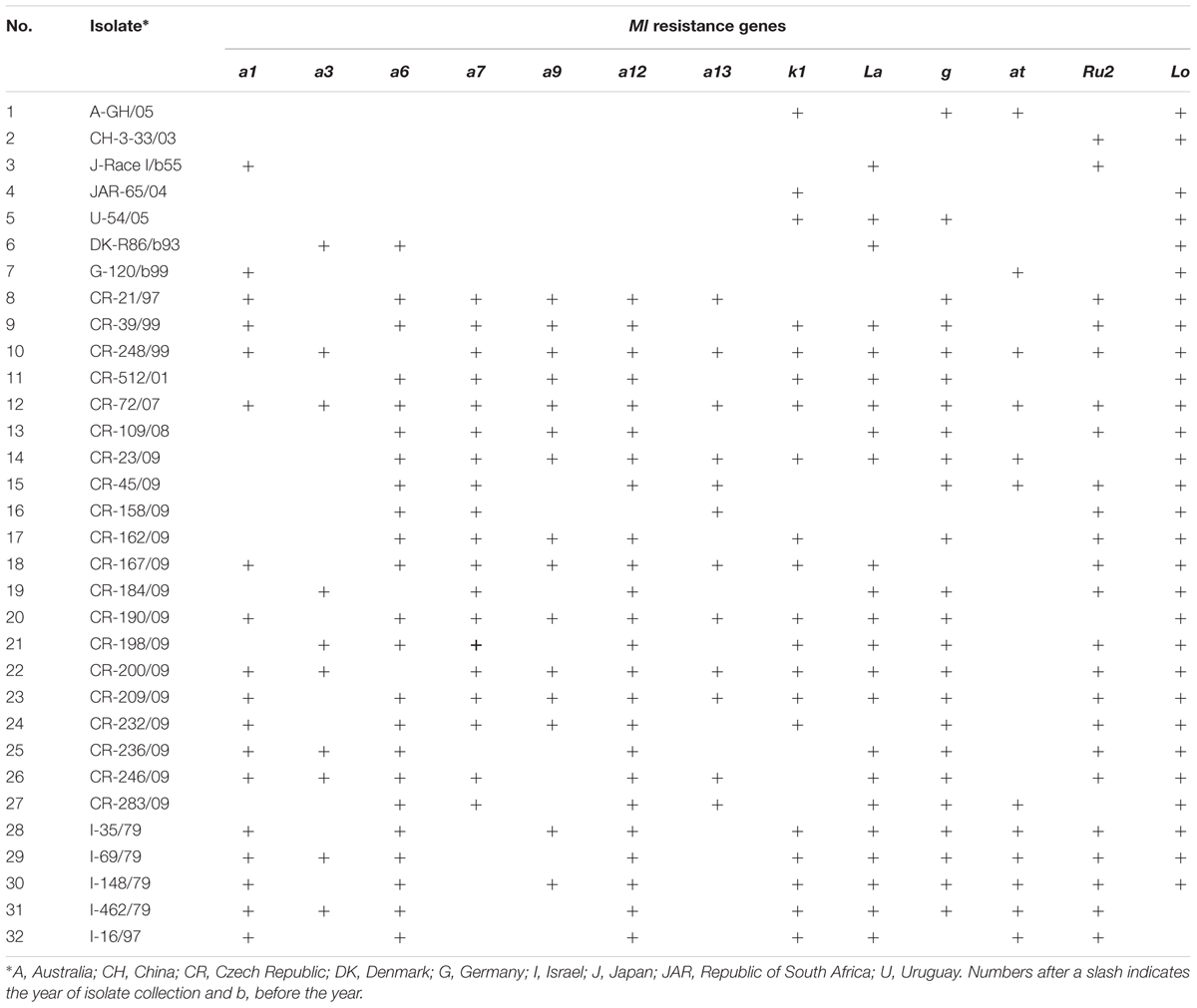
TABLE 2. Origin of 32 Blumeria graminis f. sp. hordei isolates used for resistance tests of 687 Hordeum vulgare subsp. spontaneum plant progenies and their virulence (+) against 13 Ml resistance genes.
Testing Procedure
About 30 seeds of each barley plant were sown in a pot (80 mm diameter) filled with a gardening peat substrate and placed in a mildew-proof greenhouse under natural daylight. Leaf segments 20 mm long were cut from the central part of healthy fully expanded primary leaves when the second leaves were emerging. Three segments of each plant and four segments of the susceptible B-3213 were placed with adaxial surfaces upward in a 150 mm Petri dish on water agar (0.8%) containing benzimidazole (40 mg/l) – a leaf senescence inhibitor. For each isolate, a dish with leaf segments was placed at the bottom of a metal inoculation tower and inoculated at a concentration of c. 8 conidiospores/mm2. The dishes with inoculated leaf segments were incubated at 18 ± 2.0°C under artificial light (cool-white fluorescent lamps providing 12 h light at 30 ± 5 μmol/m2/s).
