- 1State Key Laboratory of Crop Genetics and Germplasm Enhancement, College of Agriculture, Nanjing Agricultural University, Nanjing, China
- 2Jiangsu Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing, China
- 3Department of Plant Pathology and Microbiology, Texas A&M University, College Station, TX, USA
- 4Division of Biotechnology, Chonbuk National University, Iksan, South Korea
Ethylene (ET) emitted by plant tissues has been broadly reported to play important roles in plant development, response to environmental stresses and defense against certain pathogens. Recent evidence obtained from using in vitro fungal cultures exposed to ET suggested that exogenous ET may regulate the production of aflatoxin by Aspergilli. However, the function of endogenous, seed-derived ET has not been explored. In this study, we found that the maize lipoxygenase lox3 mutant, previously reported to be susceptible to Aspergillus spp., emitted greater levels of ET upon A. flavus infection, suggesting the potential involvement of endogenous ET in the susceptibility of maize to A. flavus. Supporting this idea, both colonization and conidiation of A. flavus were reduced in wild-type (WT) kernels treated with AgNO3, an ET synthesis inhibitor. There was no ET emission from non-viable kernels colonized by A. flavus, suggesting that living seed but not the fungus itself was the primary source of ET released upon infection with A. flavus. The kernels of acs2 and acs6, two ET biosynthetic mutants carrying Mutator transposons in the ACC synthase genes, ACS2 and ACS6, respectively, displayed enhanced seed colonization and conidiation, but not the levels of aflatoxin, upon infection with A. flavus. Surprisingly, both acs2 and acs6 mutant kernels emitted greater levels of ET in response to infection by A. flavus as compared with WT seed. The increased ET in single mutants was found to be due to overexpression of functional ACS genes in response to A. flavus infection. Collectively, these findings suggested that ET emitted by infected seed facilitates colonization by A. flavus but not aflatoxin production.
Introduction
Aspergillus is one of the most common mycotoxin producing fungi that contaminate a large number of crops, both pre- and post-harvest. Some of the major economic losses are caused by aflatoxin (produced by A. flavus) contamination of maize and other oil rich seed crops worldwide. Aflatoxin is one of the most potent natural carcinogens hazardous to health of humans and animals when consumed as food or feed, respectively. Unfortunately, conventional agronomic approaches have limited effectiveness for reducing mycotoxin contamination. The ideal solution would be to decrease contamination by improving genetic resistance of these crop plants. Sources of resistance that limit the ability of Aspergillus spp. to grow, reproduce and synthesize mycotoxins have been identified. However, despite significant efforts by public and private breeding programs, adequate levels of resistance have not been achieved primarily due to lacking major single resistance genes against those fungi Munkvold (2003). One strategy to accelerate resistance breeding efforts is to combine the approaches of genome-wide association analysis (GWAS) and traditional linkage mapping analysis to identify the A. flavus resistance quantitative trait loci (QTL) or genes, and closely linked markers. Recently, a linkage mapping was performed using 228 recombinant inbred lines (RILs) of maize, and a highly significant QTL that affected aflatoxin accumulation, qAA8, was mapped to chromosome 8, which can explain 6.7 to 26.8% of the phenotypic variation (Zhang et al., 2016). Several other large QTLs have also reported to be located on chromosomes one, three, four, five, and nine (Xiang et al., 2010; Warburton et al., 2011; Willcox et al., 2013). Another promising approach to aid conventional breeding efforts is to identify genes and signaling pathways regulated by these genes that underlie host resistance (or susceptibility) mechanisms. This information can be used to develop and breed beneficial alleles of these genes or genetic engineering approaches to control infection and mycotoxin production.
One of the plant-derived molecular signals that have been implicated in the regulation of aflatoxin biosynthesis is a gaseous plant hormone ethylene (ET). In higher plants, ET is initially synthesized from methionine (Met) via S-adenosyl-L-methionine (S-AdoMet) by SAM synthetase. S-AdoMet is subsequently converted to 1-aminocyclopropane-1-carboxylate (ACC) by ACC synthase (ACS, S-adenosyl-L-methionine methylthioadenosine-lyase) and finally to ET by ACC oxidase (ACO) (Yang, 1985; Kende, 1993). As a by-product of this reaction, ACS also produces 5′-methylthioadenosine (5′-MTA), which is recycled in the Yang Cycle to synthesize methionine (Met), to maintain the constant level of the cellular Met for continuous protein and ET biosynthesis (Yang, 1985; Bleecker and Kende, 2000).
Besides higher plants, ET is also synthesized by micro-organisms, including phytopathogenic fungi and bacteria (Fukuda et al., 1993), through either 2-keto-4-methylbutyric acid (KMBA) as reported for Escherichia coli (Ince and Knowles, 1986) and Cryptococcus albidus (Fukuda et al., 1989b), or via 2-oxoglutarate by Penicillium digitatum (Fukuda et al., 1989a) and Pseudomonas syringae (Nagahama et al., 1991). The fungus Penicillium citrinum can also synthesize ACC from SAM (Jia et al., 1999), while Botrytis cinerea appears to utilize the KMBA pathway to produce ET (Cristescu et al., 2002; Chague et al., 2006). However, the physiological significance of ET production by these microorganisms has not been well studied, but is hypothesized to facilitate pathogen virulence.
In plants, ET has been broadly reported to play important roles in regulating diverse physiological and defense processes, including seed germination, organ senescence, abscission and fruit ripening (Kende, 1993; Johnson and Ecker, 1998), and responses to abiotic and biotic stresses, such as wounding, chilling, drought, flooding, hypoxia, ozone damage and pathogen attack (Paul et al., 2003; Lin et al., 2009; Musungu et al., 2016). ET modulates various defense responses against pathogens either individually or in combination with other phytohormones (Guo and Ecker, 2004; Broekaert et al., 2006; Adie et al., 2007; Kendrick and Chang, 2008). For instance, the resistance of plants to the necrotrophic fungal pathogen B. cinerea was moderately enhanced by exogenous treatment of plants with ET (Diaz et al., 2002). Also, ET biosynthesis is activated when plants are challenged by pathogens; on the other hand, increased ET production is associated with enhanced defense-related gene expression (Penninckx et al., 1998; Cohn and Martin, 2005). ET acts synergistically with jasmonates (JAs) in defense responses (Penninckx et al., 1998; Ellis and Turner, 2001; Catinot et al., 2015), often accompanied by induction of clusters of genes regulated by ET or jasmonic acid (JA) (Glazebrook et al., 2003; Broekaert et al., 2006).
In addition to acting as a signaling molecule in plant defense against pathogens, ET has also been implicated in the regulation of mycotoxin production by Aspergillus spp. When ET was exogenously applied to peanuts inoculated with A. parasiticus, aflatoxin accumulation was significantly reduced (Roze et al., 2004; Gunterus et al., 2007; Huang et al., 2009). An ET generator, 2-chloroethyl phosphoric acid (CEPA, ethephon), also suppressed aflatoxin biosynthesis in A. flavus, and this suppression was probably due to the reduction of reactive oxygen species (ROS) (Huang et al., 2009). These data suggest a role for exogenous ET in modulating secondary metabolism of Aspergillus spp. However, it remains to be explored whether and how the endogenous ET derived from host plants impacts the fungal colonization and mycotoxin production. In this study, the role of seed-derived ET in maize interactions with A. flavus was elucidated by kernel bioassay using maize mutants disrupted by transposon insertions in the ACS2 and ACS6 genes.
Materials and Methods
Plant Materials and Fungal Strains
Maize acs mutants (acs2 and acs6) and lipoxygenase lox3-4 mutants were generated by Mutator-transposable element-insertional mutagenesis as described previously (Young et al., 2004; Gao et al., 2007). These acs mutants were backcrossed seven times into B73 background resulting in generation of mutants that are near-isogenic to the recurrent parent line B73. The lox3-4 mutants are at the BC5F3 genetic stage. In all experiments, lox3-4 mutants were compared to near-isogenic WTs obtained by self-pollinating WT siblings identified in the BC5F2 segregating population.
To measure the expression levels of ACS2, 6 and 7 in different organs, the samples were collected from different organs at different stages. For geminating stage, the embryos and roots were excised from the 2-day-old or 4-day-old germinated seeds, respectively. For seedling stage, roots, stems and leaves were harvested from V1 and V3 stages grown in a light shelf, respectively. For adult stage, roots, stems, leaves, pollens, tassels and ears were harvested from the matured plants. The seeds after completely matured were also harvested for gene expression study at matured stage.
Aspergillus flavus NRRL 3357 was cultured at room temperature on potato dextrose agar (PDA: Difco) as described previously (Gao et al., 2009).
Fungal Inoculation and Spore Counting on Kernels
Maize kernels in similar size were selected and weighed to ensure equivalent average seed weights across all samples. Seeds were then surface-sterilized with 100% Clorox bleach (containing 6% sodium hypochlorite) for 10–15 min and rinsed with sterilized, distilled H2O at least five times. The embryos of kernels were cut longitudinally using a razor blade to a depth of about 0.5 cm to provide an infection court for fungal inoculation. Seeds were then blotted dry with paper towel and placed in a 20-ml glass-scintillation vial (Wheaton Science, Millville, NJ, USA) and inoculated with 200 μl of conidia suspension (1 × 106/ml) of A. flavus NRRL 3357. Conidia were harvested with 0.001% Tween 20 from fungal strains grown in PDA plate. Control seeds (mock) received equal amount of 0.001% Tween 20. Four or six inoculated or mock-treated kernels were used per replicate with at least four replicates per experiment. The inoculated kernels were kept in a plastic transparent container with a wet filter paper to provide humidity and incubated with 12 h light/day at 26–29°C. Sterile, distilled H2O was added to containers as needed to maintain high humidity. Kernels were harvested at designated intervals after inoculation, either to enumerate conidia or to quantify mycotoxins.
