- 1State Key Laboratory of Subtropical Silviculture, Zhejiang Agriculture and Forestry University, Hangzhou, China
- 2State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, China
Because of the long and unpredictable flowering period in bamboo, the molecular mechanism of bamboo flowering is unclear. Recent study showed that Arabidopsis PIN1-type parvulin 1 (Pin1At) is an important floral activator and regulates floral transition by facilitating the cis/trans isomerization of the phosphorylated Ser/Thr residues preceding proline motifs in suppressor of overexpression of CO 1 (SOC1) and agamous-like 24 (AGL24). Whether bamboo has a Pin1 homolog and whether it works in bamboo flowering are still unknown. In this study, we cloned PvPin1, a homolog of Pin1At, from Phyllostachys violascens (Bambusoideae). Bioinformatics analysis showed that PvPin1 is closely related to Pin1-like proteins in monocots. PvPin1 was widely expressed in all tested bamboo tissues, with the highest expression in young leaf and lowest in floral bud. Moreover, PvPin1 expression was high in leaves before bamboo flowering then declined during flower development. Overexpression of PvPin1 significantly delayed flowering time by downregulating SOC1 and AGL24 expression in Arabidopsis under greenhouse conditions and conferred a significantly late flowering phenotype by upregulating OsMADS56 in rice under field conditions. PvPin1 showed subcellular localization in both the nucleus and cytolemma. The 1500-bp sequence of the PvPin1 promoter was cloned, and cis-acting element prediction showed that ABRE and TGACG-motif elements, which responded to abscisic acid (ABA) and methyl jasmonate (MeJA), respectively, were characteristic of P. violascens in comparison with Arabidopsis. On promoter activity analysis, exogenous ABA and MeJA could significantly inhibit PvPin1 expression. These findings suggested that PvPin1 may be a repressor in flowering, and its delay of flowering time could be regulated by ABA and MeJA in bamboo.
Introduction
The transition from vegetative to reproductive growth must start at an appropriate time in flowering plants for producing progeny and perpetuating the species. Proper timing of flowering (or “heading date" in cereals) is controlled by environmental signals (Putterill et al., 2004; Brambilla and Fornara, 2013) and internal signals (Jack, 2004; Jarillo and Piñeiro, 2011). Arabidopsis thaliana as the model plant for eudicots has four main pathways involved in flowering control: photoperiod, vernalization, autonomous, and gibberellic acid (Simpson and Dean, 2002). Rice, a short-day and the model plant species for monocots, has photoperiod and rice indeterminate 1 (RID1) pathways (Izawa, 2007; Wu et al., 2008).
Bamboo is a kind of widespread, fast-growing, renewable, and environmental-enhancing resource, whose industry contributes to providing food, building materials, and increasing the income for 2.2 billion people in the world (Chen, 2003). Bamboo products such as bamboo shoots, furniture, flooring, charcoal, beverages, and cosmetic are being used and traded by half of the world’s population (Chen, 2003). Although the bamboo industry is increasing in importance for poverty alleviation and economic development (Chen, 2003), bamboo flowering will make nothing left to these advantages because bamboo usually dies after flowering. In addition, it is difficult to analyze the phenomenon of bamboo flowering because of its unpredictability and long juvenile phase (Franklin, 2004). To surmount these problems, the genome of Phyllostachys edulis (synonym Phyllostachys heterocycla) and the transcriptomes of P. edulis, Bambusa oldhamii, B. edulis, and Dendrocalamus latiflorus have been sequenced, and numerous genes related to bamboo flowering were reported (Lin et al., 2010; Zhang et al., 2012; Peng et al., 2013; Gao et al., 2014; Shih et al., 2014; Zhao et al., 2014). As well, P. edulis and D. latiflorus contain novel miRNAs playing important roles in regulating bamboo flowering (Gao et al., 2015; Zhao et al., 2015). In addition, Louis et al. (2015) used proteomics to find that elements of stress, mobile genetics, and signal transduction cross-talk were associated with sporadic flowering of bamboo. Undoubtedly, these results provide the basis for understanding the roles of genes involved in bamboo flowering but need further experimental evidence.