Evaluations and Plant Phenotype Designations
Seven days after inoculation, RTs on the central part of the adaxial side of leaf segments were scored on a scale 0–4, where 0 = no visible mycelium or sporulation, and 4 = strong mycelial growth and sporulation on the leaf segment (Torp et al., 1978); RTs 0–2 were considered resistant. In rare cases, when slightly different RTs occurred on the three leaf segments the prevalent RT was recorded. A set of RTs provided a RTA for each Hvs plant. Based on the gene-for-gene model (Flor, 1971), a specific resistance gene of the host is matched by a virulence gene of the pathogen.
Numerical designations of Hvs phenotypes (RTAs) were based on their resistance/susceptibility patterns to the set of 32 isolates ranked in the order shown in Table 2 and divided into 10 triplets and two last isolates on their own. Each of the digits indicates susceptibility to the three isolates of the respective triplet. If susceptibility to a corresponding isolate was detected, the first isolate is given the value 1 (20), the second isolate has the value 2 (21), and the third isolate 4 (22). Therefore, each digit can have a value from 0 (no susceptibility to any of the three isolates) up to 7 (=1 + 2 + 4) denoting susceptibility to each of the three isolates. The resulting number (reverse-octal) defines the resistance/susceptibility patterns of the Hvs plants and their phenotypic classification (Gilmour, 1973; Limpert and Müller, 1994; Dreiseitl and Dinoor, 2004). The HaGiS program was used for transcription of RTAs into octal notation (Hermann et al., 1999).
Results
The set of 687 individually harvested plants derived from 146 Hvs accessions was inoculated with 32 Bgh isolates (Supplementary Table 1S). Within this set there were 211 phenotypes (i.e., different combinations of resistance and susceptibility to the Bgh isolates used). Eighty-seven phenotypes were recorded in only one plant with a phenotypic frequency of 1, while the eight most frequent phenotypes were found in 202 plants (Table 3). The most frequent phenotype (77777777773) appeared in 56 plants and was susceptible to all 32 Bgh isolates. In contrast, 46 plants showed the second most frequent phenotype (00000000000) and was resistant to all 32 pathogen isolates (Table 4).
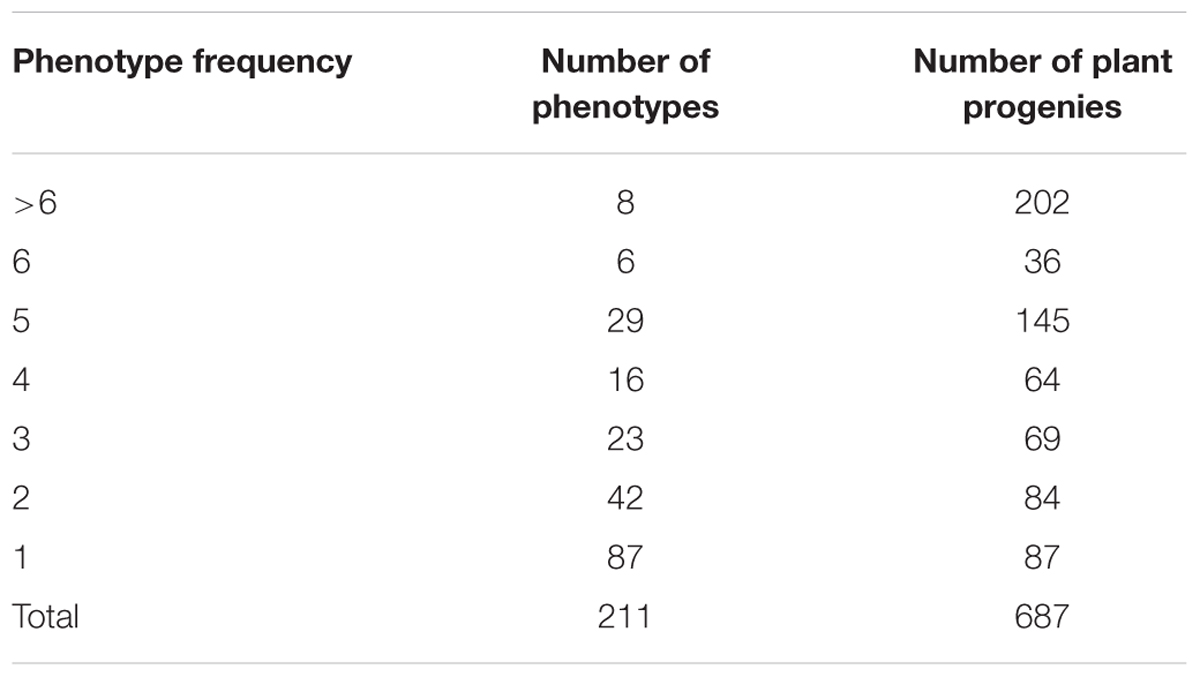
TABLE 3. Frequency of resistance phenotypes found in 687 Hordeum vulgare subsp. spontaneum plant progenies inoculated with 32 Blumeria graminis f. sp. hordei isolates.