To measure levels of conidia production, infected kernels were placed in a 20 mL glass vial with 2 mL of 0.001% Tween 20, and vortexed for 20 s to dislodge spores. The spore suspension was decanted and spores were enumerated using a hemacytometer.
Semi Quantitative RT-PCR and qRT-PCR for Gene Expression
B73 genetic background kernels were sterilized and inoculated with mock control, A. flavus. All seeds were applied with either control or fungi as described above. Control or fungi-challenged seeds were harvested at 0, 12, 24, 48, and 96 h after inoculation. Total RNA was extracted by using TRI reagent (Molecular research Center Inc., Cincinnati, OH, USA) following the manufacturer’s protocol. After extracting total RNA, these RNA samples were treated with RNase-free rDNase at 37°C for 30 min using a DNA-free kit (Ambion Inc., Austin, TX, USA). First strand cDNA synthesis (5 μg of RNA as a template for each sample) was carried out by using a First-Strand Synthesis Kit (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA) following the manufacturer’s protocol. The synthesized cDNA was diluted and equalized for all samples. The cDNA as a template was amplified with two gene-specific primers for each gene, and for the house-keeping gene ZmGAPc. The cDNA was denatured at 94°C for 5 min and amplified by following 27–32 cycles (each cycle: 45 s at 94°C, 1 min at 56°C, and 2 min at 72°C). Amplified PCR products were loaded and separated on 1.2–1.5% agarose gels.
Quantitative reverse transcription-polymerase chain reaction (qPCR) assay was performed using qRT-PCR kit purchased from Takara (Takara, Japan). Reactions were optimized for RNA and primer concentrations with each 10 μl reaction consisting of 40 ng of DNase-free RNA and 200 nM primers. qPCR analysis was performed in the ABI Prism 7000 system (Applied Biosystems, USA). The program used was as follows: 94°C for 1 min; followed by 40 cycles of 94°C for 5 s, 65°C for 15 s and 72°C for 30 s. Primers used in this study are described in Supplementary Table 1, and Tublin gene was used as internal control. The quantification of gene expression was repeated at least three times.
Quantification of Aflatoxin and Ergosterol
Infected or mock-treated kernels from each treatment were ground using a Waring blender (Waring laboratory, Torrington, CT, USA), and aflatoxin was subsequently quantified with a fluorometer using the VICAM AflaTest® USDA-FGIS procedure (VICAM, Watertown, MA, USA) with six infected kernels per replicate were frozen in liquid nitrogen until assayed. Ergosterol was extracted from infected kernels overnight with 5 ml chloroform:methanol (2:1 v/v) at room temperature as described by previous study (Woloshuk et al., 1989) with some modifications. Ergosterol was analyzed on a Shimatzu LC-20AT HPLC system (Shimatzu Scientific Instruments, Inc., Kyoto, Japan) equipped with a 4.6 U ODS column (250 mm × 4.6 mm) and a UV detector (282 nm). Quantities were calculated by comparing HPLC peak areas with ergosterol standards (Sigma). The experiment was repeated at least four times, with consistent results.
Measurement of Ethylene Produced by Kernels, Leaves and Pathogens
ET produced by infected and by control kernels was quantified as described by Gao et al. (2008) with some modifications. Briefly, the vials containing infected kernels were kept at 12 h light/day at 26–29°C and ET was measured at 1, 2, 4, and 7 days post-inoculation (dpi). Vials were sealed with screw caps with septa. The headspace gas (1 ml) was withdrawn from vials by a syringe and analyzed using gas chromatography.
To measure the ET levels produced in the leaves of B73 and acs6 mutant, the plants were grown at 25 to 28°C in commercial soil (Metro-Mix 366; Scotts-Sierra Horticultural Products, Marysville, OH, USA) under 14 h of daylight with 120 μM m−2 s−1 (Quantum Meter; Apogee Instruments, Logan, UT, USA). The seedlings were grown in long conical tubes (20.5 by 4 cm) for 2 to 3 weeks until they had three fully expanded leaves (V3 developmental stage). The leaves of B73 and acs6 were excised from the plants and immediately transferred into a 20 ml of glass scintillation vial (Wheaton Science). The vials were then tightly sealed with a plastic lid for 1 to 2 h prior to analysis to allow enough ethylene was accumulated. ET was measured as described by Gao et al. (2008).
Treatment with Ethylene Inhibitor and Precursor ACC
The pretreatment of kernels with ET inhibitor AgNO3 was performed by adding 1 ml of AgNO3 (Sigma-Aldrich, St. Louis, MO, USA) at 20 mM in 0.001% Tween-20, to the freshly wounded kernels as described above (four kernels per vial per replicate) and mixed thoroughly to ensure the complete soaking of kernels with the chemical. At least four replicates were used per treatment/genotype combination. Control kernels received the same volume of 0.001% Tween-20. The kernels were incubated for 30 min and blotted dry to remove excessive solution and were subsequently inoculated with A. flavus suspensions as described.
For the treatment with ET precursor, ACC, the kernels were pretreated with ACC (20 nM) in 0.001% Tween-20 for 30 min, followed by inoculation with 1 × 106 spores, then cultured and analyzed as described above. Pretreatment with same amount of 0.001% Tween-20 was used as control.
Results
ET Production Is Enhanced in A. flavus-Inoculated lox3-4 Mutant Kernels
We have previously reported that mutation of a maize 9-lipoxygenase gene, ZmLOX3, resulted in the increased susceptibility of kernels to the infection with A. flavus and A. nidulans (Gao et al., 2009). In an earlier study, we found that the lox3-4 mutants produced greater levels of ethylene in the roots compared to the WT (Gao et al., 2008), which prompted us to investigate whether ET production was also altered in the lox3-4 mutant kernels in response to A. flavus infection.
As shown in Figure 1, infection with A. flavus substantially increased ET emission in both near-isogenic WT and lox3-4 mutant, with the latter significantly more than the former at 7 dpi. Because lox3-4 mutant is substantially more susceptible to A. flavus colonization (Gao et al., 2009), we reasoned that one potential mechanism underlying increased susceptibility of the mutant is increased ET. To test whether volatiles including ET emitted by infected kernels impact fungal growth, sporogenesis and toxin production, we performed an in vitro plate assay by co-incubating A. flavus grown on PDA media in close proximity with maize kernels infected with A. flavus simultaneously (Figure 2A). The plates were cultured under constant dark condition for 4 days to allow for vegetative growth but prevent sporulation, then transferred to light/dark cycle (8 h dark/16 h light conditions) for another 2 days, This will allow us to investigate whether synchronizing the switch from vegetative growth to asexual reproduction is regulated by exposure to volatiles produced from infected kernels. Exposure to light/dark regimes is required for conidia production as evidenced by rapid green pigmentation of fungal hyphal mass in Aspergilli spp. (Rodriguez-Romero et al., 2010; Ruger-Herreros et al., 2011). Conidia and aflatoxin produced by the fungal cultures in response to volatiles emitted by the infected WT or lox3-4 mutant kernels were measured. As shown in Figures 2B,C, after expose to light for 2 days, in contrast to the colonies co-incubated with infected WT kernels, fungal colonies exposed to volatiles emitted by infected lox3-4 kernels produced greater levels of conidia as evidenced by green pigmentation of fungal colonies and conidia enumeration. However, aflatoxin content produced by A. flavus was not different among the plates supplemented with lox3-4 or WT.
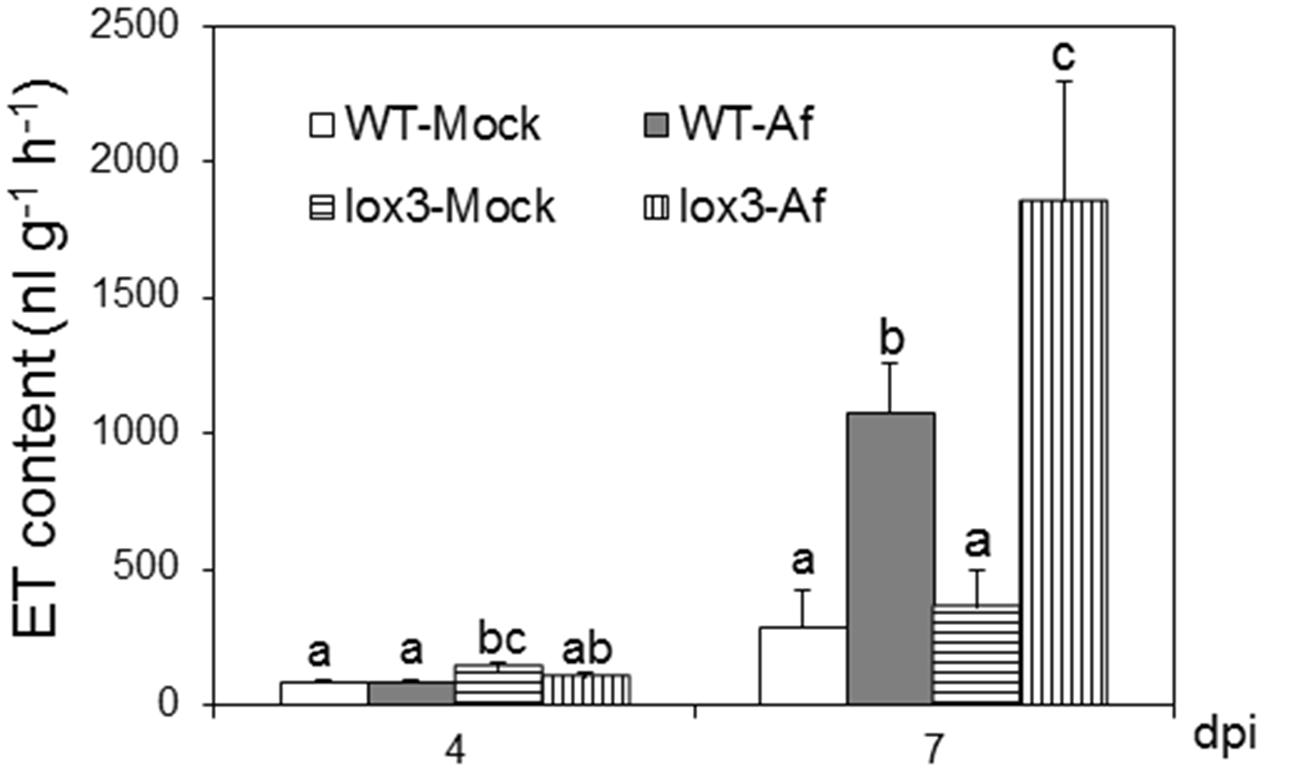
FIGURE 1. Ethylene production on lox3-4 mutants upon inoculation with A. flavus NRRL 3357. The kernels were inoculated with 1 × 106 spores and cultured in a 20 ml of glass scintillation vial (Wheaton Science) at 29°C under 12L/12D for 4 and 7 days. ET was measured by withdrawing 1 mL volume of headspace gas with a syringe and injecting into a digital gas chromatograph (Photovac 10 plus, PerkinElmer, Inc., Norwalk, CT, USA) The values are the mean ± SD of three replicates (four vials each treatment each genotype), with six kernels in each vial. Different letters above bars denote significant differences (P < 0.05, ANOVA) between genotypes and treatments within same time point.