The function of flowering genes has been heavily investigated in both Arabidopsis and rice; the results can provide some enlightenment on bamboo flowering. Recent study of Arabidopsis PIN1-type parvulin 1 (Pin1At) showed phosphorylation-dependent prolyl cis/trans isomerization of key transcription factors as an important flowering regulatory mechanism (Wang et al., 2010). In the 1980s, peptidylprolyl cis/trans isomerases (PPIases) were discovered (Fischer et al., 1983). Peptidylprolyl cis/trans isomerases act as enzymes catalyzing incongruous cis/trans isomerization of the peptide bonds preceding a proline residue to assist the client protein folding and restructuring (Kiefhaber et al., 1990; Hunter, 1998). There are four subfamilies of PPIases: FK506 binding proteins, cyclophilins, parvulins, and PP2A phosphatase activator (Lu and Zhou, 2007). Pin1, a member of the parvulin family of PPIases, is unique among the parvulin family because it functions by specifically recognizing phosphorylated Ser/Thr residues preceding proline (pSer/Thr-Pro) and catalyzing the conformational change of the phosphorylated substrates (Ranganathan et al., 1997; Hsu et al., 2001; Pastorino et al., 2006). Protein structure analysis showed that Pin1 in humans comprises an N-terminal WW regulatory domain and a C-terminal PPIase domain (Schiene-Fischer, 2015), and both domains can bind specifically to phospho-Ser/Thr-Pro-containing sequences (Yaffe et al., 1997; Lu et al., 1999). Pin1’s regulation of phosphorylation-dependent prolyl cis/trans isomerization has been found essential for cell growth and division, DNA repair, apoptosis, and transcription (Hanes et al., 1989; Hani et al., 1995; Lu et al., 1996).
Pin1At was the first identified PIN1-type PPIase from Arabidopsis (Landrieu et al., 2000; He et al., 2004); since then, several Pin1 plant homologs from Glycine max, Lycopersicon esculentum, and Malus domestica have been cloned (Landrieu et al., 2000; Metzner et al., 2001; Yao et al., 2001; Wang et al., 2010). Unlike Pin1 in human and its homolog in yeast (Hanes et al., 1989; Hani et al., 1995; Lu et al., 1996), PIN1-type PPIases in plants have only one PPIase domain with four additional amino acids but without a WW domain (Yao et al., 2001), and except Pin1At, their function is still unknown.
Bamboo has unique characteristics in flowering. To determine whether bamboo has a Pin1 homolog and whether it works in bamboo flowering, we isolated a Pin1 homolog from Phyllostachys violascens (Lei bamboo) and named it PvPin1. Lei bamboo is widely distributed in southern China and has high economic value because of its delicious shoots. The income for intensively managed Lei bamboo forest is about 20 times that for rice (Song et al., 2011). However, shoot production of Lei bamboo forest decreases sharply during flowering. PvPin1 from Lei bamboo was studied by analyzing its sequence structure, expression pattern, and phenotypes of transgenic Arabidopsis and rice. Unlike Pin1At, which can promote flowering, PvPin1 delayed flowering. In addition, PvPin1 could be regulated by abscisic acid (ABA) and methyl jasmonate (MeJA). PvPin1 might act as a flowering repressor in bamboo by responding to ABA and MeJA. Our data lay a good foundation for bamboo flowering and provide a basis for understanding bamboo flowering and provide a basis for developing technologies to inhibit it.