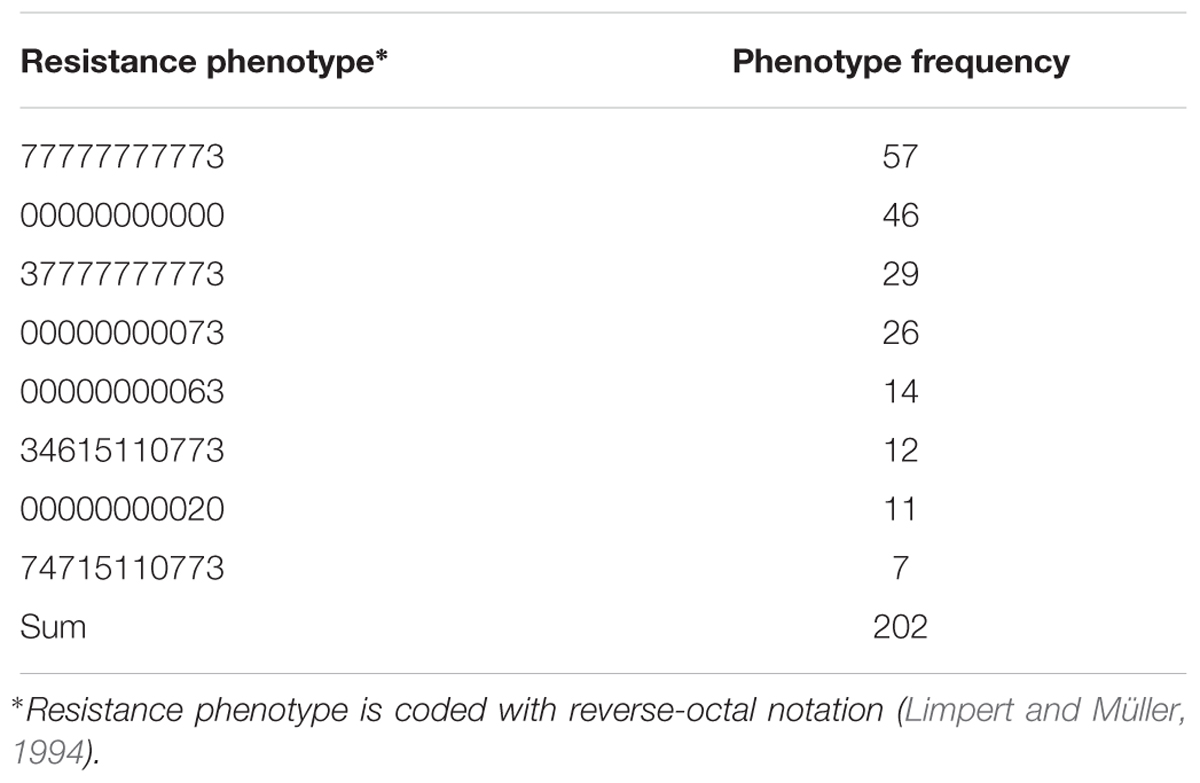
TABLE 4. The eight most common resistance phenotypes found among 687 individually harvested Hordeum vulgare subsp. spontaneum plants inoculated with 32 Blumeria graminis f. sp. hordei isolates.
Eleven phenotypes (000000000xx), comprising 127 plants (18.5% of 687 total plants tested), were resistant to all non-Israeli Bgh isolates including four of the eight most frequent (00000000000, 00000000073, 00000000063, and 00000000020) (Table 4). At least 50% of plants of 25 accessions (17.1% of the 146 tested accessions) were resistant to all non-Israeli isolates and there were 100 resistant plants among the 120 plants from these accessions (Table 5). The remaining 27 resistant plants were found in 22 other accessions.