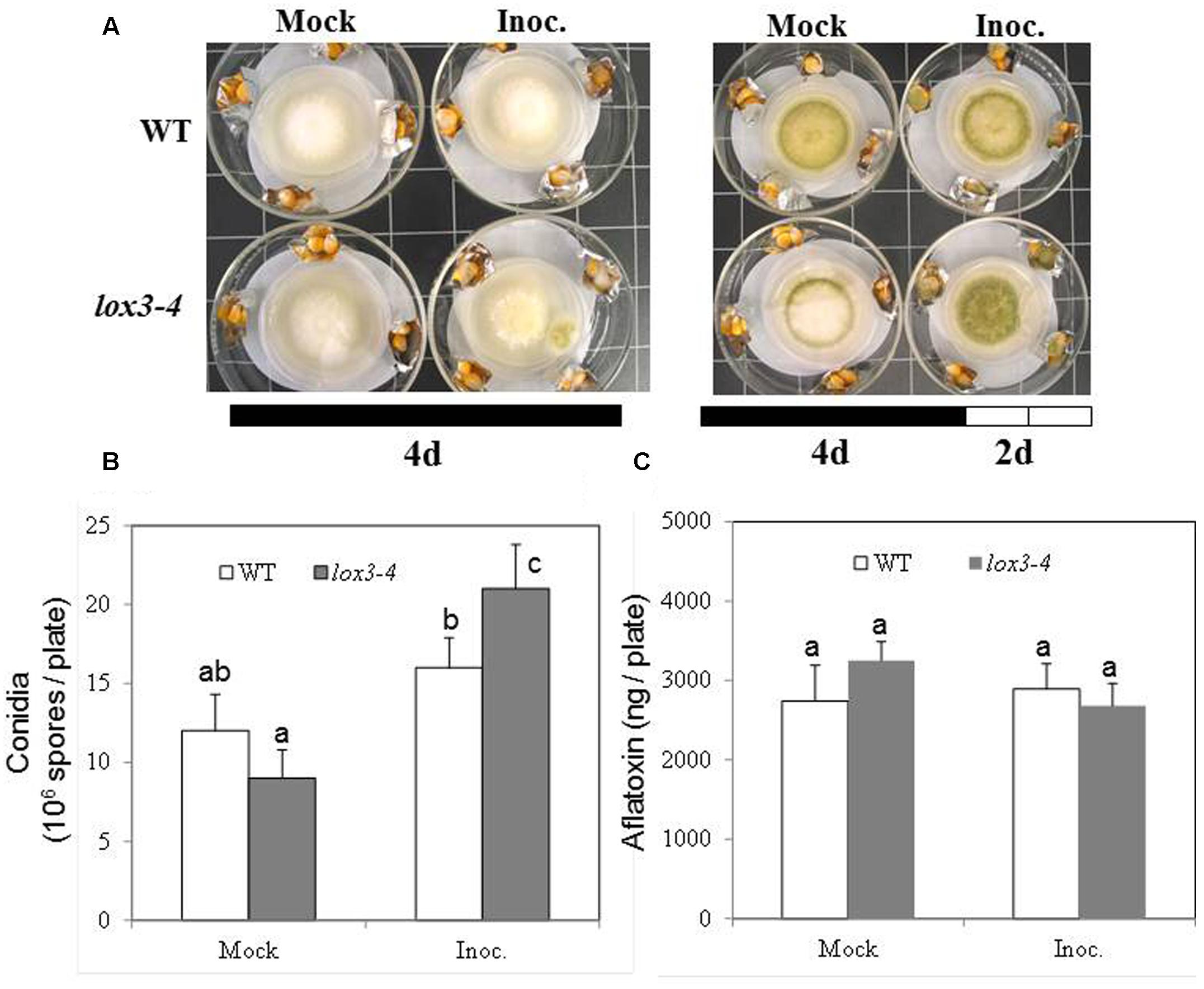
FIGURE 2. Kernel plate assay for testing the effect of potent gas produced by lox3-4 kernels inoculated with A. flavus on the fungal growth, sporulation, and aflatoxin production by A. flavus grown on a plate. WT and lox3-4 kernels were inoculated with 1 × 106 spores and placed in a large petri dish, containing a smaller petri dish where the A. flavus was growing on the PDA media (A). (A) The infected kernels were co-incubated with the fungal plate for either 4 days under continuous darkness or 4 days continuous darkness then moving to the continuous light for an additional 2 days. (B) The conidia, and (C) aflatoxin produced from the fungi grown on the PDA plates inside the small petri dishes was determined, respectively, at 6 days post culture (4 days dark+2 days light). The values are the mean ± SD of three replicates, and different letters above bars denote significant differences (P < 0.05, ANOVA) between genotypes and treatments.
Ethylene Biosynthetic Genes Are Differentially Induced in Seed in Response to Infection with A. flavus
To investigate the role of ET and ET-biosynthesis genes in the regulation of seed colonization of maize by A. flavus, we inoculated the WT kernels (B73 inbred line) with fungal spore suspension of A. flavus at 1 × 106 spores/ml or H2O as a mock control, and the transcript levels of three ET biosynthesis genes, ZmACS2, ZmACS6, and ZmACO31 (ACC oxidase 31), were quantified in either mock-treated or A. flavus inoculated kernels at different time points post treatments. While the expression of ZmACO31 was found to be induced in the kernels infected with A. flavus at 24 and 48 hpi, the transcriptional level of ZmACS2 and ZmACS6 was induced at 96 and 24 hpi, respectively (Figure 3). These results suggest that seed respond to pathogen infection by increased synthesis of ET.
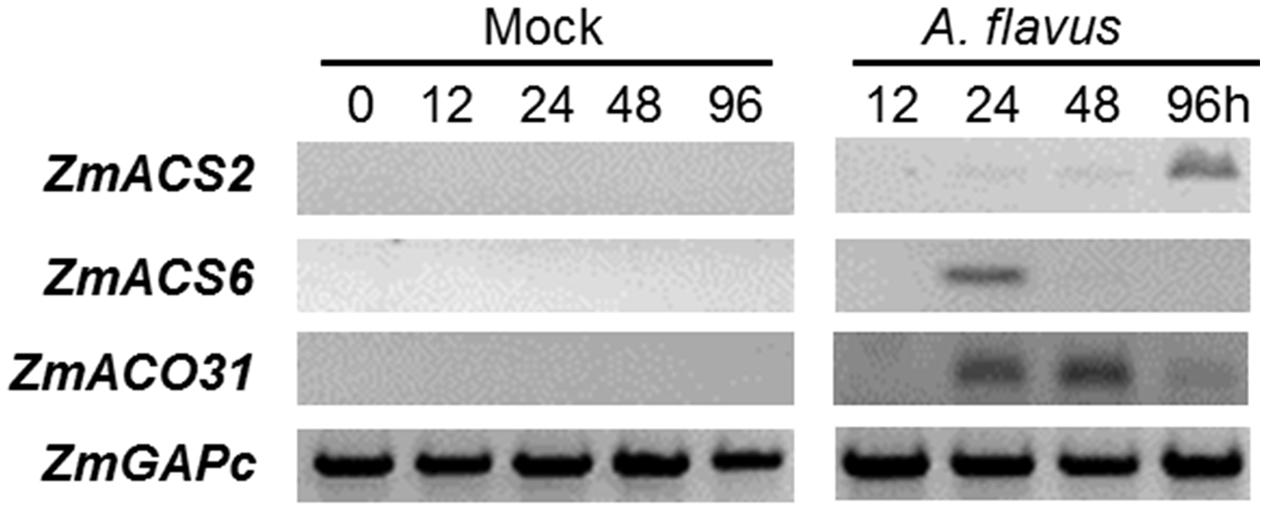
FIGURE 3. Genes encoding ethylene biosynthesis enzymes, ACC synthase (ZmACS) and ACC oxidase (ZmACO) are induced in maize seed in response to A. flavus infection. The WT kernels (B73 inbred line) were inoculated with A. flavus or mock-inoculated, incubated and harvested at 0, 12, 24, 48, and 96 h post-inoculation. The expression of two ACS genes, ZmACS2 and ZmACS6, and ZmACO31 were tested. Expression of a house-keeping gene ZmGAPc was used as an internal standard in this experiment.
To test whether ET biosynthesis genes are differentially regulated in seeds compared to other tissues, we examined the expression levels of ACS2, 6 and 7 in different tissues at different developmental stages. While all three genes are found to be differentially expressed in different tissues, the expression levels are relatively higher in the tissues from mature plants compared to young seedling tissues. Expression levels of ACS2, 6 and 7 in the mature dry seeds are much lower than other tissues at mature stages (Supplementary Figures S1A–C). This data corresponds to our finding that ACSs are not expressed in the uninfected seeds at detectable levels, but induced to higher levels upon pathogen attack (Figure 3). Unlike seeds, ethylene is normally produced in the vegetative tissues, particularly at mature stages. This could explain that plants likely deploy different ACS genes to produce ET in diverse tissues and under pathogen attack.