Results
Isolation of PvPin1 Gene
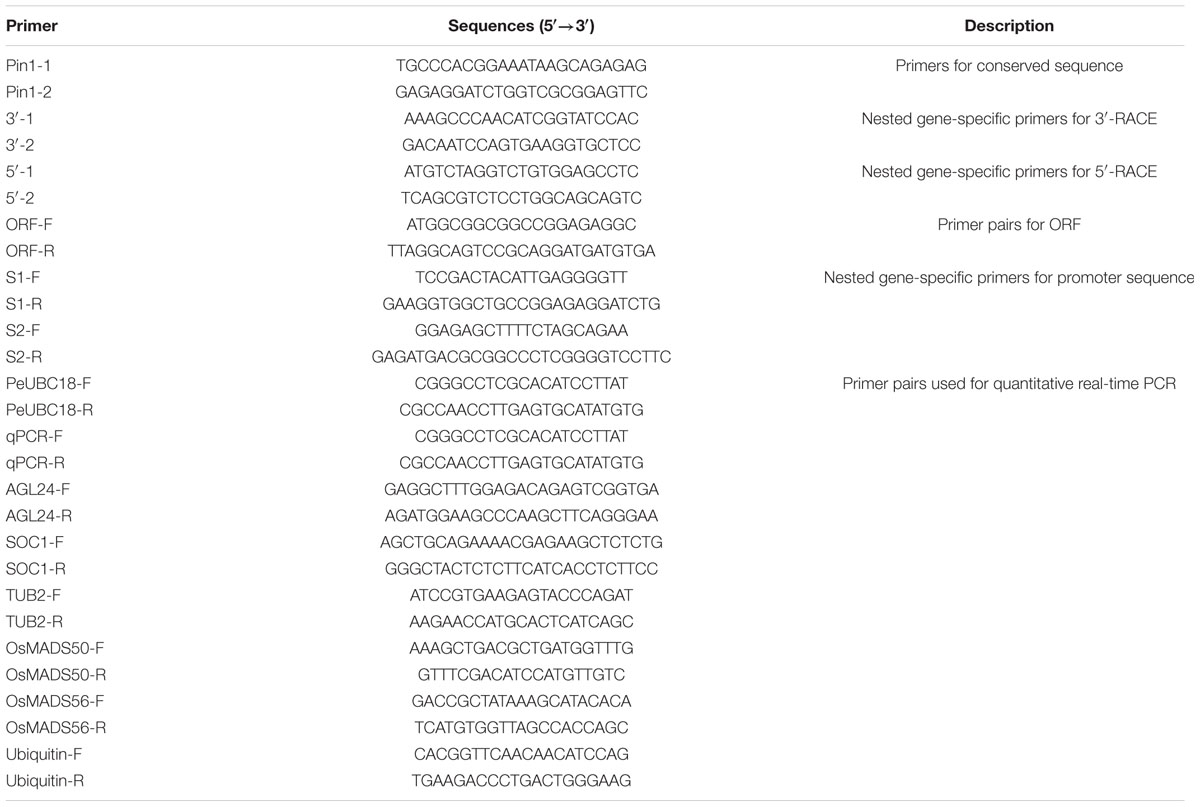
To isolate a Pin1-like gene from P. violascens, the amino acid sequences of Pin1 homologs from grass family plants were compared and the primers from the conserved regions were designed. Then a 300-bp fragment of the PvPin1 gene was amplified from P. violascens. Using gene-specific primers, a 744-bp cDNA sequence of Pin1-like was isolated from P. violascens by using 3′ and 5′ rapid amplification of cDNA ends (RACE) and designated as PvPin1. DNA sequencing analysis showed that the 744-bp cDNA contained a complete open reading frame (ORF) encoding a polypeptide of 122 amino acids. Based on the cDNA sequence of PvPin1, a 3078-bp genomic DNA sequence was cloned. Comparison of the genomic DNA sequence and ORF sequences showed that PvPin1 had two exons (248 and 121 bp) and one intron (2709 bp) (Figure 1A), which was same as Pin1At (Landrieu et al., 2000) and Pin1-like in rice (NCBI Oryza sativa Japonica Group Annotation Release 101). Amino acid sequence alignment revealed that the PvPin1 protein had the theoretical values of 7.97/13148.7 pI/Mw. Secondary structure analysis with SOPMA indicated that the putative PvPin1 protein contained an alpha helix (38.52%), a beta turn structure (16.39%), and a random coil (38.52%). Sequence comparison of the PvPin1 protein with its homologs in other plants showed that the catalytic core was well conserved and contained only a PPIase domain with four additional amino acids (Figure 1B).