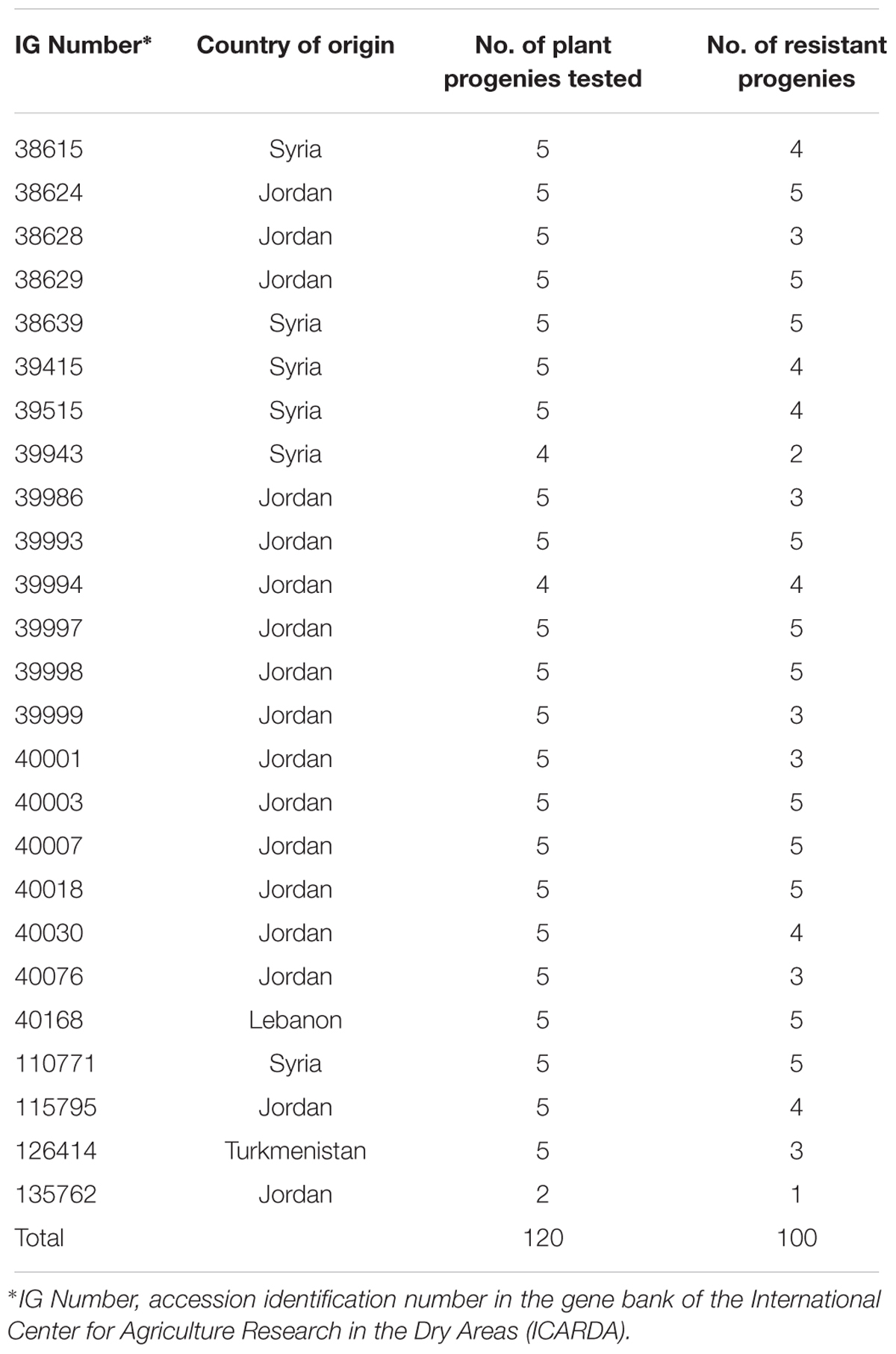
TABLE 5. Origin of 25 Hordeum vulgare subsp. spontaneum accessions for which at least half of the plant progenies were resistant to all 27 non-Israeli Blumeria graminis f. sp. hordei isolates.
Sixty-four of the 146 accessions (43.8%) originated from collections in Syria and 50 (34.2%) from Jordan (Table 1). Of the 25 accessions for which at least 50% of plants were resistant to all non-Israeli isolates, six originated from Syria (9.4% of Syrian accessions) and 17 from Jordan (34.0% of Jordanian accessions) (Table 5).
A total of 128 accessions were represented by five tested plants. Plants with a single identical phenotype were found in 47 of these accessions (36.7%; Table 6), and all of the plants from 11 accessions were resistant to every non-Israeli Bgh isolates (Table 5). Only four accessions (38636, 40038, 40095, and 110823) were fully susceptible. In contrast, in two accessions (116131 and 117892), each plant exhibited a different phenotype. Plants from each of the remaining 79 accessions displayed from two to four phenotypes (Table 6).