Colonization, Sporulation and Mycotoxin Production by A. flavus Grown on the acs Mutant Kernels
To examine the role of seed-derived ET in colonization and conidiation of A. flavus, we performed kernel assays using previously reported ET biosynthesis mutants that carry the Mutator transposons in the ACC synthase genes ZmACS2 and ZmACS6, respectively (Young et al., 2004). The knockout mutants for the remaining maize ACS gene family member, ZmACS7, was not available for this study. The loss of ZmACS6 expression in the acs6 mutant resulted in a reduction of 90% of foliar ethylene, while ethylene evolution from the acs2 mutants was only 55% of the levels produced in WT leaves (Young et al., 2004). We reasoned that these two mutants are excellent tools to investigate the role of ET in resistance to A. flavus. As shown in Figure 4A, we found that infection of both acs2 and acs6 mutants with A. flavus resulted in greater levels of seed colonization compared to WT kernels at both 3.5 and 7 dpi. Increased biomass of the fungus grown in the mutant seed was also demonstrated by increased amount of the fungus-specific lipid, ergosterol (Figure 4B). The enhanced colonization of the acs mutants was supported by increased number of conidia produced by the fungus on both mutants (Figure 4C). Unexpectedly, while fungal vegetative growth on acs2 and acs6 mutants was increased, aflatoxin B1 levels remained similar in all maize genotypes (Figure 4D). These results suggest that the ACS2 and ACS6 genes function in maize responses to A. flavus colonization and spore production but are irrelevant to the fungal ability to produce aflatoxin.
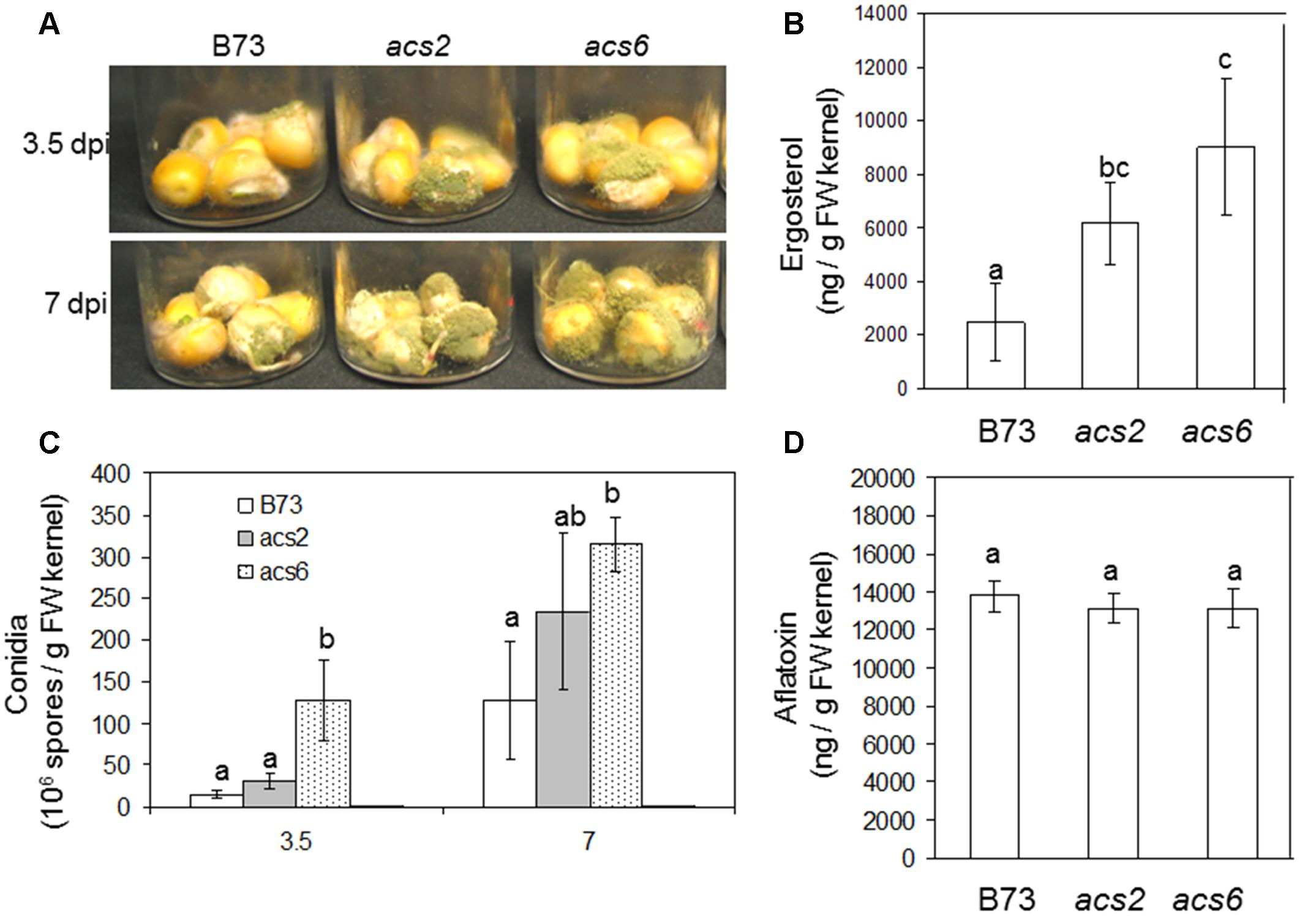
FIGURE 4. Kernel infection bioassay of the acs mutants by A. flavus. (A) Infected kernels of WT and acs mutants were cultured in glass vials and incubated for 3.5 and 7 days. (C) Enumeration of conidia in the acs mutants and WT (B73) following inoculation with A. flavus. (D) Aflatoxin and (B) ergosterol was measured at 7 days post-inoculation (dpi), respectively. The acs2, acs6 single mutants were generated by back crossing to B73 seven times (BC7 stage). The values are the mean ± SD of four replicates. Different letters above bars denote significant differences (P < 0.05, ANOVA) analyzed by the SPSS program (SPSS Inc., Chicago, IL, USA) between the host genotypes within same time point. Similar results were obtained in three independent experiments. dpi, days post infection.
ET Production Is Induced in the acs Mutants Infected with A. flavus
Expression pattern of ET biosynthesis genes after inoculation with A. flavus (Figure 3) and greater colonization of acs mutant kernels by A. flavus (Figure 4) suggested that ET could be a key factor in the regulation of seed colonization by A. flavus.
To confirm that acs mutants are true knock-out alleles, we measured ET levels in B73 and acs6 mutant leaves. As shown in Supplementary Figure S1D, almost no ET was detected in the leaves of the acs6 mutant, confirming the mutant is indeed a true knock-out mutant. To further test whether ET production is altered in kernels of the acs mutants in response to A. flavus infection, we measured ET emission in WT and the acs mutants kernels in response to infection by A. flavus. ET was quantified from the headspace of the vials containing WT or the acs mutant kernels at 7 dpi. While infected WT kernels emitted only slightly greater levels of ET, unexpectedly, ET levels were strongly enhanced in the kernels of both acs2 and acs6 mutants, with the significantly higher levels in acs6 mutants (Figure 5). This is in sharp contrast to the reported reduced ET production levels in the leaves of the two mutants (Young et al., 2004). Non-infected desiccated seed of the mutants or WT did not emit any detectable ET (data not shown).
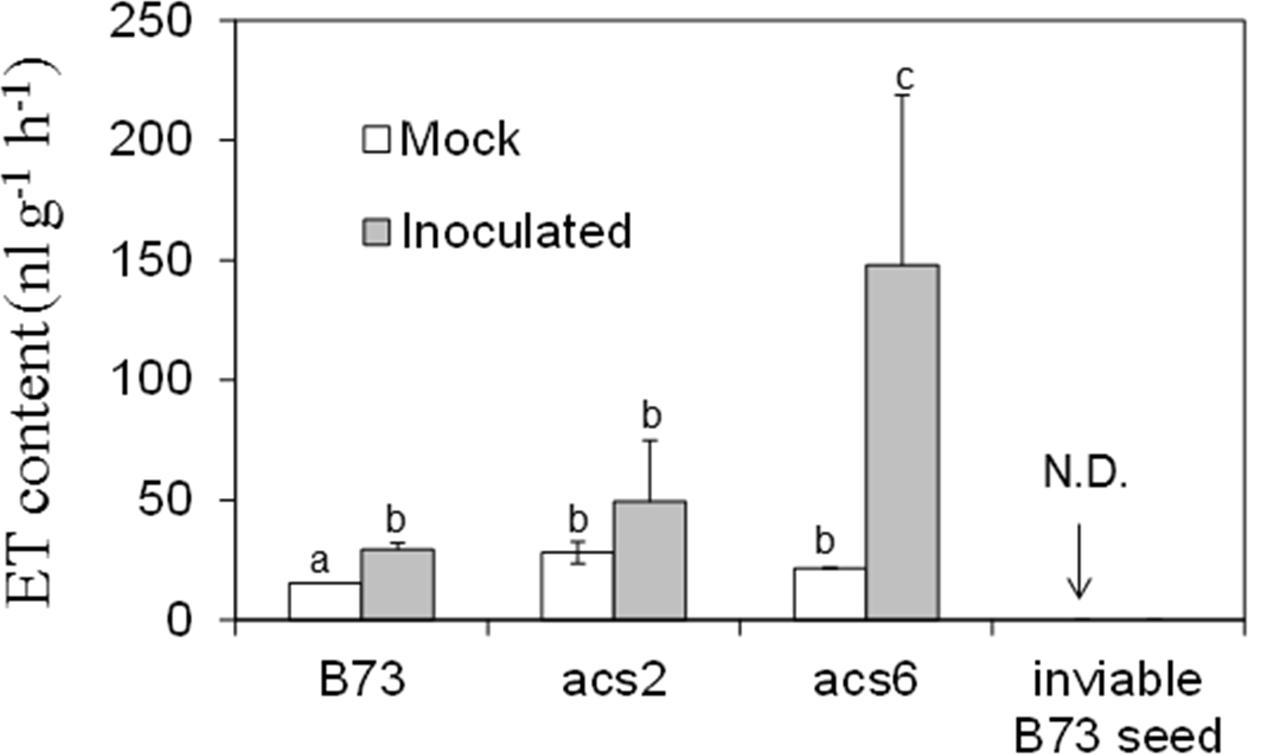
FIGURE 5. Ethylene production on acs mutants upon inoculation with A. flavus NRRL 3357. The kernels were inoculated with 1 × 106 spores and cultured in a 20 ml of glass scintillation vial (Wheaton Science) at 29°C under 12L/12D for 7 days. The vial was then tightly sealed with a plastic lid for 1 to 2 h prior to analysis to allow enough ethylene was accumulated. ET was measured by withdrawing 1 mL volume of headspace gas with a syringe and injecting into a digital gas chromatograph (Photovac 10 plus, PerkinElmer, Inc., Norwalk, CT, USA) The values are the mean ± SD of four replicates (four vials each treatment each genotype), with six kernels in each vial. Different letters above bars denote significant differences (P < 0.05, ANOVA) analyzed by SPSS program (SPSS Inc., Chicago, IL, USA) between the host genotypes within same time point. Similar results were obtained in three independent experiments. dpi, days post infection. N.D., not detectable.