TABLE 6. Number of resistance phenotypes found among 128 accessions of Hordeum vulgare subsp. spontaneum inoculated with 32 Blumeria graminis f. sp. hordei isolates; five plant progenies were tested from each accession.
Testing the set of Hvs plants with 32 Bgh isolates yielded 21,984 RTs. RT2 was the most frequent in 7,769 (=35.3%) Hvs–Bgh interactions. In contrast, RT0 was the least frequent and was only found in 1,178 (5.4%) interactions (Table 7). This RT denotes the greatest resistance (immunity) and was most frequently observed on plants inoculated with non-European Hvv isolates (mean of 85 RT0 per isolate). Inoculation with these isolates resulted in the lowest determined frequency recorded (mean of 117) for RT4 (fully susceptible). Conversely, the lowest frequency for RT0 was found among plants inoculated with Hvs isolates from Israel (mean of 7). Inoculation with these isolates also had the highest frequency (mean of 139) for RT4. RT1 (complete resistance), was also most often seen on plants inoculated with non-European Hvv isolates (83) and least frequently observed on plants inoculated with isolates found on Hvs in Israel (12).
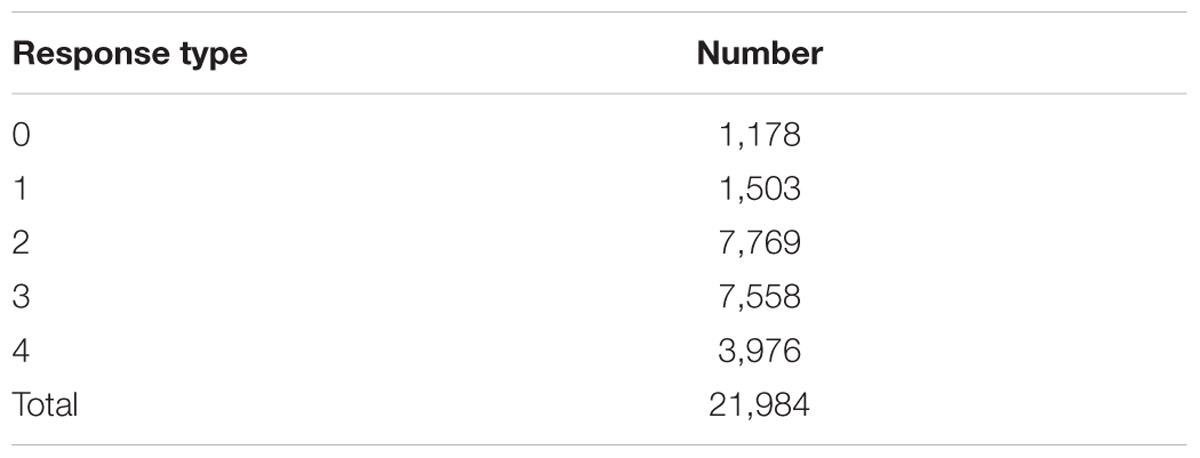
TABLE 7. Number of response types developed in 687 Hordeum vulgare subsp. spontaneum plant progenies after inoculation with 32 Blumeria graminis f. sp. hordei isolates.
Discussion
In this study, 127 out of 687 Hvs plants were resistant to all 27 non-Israeli Bgh isolates. This contrasts with previous reports describing the response of Hvv lines to Bgh. For example, seven out of 159 local landraces of the Spanish core collection were resistant to seven Bgh isolates (Silvar et al., 2009). Furthermore, only one out of 147 Chinese varieties was resistant to all 32 isolates of Bgh (Dreiseitl and Yang, 2007), and no powdery mildew resistance to 28 isolates was found among 129 genotypes obtained from the Serbian barley gene bank (Šurlan-Momirović et al., 2016). Thus, Hvs is recognized as a good source of broadly acting resistance to Bgh.
In the present investigation, there was great diversity of Hvs resistance to Bgh; 211 phenotypes were recorded among 146 accessions. One hundred and twenty-eight accessions were represented by five tested plants. All plants within each of 47 accessions were homogeneous, had identical phenotypes and in the set of 47 accessions 38 different phenotypes were recorded.
In contrast, 81 heterogeneous accessions always exhibited more than one phenotype (mean of 2.63 phenotypes). Hence, Hvs diversity of resistance to Bgh has developed naturally during long-term co-evolution of these organisms (Fischbeck et al., 1976; Moseman et al., 1981; Dreiseitl and Dinoor, 2004). It is likely that the high resistance diversity of accessions housed in gene banks might also has arisen first, from the original collection of bulked samples that were more likely to be heterogeneous, second, as a result of the open flowering nature of Hvs (Brown et al., 1978) leading to frequent out-pollination in multiplication plots, and third, from mechanical admixtures during successive cycles of seed increase (Dreiseitl, 2007).