Seed but not the Fungus Is Responsible for Induced ET Emission
Because the acs2 and acs6 infected seed emitted greater than WT levels of ET, we tested whether ET was produced by A. flavus rather than by the host (or in addition to the host). We reasoned that autoclaved, and thus non-viable, seed will not be able to synthesize ET, while the fungus that colonizes such seed may produce ET. In contrast to viable seed, no ET emission could be detected when non-viable kernels were infected by A. flavus (Figure 5). These data suggest that ET is originated from maize host but not the fungus.
Effects of an ET Inhibitor on the Colonization and Sporulation of A. flavus
To confirm the role of ET in the kernel-A. flavus interaction by a pharmacological approach, the potent ET inhibitor, AgNO3, was applied to the kernels prior to inoculation. The pretreatment with the inhibitor reduced fungal colonization of both WT and the single acs mutants, as evidenced by the reduced number of conidia (Figures 6A,B). In the WT kernels, the inhibitor pretreatment resulted in reduced production of aflatoxin, which correlated with the lower fungal growth (Figure 6C). However, pretreatment with the inhibitor did not appear to have an effect on the levels of aflatoxin in both acs mutants (Figure 6C).
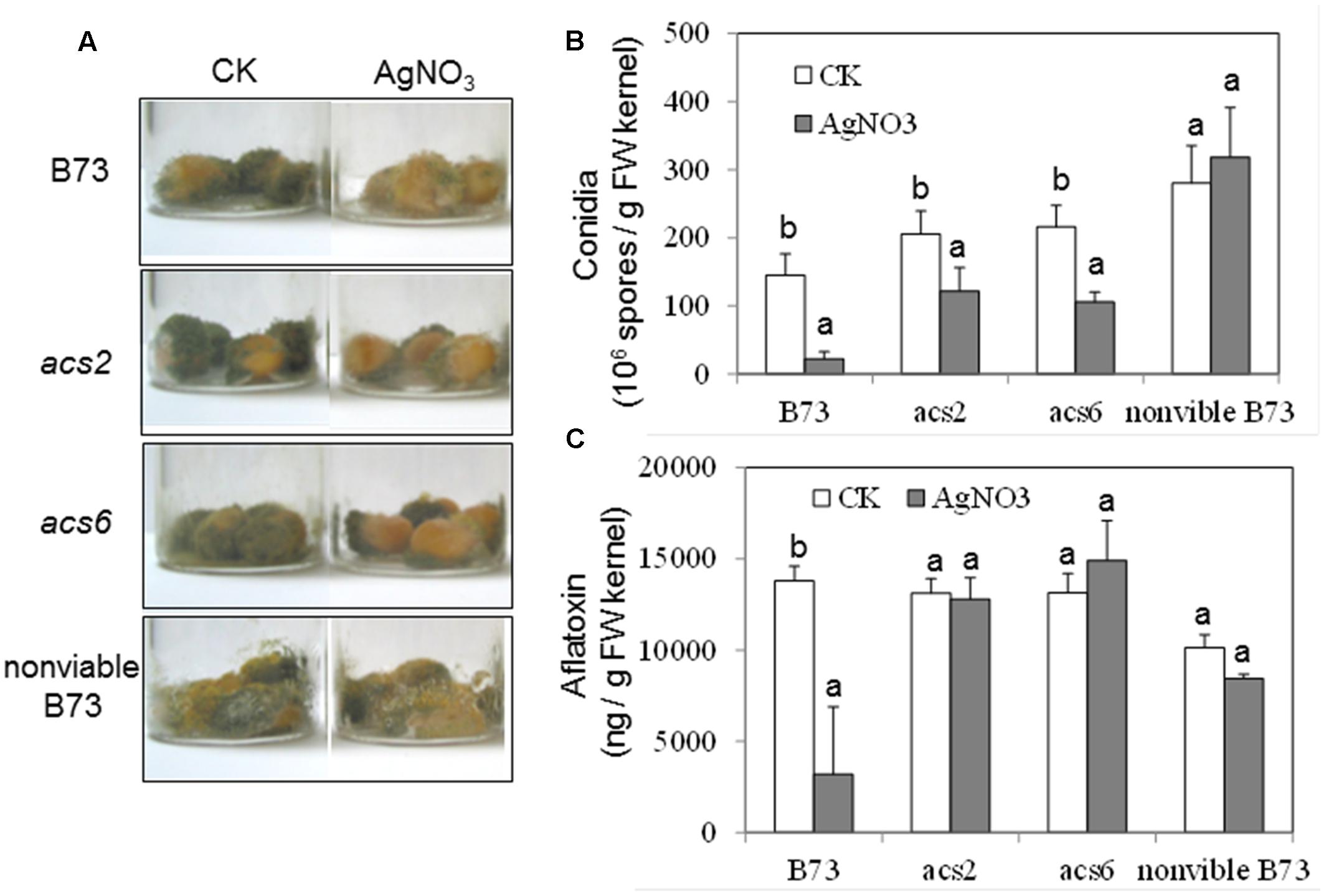
FIGURE 6. Effect of ethylene inhibitor AgNO3 on the colonization, conidiation and aflatoxin production of A. flavus NRRL 3357 on acs mutants. (A) The kernels were pretreated with AgNO3 (20 mM) 30 min, followed by inoculation with 1 × 106 spores, then cultured in a 20 ml of glass scintillation vial (Wheaton Science) at 29°C under 12L/12D for 7 days. (B) Conidiation of acs mutants upon infection of A. flavus NRRL 3357. The values are the mean ± SD of four replicates (four vials each treatment each genotype), with six kernels in each vial. (C) Aflatoxin B1 was quantified using Vicam Aflatest (Vicam, Watertown, MA, USA), according to the USDA-FGIS protocol. The values are the mean ± SD of four replicates (four vials each treatment each genotype), with six kernels in each vial. Different letters above bars denote significant differences (P < 0.05, ANOVA) analyzed by SPSS program within same host genotypes. Similar results were obtained in at least three independent experiments. dpi, days post infection.
We also tested whether the inhibitor treatment had any unintended effect on the ability of A. flavus to colonize seed, and produce conidia and aflatoxin in the infected, non-viable seeds. As shown in Figure 6A, the ability of A. flavus to colonize kernels and to produce conidia was not affected by AgNO3 in the non-viable seeds, compared to control non-treated seeds (Figures 6A,B). These data suggested that AgNO3 treatment itself unlikely affected fungal development and secondary metabolism (e.g., toxin production) directly, and that the inhibitor effects observed on the living seed were due to the suppression of ET effect in the host seed.
To test whether ET precursor, ACC, could itself promote pathogenicity of A. flavus in maize kernels, we pretreated kernels of B73, acs2 and acs6 mutant seeds with ACC, followed by inoculation with A. flavus. While we found that both acs2 and acs6 displayed enhanced conidiation of A. flavus on mutant kernels compared to B73, ACC treatment showed increased but not statistically significantly higher number of conidia produced by A. flavus, compared to the mock treatment (Supplementary Figures S2A,B). However, pretreatment with ACC treatment could significantly increase the ergosterol content in B73 seeds, compated to the control (Supplementary Figure S2C), supporting our findings that ET might serve as a susceptibility factor for fungal growth. The effect of ACC on ergosterol was not observed in either acs mutant. This could be due to possible saturation of the seed capability of converting ACC to yet additional ET in the acs mutant kernels in response to A. flavus.
Expression of ET Biosynthetic Genes in WT and acs6 Mutants upon Infection with A. flavus
Since the acs mutants, produced more ET compared to WT, in response to the infection with A. flavus, we hypothesized that the increased ET levels observed in the single mutants might be due to the overexpression of other ACS genes. As shown in Figures 7A,B, the expression of both ACS2 and ACS6 was moderately increased in WT seed upon A. flavus infection, while ACS2 expression, but not ACS7, was strongly induced to a much higher levels in the acs6 mutant compared to that in WT seeds, at 4 dpi. In the acs2 mutant seed, ACS6 expression was induced to a higher level upon infection, compared to that in WT seed (Figure 7C), at 4 dpi. However, the expression of ACS7 was enhanced earlier, at 2 dpi, in the acs2 mutant seed, compared to that in WT, where it was induced to a higher level at 4 dpi (Figure 7D). In support of this finding, the expression of both ACS2 and ACS7 was significantly higher in the lox3-4 mutants after infection with A. flavus at 4 dpi, compared to that in WT (Figures 8A,B). Although the expression of ACS6 was also strongly increased in lox3-4 mutant, however, that numerical increase was not statistically different (Figure 8C).
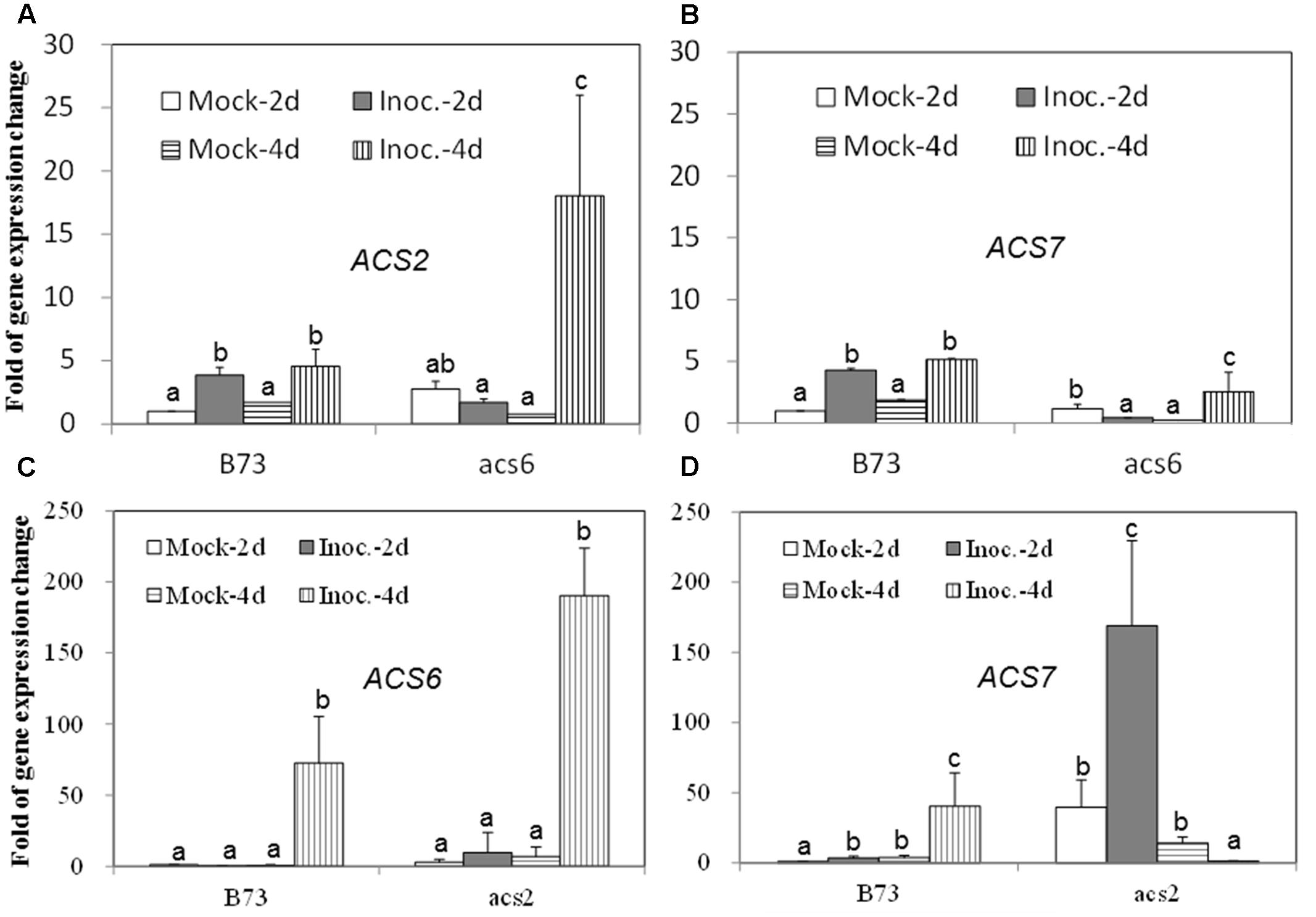
FIGURE 7. The expression of ET biosynthetic genes, ACS2, ACS6 and ACS7, in the WT and acs mutant kernels infected with A. flavus. The kernels were inoculated with 1 × 106 spores and cultured in a 20 ml of glass scintillation vial (Wheaton Science) at 29°C under 12L/12D for 2 and 4 days, and gene expression levels of ACS2 (A) and ACS7 (B) in B73 vs. acs6 mutants, and ACS6 (C) and ACS7 (D) in B73 vs. acs2 mutants, respectively, were quantified using qRT-PCR. Different letters above bars denote significant differences (P < 0.05, ANOVA) analyzed by the SPSS program between different treatments within the host genotypes. Similar results were obtained in three independent experiments.
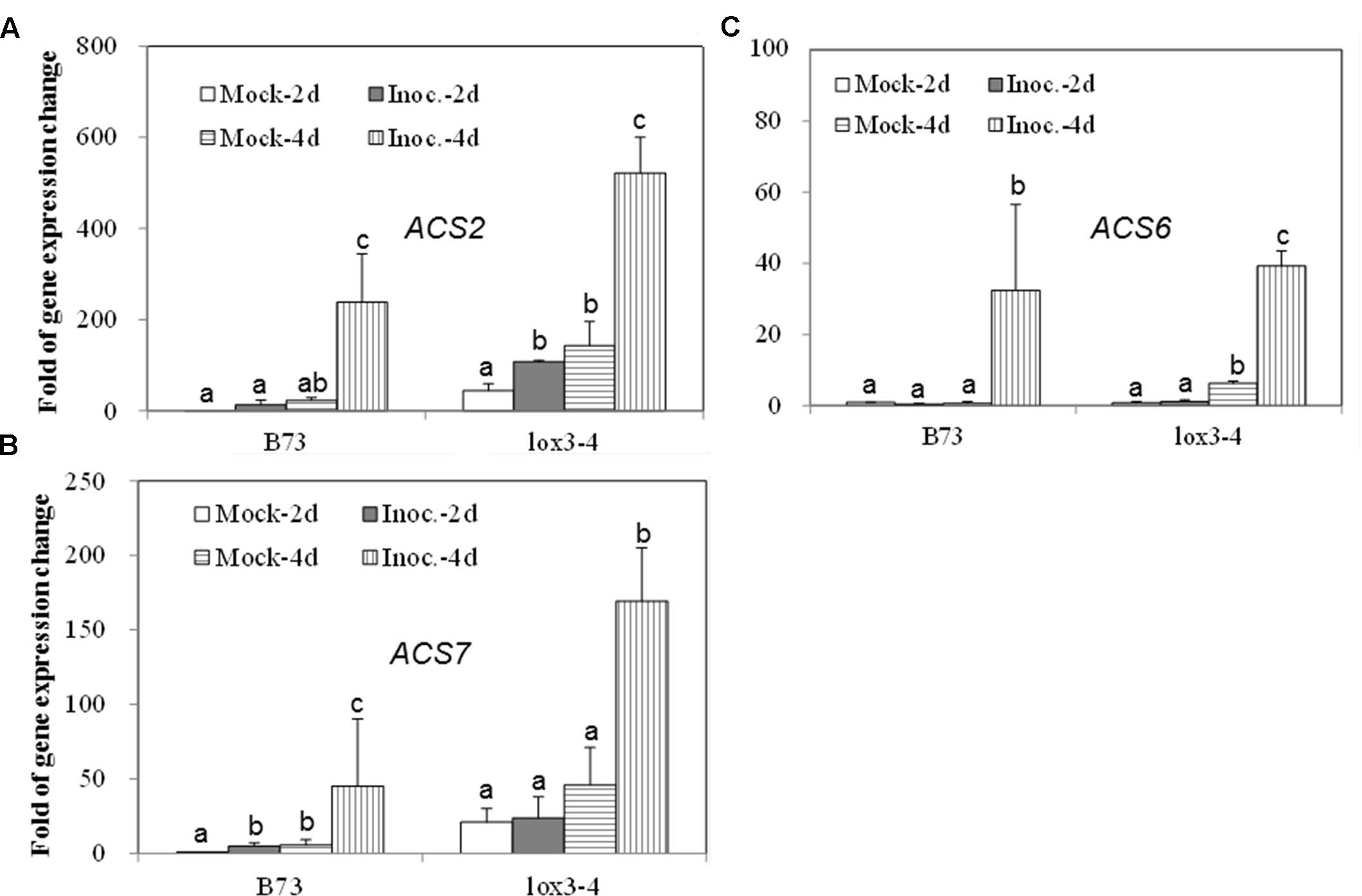
FIGURE 8. The expression of ET biosynthetic genes, ACS2, ACS6 and ACS7, in the WT and lox3-4 mutant kernels infected with A. flavus. The kernels were inoculated with 1 × 106 spores and cultured in a 20 ml of glass scintillation vial (Wheaton Science) at 29°C under 12L/12D for 2 and 4 days, and gene expression levels of ACS2 (A), ACS6 (B) and ACS7 (C) in B73 vs. lox3-4 mutants, respectively, were quantified using qRT-PCR. Different letters above bars denote significant differences (P < 0.05, ANOVA) analyzed by the SPSS program between different treatments within the host genotypes. Similar results were obtained in three independent experiments.
Discussion
Aspergillus flavus colonizes the same niche, i.e., maize kernels, but is evolutionarily and ecologically distinct from Fusarium spp. Kernel assays showed consistently that acs mutants were more susceptible to A. flavus. The increased colonization and sporulation of A. flavus on acs mutant kernels was associated with the enhanced ET production, especially in acs6 (Figure 5). Here we also demonstrated that susceptible to Aspergilli spp. lox3-4 mutant (Gao et al., 2009), also produced greater levels of ET upon infection with A. flavus compared to WT and provides a support for presumptive role of ET in facilitating A. flavus pathogenesis (Figure 1). Coincidently, lox3-4 mutant was also reported to produce greater levels of ET in the roots, suggesting that 9-oxylipins produced by the LOX3 isoform are potent inhibitors of ET synthesis in both seeds and roots.