Once RTAs reach a certain complexity the data are collated. Mathematical codes are the most suitable parameter, especially octal notation and separation data into triplets (Gilmour, 1973; Limpert and Müller, 1994). Therefore, the use of this notation for classifying pathotypes (physiologic races) has been recommended (Limpert et al., 1994). Since host–parasite relationships relate to compatibility responses, it was proposed to use octal notation also for classifying host resistances (Dreiseitl and Dinoor, 2004).
The present study set was composed of accessions selected from 363 accessions that had been tested previously and were heterogeneous in their response to well-characterized Bgh isolates. Forty-seven accessions had homogeneous phenotypes for all five tested plants. In previous tests, standard seed samples were used and each accession was composed of about 30 plants. However, in this report testing of “only” five plant progenies did not establish that these 47 accessions were heterogeneous. Testing more plants would result in reduced numbers of homogeneous accessions caused by fewer impurities arising from cross-pollination or mechanical admixtures.
The region where Hvs naturally occurs is subject to annual Bgh epidemics. As a result of co-evolution, the local pathogen population has adapted to specific resistances (Dreiseitl et al., 2006) that originate in this area and which were, or still are, fully effective worldwide (Jahoor and Fischbeck, 1987; Dreiseitl and Bockelman, 2003). The five pathogen isolates originating from collections in Israel, therefore, provide a deeper understanding of the diversity of resistance in the evaluated accessions. Although the RTAs suggest that the resistances are not durable, their effectiveness in other regions should be high.
All the recorded RTAs were compared with RTAs of standard barley genotypes with known resistance genes in order to identify resistance genes in the Hvs accessions. Nevertheless, this was satisfactory only in those accessions with RTAs composed of just one or a few avirulent RTs. Twenty-nine plants had the 37777777773 phenotype, which is resistant to the Japanese isolate Race I (Hiura and Heta, 1955). This phenotype can be conditioned by the presence of the following two resistance genes – Mla8, which is already well characterized, produced RT0 in 13 plants, while a lesser known resistance, originally designated Ml(Ch) and present in many Chinese Hvv accessions (Dreiseitl and Yang, 2007), expressed RT2 in 16 plants. The chromosomal location of the latter gene is unknown and some accessions with Mla8 might contain both resistances. In some plants, one or two of these genes may be present along with unidentified gene(s) that mask the corresponding responses to the pathogen. Their combined presence cannot, therefore, be established using conventional screening tests.
Immunity (RT0) to Race I was found in four plants of accession 38615 and six plants from five other accessions, although these genotypes also showed RT0 to two Israeli isolates. This set of three avirulences typifies the presence of the Ml(Lo) (Dreiseitl, 2011b). All the above 10 plants had a range of avirulent responses (mostly RT2) following inoculation with some of the other 29 isolates suggesting the presence of unidentified resistances in addition to Ml(Lo).
The two most important basic requirements for genetic resistance of hosts to pathogens are effectiveness and durability. Effectiveness can be defined by the host response to individual pathotypes – e.g., the effectiveness of Mla8 against the Japanese Race I is very high (RT0). However, it is more appropriate to characterize effectiveness at the population level. For example, Mla8 is ineffective against the European Bgh population is zero because no isolate avirulent to Mla8 was present in the population. The minimum requirement for durability is that resistance is maintained throughout a host’s lifespan.