Taken together, the findings from this study suggest that seed derived ET facilitate seed colonization and conidiation by A. flavus. Our results did not provide conclusive evidence as to whether host-derived ET has a role in the regulation of aflatoxin biosynthesis. While fungal colonization and spore production by A. flavus were increased in the acs mutant kernels, aflatoxin production was not impacted in acs2 or acs6 (Figure 4), suggesting that ET function differentially in the regulation of fungal growth and secondary metabolite production. It appears, however, that the increased colonization of acs mutants may be responsible for the same level of aflatoxin produced by relatively lower fungal biomass in WT seed.
It was intriguing to find that seed-derived ET upon the infection stimulates the growth of A. flavus, whereas did not show any impact on aflatoxin production in corn seeds. This contrasts with the studies showing that exogenously applied ET could inhibit aflatoxin production by A. parasiticus in peanuts (Roze et al., 2004; Gunterus et al., 2007; Huang et al., 2009). ET and its generator, ethephon, also suppressed aflatoxin biosynthesis in A. flavus in vitro, which might be due to ROS reduction (Huang et al., 2009). There are three possible reasons for this discrepancy. First, the amount of seed-derived endogenous ET that we detected is within the range of nl/L, which was far below the amount of ET that previous studies used (1∼100ppm, within a range of ul/L) (Roze et al., 2004; Gunterus et al., 2007; Huang et al., 2009). Another possible reason is that endogenous ET at low threshold produced by host upon infection could serve as signaling molecule facilitating A. flavus vegetative growth, but not aflatoxin synthesis. There is also a possibility that ET role in the pathogenicity is host-dependent (peanut vs. maize). Further studies are required to investigate the precise function and mechanisms of ET in different pathosystems.
One of the intriguing findings of this study is that ET production in response to A. flavus infection was not reduced, as expected, in both acs2 and acs6 mutant kernels (Figure 5). On the contrary, ET was increased in acs2 and even higher in acs6 mutants. Further analyses of why the single mutants produce greater levels of ET revealed that they overexpress other members of the ACS gene family. Maize ACS gene family consists of at least three members, ZmACS2, its closely related paralog ZmACS7, and a distantly related ZmACS6 (Gallie and Young, 2004; Young et al., 2004). ACS2 and to a lesser extent ACS7 transcript abundance was enhanced in the acs6 mutant seed upon the infection with the fungus, and acs2 mutant overexpressed the ACS6 gene. Interestingly, it has been shown in an unrelated study that maize acs6 mutants overexpress ZmACS2 gene while expression of the ZmACS7 gene was reduced (Young et al., 2004). Similar to our finding that single acs mutants overproduce ET, previous study showing that the Arabidopsis eto2 mutant, which produces 20-fold greater levels of ET, is deficient in the ACS5 gene expression supports the idea of antagonistic interaction between ET-producing enzyme isoforms or the ability of the functional genes to compensate for the lack of a missing gene family member (Vogel et al., 1998).
Previous reports showed that A. parasiticus produces ET on solid culture medium (Roze et al., 2004). In this study, however, no fungus-derived ET was detected in either fungal culture plates (data not shown) or from the fungus grown on non-viable seeds (Figure 5), suggesting that A. flavus is not the source of increased ET in the assays with living seeds. This discrepancy may be explained by different timing of ET sampling. For example, it has been reported that A. flavus produced much less ET compared to A. parasiticus, and the emission of ET by the former occurred at the early period of their growth, after which the ET level declined to the level that was not detectable in the system (Sharma et al., 1985). Alternatively, even if A. flavus produces ET when grown on a medium, this production may be inhibited on a plant tissue. This scenario was reported in B. cinerea-tobacco pathosystem, where B. cinerea produced easily detectable levels of ET in vitro but did not produce any ET in planta (Chague et al., 2006).
Our findings in this study provide strong evidence that ET biosynthetic enzymes ZmACS2 and ZmACS6 and their final product ET directly or indirectly play a major role in governing the outcomes of seed interactions with the mycotoxin-producing fungi. The phenotypes of the two mutants resemble closely the disease phenotypes previously reported for the 9-lipoxygenase mutant, lox3-4, except that F. verticillioides grew equally well on both the lox3-4 mutant and B73, but produced up to 200-fold lower fumonisin and threefold lower conidia levels (Gao et al., 2007). Supporting the hypothesis that ET may facilitate fungal growth and conidia production is our finding that A. flavus-infected lox3-4 mutants emitted greater levels of ET compared to WT (Figure 1). Similar to ET positive correlation with fungal growth and conidiation shown in this study, in the lox3-4 studies increased colonization by A. flavus of lox3-4 mutant and greater spore and mycotoxin production by F. verticillioides grown on WT kernels correlated with the increased accumulation of fatty acids (Gao et al., 2009). Therefore, we propose that both fungi exploit not only ET but also lipids to facilitate their virulence. It is possible that the two groups of signals, ZmLOX3-dependent lipid-derived molecules and ZmACS2/6-dependent ET, may act synergistically or are interdependent. The precise mechanism of their interactions, if any, is not clear and needs to be further examined by using the lox3 and the acs double and triple mutants being constructed in our laboratories.
Distribution of Materials
Novel materials described in this publication may be available for non-commercial research purposes upon acceptance and signing of a material transfer agreement. In some cases such materials may contain or be derived from materials obtained from a third party. In such cases, distribution of material will be subject to the requisite permission from any third-party owners, licensors, or controllers of all or parts of the material. Obtaining any permission will be the sole responsibility of the requestor. Plant germplasm will not be made available except at the discretion of the owner and then only in accordance with all applicable governmental regulations.
Author Contributions
XG and MK designed the research. SW, Y-SP, YY, TI, and XG performed research. SW, Y-SP, EB, XG, and MK performed data analysis. Y-SP, XG, and MK drafted the article. XG and MK performed critical revision of the article. XG and MK carried out final approval of the final version.
Funding
This research was supported by National Science Foundation grants NSF IOB-0544428 and NSF IOS-0951272 to MK, and grants from NSFC (No. 31471508, No. 31671702), The National Key Research and Development Program of China (No. 2016YFD0101002), Natural Science Foundation of Jiangsu Province (No. BK20141370) and Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP) to XG.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank Dr. Daniel R. Gallie (University of California at Riverside) for his kind gift of the seeds of acs2 and acs6 mutants used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00415/full#supplementary-material
FIGURE S1 | Expression levels of ACS2, ACS6 and ACS7 in different maize organs from different growth stages (A–C), and ET levels in the leaves of acs6 mutant (D). Expression levels of ACS2 (A), ACS6 (B) and ACS7 (C) was examined by qRT-PCR in different maize organs from different growth stages. Eg, embryo from geminating seeds (2-day-old); Rg, root from geminating seeds (4-day-old); R1, root at V1 stage; S1, stem at V1 stage; L1, leaf at V1 stage; R3, root at V3 stage; S3, stem at V3 stage; L3, leaf at V3 stage; Rf, root at flowering time; Sf, stem at flowering time; Lf, leaf at flowering time; T, tassel; E, ear; P, pollen; K, kernel. (D) ET levels in the leaves of B73 and acs6 mutant. The plants were grown as described in Section “Materials and Methods.” The leaves of B73 and acs6 were excised from the plants and immediately transferred into a 20 ml of glass scintillation vial (Wheaton Science). The vial was then tightly sealed with a plastic lid for 1 to 2 h prior to analysis to allow sufficient levels of ethylene to accumulate. ET was measured by gas chromatograph as described. The values are the mean ± SD of four replicates (four vials each treatment each genotype), with three leaf segments in each vial.
FIGURE S2 | Effect of ethylene precursor ACC on the colonization, conidiation, and ergosterol production by A. flavus NRRL 3357 on acs mutants. (A) The kernels were pretreated with ACC (20 nM) for 30 min, followed by inoculation with 1 × 106 spores, then cultured in a 20 ml of glass scintillation vial (Wheaton Science) at 29°C under 12L/12D for 7 days. CK is the control by pretreatment with same amount of 0.001% Tween-20 as that used for inoculum of A. flavus. (B) Conidiation observed on the acs mutants upon infection of A. flavus NRRL 3357. The values are the mean ± SD of at least four replicates (four vials each treatment each genotype), with four kernels in each vial. Different letters above bars denote significant differences (P < 0.05, ANOVA) analyzed by SPSS program over B73-CK. Similar results were obtained in two independent experiments. (C) Ergosterol content was measured at 7 days post-inoculation (dpi).
References
Adie, B., Chico, J. M., Rubio-Somoza, I., and Solano, R. (2007). Modulation of plant defenses by ethylene. J. Plant Growth Regul. 26, 160–177. doi: 10.1016/j.jplph.2014.12.013
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Broekaert, W. F., Delaure, S. L., De Bolle, M. F. C., and Cammue, B. P. A. (2006). The role of ethylene in host-pathoven interactions. Annu. Rev. Phytopathol. 44, 393–416. doi: 10.1146/annurev.phyto.44.070505.143440
Catinot, J., Huang, J. B., Huang, P. Y., Tseng, M. Y., Chen, Y. L., Gu, S. Y., et al. (2015). ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate - and ethylene-responsive defence genes. Plant Cell Environ. 38, 2721–2734. doi: 10.1111/pce.12583
Chague, V., Danit, L. V., Siewers, V., Gronover, C. S., Tudzynski, P., Tudzynski, B., et al. (2006). Ethylene sensing and gene activation in Botrytis cinerea: a missing link in ethylene regulation of fungus-plant interactions? Mol. Plant Microbe Interact. 19, 33–42. doi: 10.1094/MPMI-19-0033
Cohn, J. R., and Martin, G. B. (2005). Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 44, 139–154. doi: 10.1111/j.1365-313X.2005.02516.x
Cristescu, S. M., De Martinis, D., Hekkert, S. T., Parker, D. H., and Harren, F. J. M. (2002). Ethylene production by Botrytis cinerea in vitro and in tomatoes. Appl. Environ. Microbiol. 68, 5342–5350. doi: 10.1128/AEM.68.11.5342-5350.2002
Diaz, J., ten Have, A., and van Kan, J. A. L. (2002). The role of ethylene and wound signaling in resistance of tomato to Botrytis cinerea. Plant Physiol. 129, 1341–1351. doi: 10.1104/pp.001453
Ellis, C., and Turner, J. G. (2001). The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13, 1025–1033. doi: 10.1105/tpc.13.5.1025
Fukuda, H., Kitajima, H., Fujii, T., Tazaki, M., and Ogawa, T. (1989a). Purification and some properties of a novel ethylene-forming enzyme produced by Penicillium digitatum. FEMS Microbiol. Lett. 59, 1–5. doi: 10.1111/j.1574-6968.1989.tb03072.x
Fukuda, H., Takahashi, M., Fujii, T., Tazaki, M., and Ogawa, T. (1989b). An Nadh-Fe(III)Edta Oxidoreductase from Cryptococcus albidus - an enzyme involved in ethylene production In vivo. FEMS Microbiol. Lett. 60, 107–111.