In summary, genetic resistance to Bgh can be divided into qualitative resistance conditioned by major genes of specific resistance (Jørgensen, 1994), quantitative resistance conditioned by minor genes (Niks et al., 2015), and durable mlo resistance gene (Jørgensen, 1992). The latter has not yet been found in Hvs. Many specific resistance genes have been detected in Hvs (Dreiseitl, 2014) and many others can be hypothesized based on the present findings. However, using specific resistances to Bgh singly is not recommended for breeding barley varieties. Nevertheless, they can be used in varieties when combined with mlo. This is a useful strategy since the pathogen can gradually adapt even to partial virulence to this durable resistance gene (Schwarzbach, 1987) and erode its effectiveness. In such combinations, an effective specific resistance can prevent the erosion of mlo efficacy. A new method may also be devised that will improve the durability of specific resistances (Brown, 2015). Detecting specific resistances is, therefore, important first, for their potential use in breeding programs; second, for easier detection of minor genes that are frequently masked by specific resistance genes; third for increasing our knowledge of the occurrence and role of resistance in natural ecosystems.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Funding
The article was generated within project no. RO0211 supported by the Ministry of Agriculture of the Czech Republic.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
Excellent technical assistance of Mrs. Dagmar Krejčířová is gratefully acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00202/full#supplementary-material
References
Bothmer, R., von Sato, K., Komatsuda, T., Yasuda, S., and Fischbeck, G. (2003). “The domestication of cultivated barley,” in Diversity in Barley (Hordeum vulgare), eds R. von Bothmer, T. van Hintum, H. Knüpffer, and K. Sato (Amsterdam: Elsevier), 9–27. doi: 10.1016/S0168-7972(03)80004-X
Brown, A. H. D., Zohary, D., and Nevo, E. (1978). Outcrossing rates and heterozygosity in natural populations of Hordeum spontaneum Koch in Israel. Heredity 41, 49–62. doi: 10.1038/hdy.1978.63
Brown, J. K. M. (2015). Durable resistance of crops to disease: a Darwinian perspective. Annu. Rev. Phytopathol. 53, 513–539. doi: 10.1146/annurev-phyto-102313-045914
Dai, F., Nevo, E., Wu, D., Comadran, J., Zhou, M., Qiu, L., et al. (2012). Tibet is one of the centers of domestication of cultivated barley. Proc. Natl. Acad. Sci. U.S.A. 109, 16969–16973. doi: 10.1073/pnas.1215265109
Dinoor, A., and Eshed, N. (1990). “Plant diseases in natural population of wild barley (Hordeum spontaneum),” in Pests, Pathogens and Plant Communities, eds J. J. Burdon and S. R. Leather (Oxford: Blackwell Scientific Publications), 169–186.
Dreiseitl, A. (2007). Powdery mildew resistance in winter barley cultivars. Plant Breed. 126, 268–273. doi: 10.1007/BF01184924
Dreiseitl, A. (2011a). Differences in powdery mildew epidemics in spring and winter barley based on 30-year variety trials. Ann. Appl. Biol. 159, 49–57. doi: 10.1111/j.1744-7348.2011.00474.x
Dreiseitl, A. (2011b). Dissimilarity of barley powdery mildew resistances Heils Hanna and Lomerit. Czech J. Genet. Plant Breed. 47, 95–100.
Dreiseitl, A. (2014). The Hordeum vulgare subsp. spontaneum - Blumeria graminis f. sp. hordei pathosystem: its position in resistance research and breeding applications. Eur. J. Plant Pathol. 138, 561–568. doi: 10.1007/s10658-013-0266-8
Dreiseitl, A., and Bockelman, H. E. (2003). Sources of powdery mildew resistance in a wild barley collection. Genet. Resour. Crop Evol. 50, 345–350. doi: 10.1023/A:1023953819787
Dreiseitl, A., and Dinoor, A. (2004). Phenotypic diversity of barley powdery mildew resistance sources. Genet. Resour. Crop Evol. 51, 251–258. doi: 10.1023/B:GRES.0000024010.12369.b3
Dreiseitl, A., Dinoor, A., and Kosman, E. (2006). Virulence and diversity of Blumeria graminis f.sp. hordei in Israel and in the Czech Republic. Plant Dis. 90, 1031–1038. doi: 10.1094/PD-90-1031
Dreiseitl, A., and Yang, J. (2007). Powdery mildew resistance in a collection of Chinese barley varieties. Genet. Resour. Crop Evol. 54, 259–266. doi: 10.1007/s10722-005-3810-3
Eyal, Z., Yurman, R., Moseman, J. G., and Wahl, I. (1973). Use of mobile nurseries in pathogenicity studies of Erysiphe graminis hordei on Hordeum spontaneum. Phytopathology 63, 1330–1334. doi: 10.1094/Phyto-63-1330
Fischbeck, G., Schwarzbach, E., Sobel, Z., and Wahl, I. (1976). Mildew resistance in Israeli populations of 2-rowed wild barley (Hordeum spontaneum). Z. Pflanzenzüchtung 76, 163–166.
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev-phyto-072910-095339
Gilmour, J. (1973). Octal notation for designating physiologic races of plant pathogens. Nature 242:620. doi: 10.1038/242620a0
Hermann, A., Löwer, C. F., and Schachtel, G. A. (1999). A new tool for entry and analysis of virulence data for plant pathogens. Plant Pathol. 48, 154–158. doi: 10.1046/j.1365-3059.1999.00325.x
Hiura, U., and Heta, H. (1955). Studies on the disease-resistance in barley. III. Further studies on the physiologic races of Erysiphe graminis hordei in Japan. Ber. Ohara Inst. Landwirtschaftliche Biol. 10, 135–156.