Fukuda, H., Ogawa, T., and Tanase, S. (1993). Ethylene production by microorganisms. Adv. Microb. Physiol. 35, 275–306. doi: 10.1016/S0065-2911(08)60101-0
Gallie, D. R., and Young, T. E. (2004). The ethylene biosynthetic and perception machinery is differentially expressed during endosperm and embryo development in maize. Mol. Genet. Genomics 271, 267–281. doi: 10.1007/s00438-004-0977-9
Gao, X. Q., Brodhagen, M., Isakeit, T., Brown, S. H., Gobel, C., Betran, J., et al. (2009). Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Mol. Plant Microbe Interact. 22, 222–231. doi: 10.1094/MPMI-22-2-0222
Gao, X. Q., Shim, W. B., Gobel, C., Kunze, S., Feussner, I., Meeley, R., et al. (2007). Disruption of a maize 9-lipoxygenase results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Mol. Plant Microbe Interact. 20, 922–933. doi: 10.1094/MPMI-20-8-0922
Gao, X. Q., Starr, J., Gobel, C., Engelberth, J., Feussner, I., Tumlinson, J., et al. (2008). Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol. Plant Microbe Interact. 21, 98–109. doi: 10.1094/MPMI-21-1-0098
Glazebrook, J., Chen, W. J., Estes, B., Chang, H. S., Nawrath, C., Metraux, J. P., et al. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217–228. doi: 10.1046/j.1365-313X.2003.01717.x
Gunterus, A., Roze, L. V., Beaudry, R., and Linz, J. E. (2007). Ethylene inhibits aflatoxin biosynthesis in Aspergillus parasiticus grown on peanuts. Food Microbiol. 24, 658–663. doi: 10.1016/j.fm.2006.12.006
Guo, H. W., and Ecker, J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7, 40–49. doi: 10.1016/j.pbi.2003.11.011
Huang, J. Q., Jiang, H. F., Zhou, Y. Q., Lei, Y., Wang, S. Y., and Liao, B. S. (2009). Ethylene inhibited aflatoxin biosynthesis is due to oxidative stress alleviation and related to glutathione redox state changes in Aspergillus flavus. Int. J. Food Microbiol. 130, 17–21. doi: 10.1016/j.ijfoodmicro.2008.12.027
Ince, J. E., and Knowles, C. J. (1986). Ethylene formation by cell-free-extracts of Escherichia coli. Arch. Microbiol. 146, 151–158. doi: 10.1007/BF00402343
Jia, Y. J., Kakuta, Y., Sugawara, M., Igarashi, T., Oki, N., Kisaki, M., et al. (1999). Synthesis and degradation of 1-aminocyclopropane-1-carboxylic acid by Penicillium citrinum. Biosci. Biotechnol. Biochem. 63, 542–549. doi: 10.1271/bbb.63.542
Johnson, P. R., and Ecker, J. R. (1998). The ethylene gas signal transduction pathway: a molecular perspective. Annu. Rev. Genet. 32, 227–254. doi: 10.1146/annurev.genet.32.1.227
Kende, H. (1993). Ethylene biosynthesis. Annu. Rev. Plant Phys. 44, 283–307. doi: 10.1146/annurev.pp.44.060193.001435
Kendrick, M. D., and Chang, C. (2008). Ethylene signaling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 11, 479–485. doi: 10.1016/j.pbi.2008.06.011
Lin, Z. F., Zhong, S. L., and Grierson, D. (2009). Recent advances in ethylene research. J. Exp. Bot. 60, 3311–3336. doi: 10.1093/jxb/erp204
Munkvold, G. P. (2003). Cultural and genetic approaches to managing mycotoxins in maize. Annu. Rev. Phytopathol. 41, 99–116. doi: 10.1146/annurev.phyto.41.052002.095510
Musungu, B. M., Bhatnagar, D., Brown, R. L., Payne, G. A., OBrian, G., Fakhoury, A. M., et al. (2016). A network approach of gene co-expression in the Zea mays/Aspergillus flavus pathosystem to map host/pathogen interaction pathways. Front. Genet. 7:206. doi: 10.3389/fgene.2016.00206
Nagahama, K., Ogawa, T., Fujii, T., Tazaki, M., Tanase, S., Morino, Y., et al. (1991). Purification and properties of an ethylene-forming enzyme from Pseudomonas syringae Pv Phaseolicola-Pk2. J. Gen. Microbiol. 137, 2281–2286. doi: 10.1099/00221287-137-10-2281
Paul, C., Naidoo, G., Forbes, A., Mikkilineni, V., White, D., and Rocheford, T. (2003). Quantitative trait loci for low aflatoxin production in two related maize populations. TAG Theor. Appl. Genet. 107, 263–270. doi: 10.1007/s00122-003-1241-0
Penninckx, I. A. M. A., Thomma, B. P. H. J., Buchala, A., Metraux, J. P., and Broekaert, W. F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. doi: 10.1105/tpc.10.12.2103
Rodriguez-Romero, J., Hedtke, M., Kastner, C., Muller, S., and Fischer, R. (2010). Fungi, hidden in soil or up in the air: light makes a difference. Annu. Rev. Microbiol. 64, 585–610. doi: 10.1146/annurev.micro.112408.134000
Roze, L. V., Calvo, A. M., Gunterus, A., Beaudry, R., Kall, M., and Linz, J. E. (2004). Ethylene modulates development and toxin biosynthesis in Aspergillus possibly via an ethylene sensor-mediated signaling pathway. J. Food Prot. 67, 438–447. doi: 10.4315/0362-028X-67.3.438
Ruger-Herreros, C., Rodriguez-Romero, J., Fernandez-Barranco, R., Olmedo, M., Fischer, R., Corrochano, L. M., et al. (2011). Regulation of conidiation by light in Aspergillus nidulans. Genetics 188, 809–822. doi: 10.1534/genetics.111.130096
Sharma, A., Padwaldesai, S. R., and Nadkarni, G. B. (1985). Possible implications of reciprocity between ethylene and aflatoxin biogenesis in Aspergillus flavus and Aspergillus parasiticus. Appl. Environ. Microbiol. 49, 79–82.
Vogel, J. P., Woeste, K. E., Theologis, A., and Kieber, J. J. (1998). Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. U.S.A. 95, 4766–4771. doi: 10.1073/pnas.95.8.4766
Warburton, M. L., Brooks, T. D., Windham, G. L., and Williams, W. P. (2011). Identification of novel QTL contributing resistance to aflatoxin accumulation in maize. Mol. Breed. 27, 491–499. doi: 10.1007/s11032-010-9446-9
Willcox, M. C., Davis, G. L., Warburton, M. L., Windham, G. L., Abbas, H. K., Betran, J., et al. (2013). Confirming quantitative trait loci for aflatoxin resistance from Mp313E in different genetic backgrounds. Mol. Breed. 32, 15–26. doi: 10.1007/s11032-012-9821-9
Woloshuk, C. P., Seip, E. R., Payne, G. A., and Adkins, C. R. (1989). Genetic transformation system for the aflatoxin-producing fungus Aspergillus flavus. Appl. Environ. Microbiol. 55, 86–90.
Xiang, K., Zhang, Z. M., Reid, L. M., Zhu, X. Y., Yuan G.-S., and Pan, G. T. (2010). A meta-analysis of QTL associated with ear rot resistance in maize. Maydica 55, 281–290.
Young, T. E., Meeley, R. B., and Gallie, D. R. (2004). ACC synthase expression regulates leaf performance and drought tolerance in maize. Plant J. 40, 813–825. doi: 10.1111/j.1365-313X.2004.02255.x
Keywords: Aspergillus flavus, ethylene, maize, colonization, aflatoxin
Citation: Wang S, Park Y-S, Yang Y, Borrego EJ, Isakeit T, Gao X and Kolomiets MV (2017) Seed-Derived Ethylene Facilitates Colonization but Not Aflatoxin Production by Aspergillus flavus in Maize. Front. Plant Sci. 8:415. doi: 10.3389/fpls.2017.00415
Received: 16 November 2016; Accepted: 10 March 2017;
Published: 28 March 2017.
Edited by:
Péter Poór, University of Szeged, HungaryCopyright © 2017 Wang, Park, Yang, Borrego, Isakeit, Gao and Kolomiets. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiquan Gao, xgao@njau.edu.cn Michael V Kolomiets, kolomiets@tamu.edu
†These authors have contributed equally to this work.
 Shi Wang1,2†
Shi Wang1,2† Yong-Soon Park3,4†
Yong-Soon Park3,4† Yang Yang
Yang Yang Eli J. Borrego
Eli J. Borrego Xiquan Gao
Xiquan Gao Michael V. Kolomiets
Michael V. Kolomiets