Jahoor, A., and Fischbeck, G. (1987). Sources of resistance to powdery mildew in barley lines derived from Hordeum spontaneum collected in Israel. Plant Breed. 99, 274–281. doi: 10.1111/j.1439-0523.1987.tb01182.x
Jørgensen, J. H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63, 141–152. doi: 10.1007/BF00023919
Jørgensen, J. H. (1994). Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 13, 97–119. doi: 10.1080/07352689409701910
Kølster, P., Munk, L., Stølen, O., and Løhde, J. (1986). Near-isogenic barley lines with genes for resistance to powdery mildew. Crop Sci. 26, 903–907. doi: 10.2135/cropsci1986.0011183X002600050014x
Limpert, E., Clifford, B., Dreiseitl, A., Johnson, R., Müller, K., Roelfs, A., et al. (1994). Comparing systems of designation of pathotypes of plant pathogens. J. Phytopathol. 140, 359–362. doi: 10.1111/j.1439-0434.1994.tb00618.x
Limpert, E., and Müller, K. (1994). Designation of pathotypes of plant pathogens. J. Phytopathol. 140, 346–358. doi: 10.1111/j.1439-0434.1994.tb00617.x
McDonald, B. A., and Linde, C. (2002). Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. doi: 10.1146/annurev.phyto.40.120501.101443
Moseman, J. G., Baenzinger, P. S., and Kilpatrick, R. A. (1981). Genes conditioning resistance of Hordeum spontaneum to Erysiphe graminis f. sp. hordei. Crop Sci. 21, 229–232. doi: 10.2135/cropsci1981.0011183X002100020006x
Nevo, E. (2013). “Evolution of wild barley and barley improvement. Advance in barley sciences,” in Proceedings of the 11th International Barley Genetics Symposium, eds G. Zhang, C. Li, and X. Liu (Dordrecht: Zhejiang University Press and Springer Science + Business Media), 1–23. doi: 10.1007/978-94-007-4682-4_1
Niks, R. E., Xiaoquan, Q., and Thierry, C. M. (2015). Quantitative resistance to biotrophic filamentous plant pathogens: concepts, misconceptions, and mechanisms. Annu. Rev. Phytopathol. 53, 445–470. doi: 10.1146/annurev-phyto-080614-115928
Pickering, R., Ruge-Wehling, B., Johnston, P. A., Schweizer, G., Ackermann, P., and Wehling, P. (2006). The transfer of a gene conferring resistance to scald (Rhynchosporium secalis) from Hordeum bulbosum into H-vulgare chromosome 4HS. Plant Breed. 125, 576–579. doi: 10.1111/j.1439-0523.2006.01253.x
Schwarzbach, E. (1987). “Shifts to increased pathogenicity on mlo varieties,” in Integrated Control of Cereal Mildews: Monitoring the Pathogen, eds M. S. Wolfe and E. Limpert (Dordrecht: Martinus Nijhoff Publishers), 5–7.
Shtaya, M. J. Y., Sillero, J. C., Flath, K., Pickering, R., and Rubiales, D. (2007). The resistance to leaf rust and powdery mildew of recombinant lines of barley (Hordeum vulgare L.) derived from H-vulgare x H-bulbosum crosses. Plant Breed. 126, 259–267. doi: 10.1111/j.1439-0523.2007.01328.x
Silvar, C., Casas, A. M., Kopanhe, D., Habekuss, A., Schweizer, G., Gracia, M. P., et al. (2009). Screening the Spanish barley core collection for disease resistance. Plant Breed. 129, 45–52. doi: 10.1111/j.1439-0523.2009.01700.x
Šurlan-Momirović, G., Flath, K., Silvar, C., Branković, G., Kopahnke, D., Knežević, D., et al. (2016). Exploring the Serbian GenBank barley (Hordeum vulgare L. subsp vulgare) collection for powdery mildew resistence. Genet. Resour. Crop Evol. 63, 275–287. doi: 10.1007/s10722-015-0246-2
Torp, J. H., Jensen, P., and Jørgensen, J. H. (1978). Powdery Mildew Resistance Genes in 106 Northwest European Spring Barley Varieties. Copenhagen: Royal Veterinary and Agricultural University, 75–102.
Keywords: Blumeria graminis f. sp. hordei, diversity of resistances, Hordeum vulgare subsp. spontaneum, phenotypes, response type arrays
Citation: Dreiseitl A (2017) Heterogeneity of Powdery Mildew Resistance Revealed in Accessions of the ICARDA Wild Barley Collection. Front. Plant Sci. 8:202. doi: 10.3389/fpls.2017.00202
Received: 13 July 2016; Accepted: 02 February 2017;
Published: 14 February 2017.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Javier Sanchez, University of Zurich, SwitzerlandPatrick Schweizer, Institute of Plant Genetics and Crop Plant Research (LG), Germany
Copyright © 2017 Dreiseitl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonin Dreiseitl, dreiseitl@vukrom.cz
 Antonin Dreiseitl
Antonin Dreiseitl