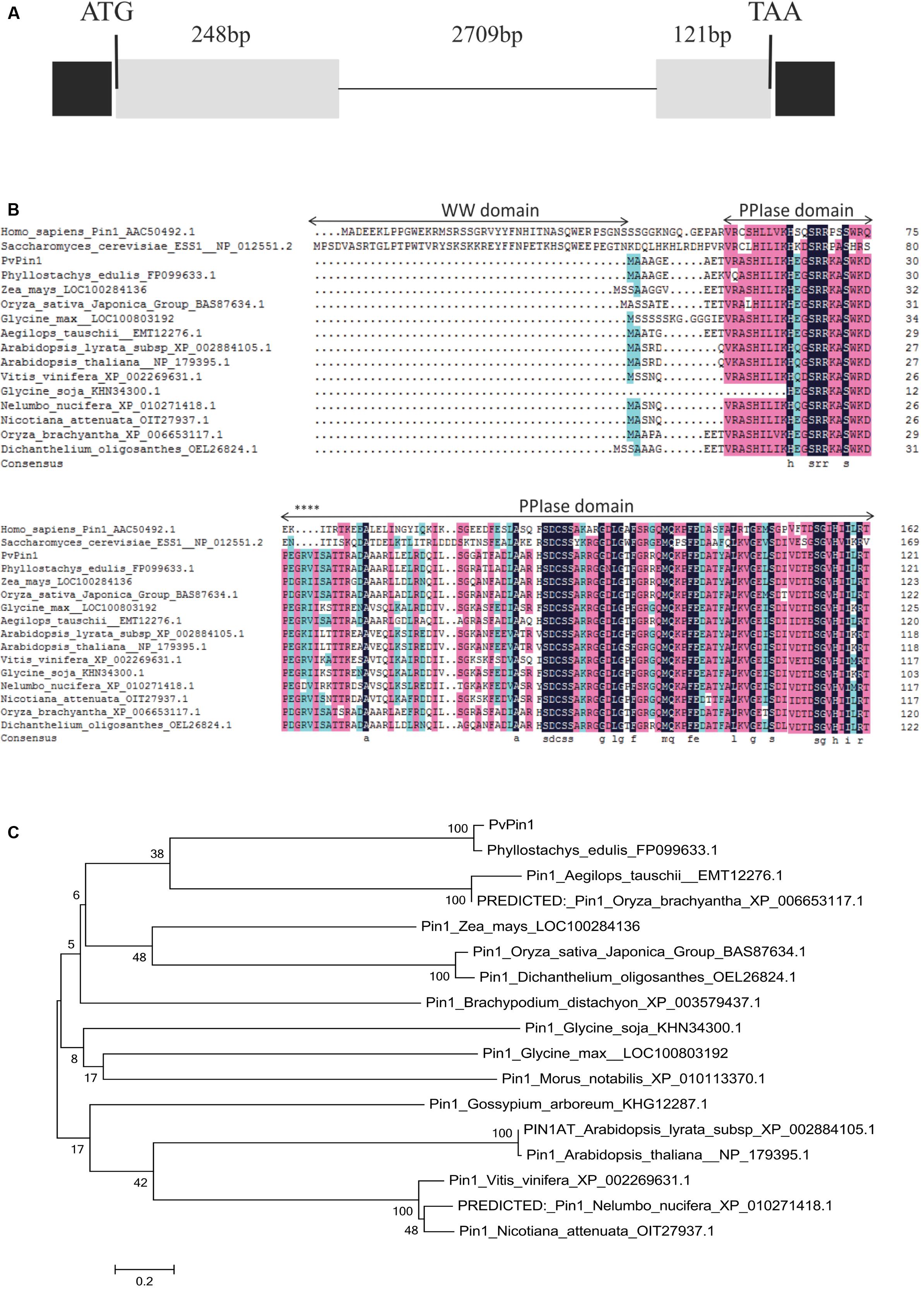
FIGURE 1. Genomic organization of the PvPin1, protein sequence similarities, and phylogenetic analysis of PvPin1 protein. (A) Genomic organization of PvPin1 showing untranslated (black boxes) and translated (gray boxes) regions. (B) Alignment of amino acid sequences for PvPin1 and its homologs from other plant species. The PPIase and WW domains are overlined. Asterisks indicate four unique amino acids in plant homologs. (C) Phylogenetic analysis of PvPin1 protein. The phylogenetic tree was generated by using MEGA 5.0 and shows branch lengths proportional to distances.
The sequences of Pin1-like from more than 16 plant species were downloaded from NCBI. Phylogenetic comparison of the PvPin1 protein with homologs in other plants species showed that PvPin1 belongs to the monocots clade and is closely related to Pin1-like proteins from O. sativa, O. brachyantha, Dichanthelium oligosanthes, Zea mays, Aegilops tauschii, and Brachypodium distachyon, especially Pin1-like in P. edulis, which is the affinis species of P. violascens, having the highest identity (94.26%) with PvPin1 (Figure 1C).
Expression Pattern of PvPin1
RT-qPCR was used to characterize the expression pattern of PvPin1 in young and mature leaf, floral bud, culm, bamboo shoot, and rhizome tissue in flowering bamboo plants. Although PvPin1 transcripts were detectable in almost all tested organs, its expression was highest in young leaves and lowest in floral bud (Figure 2A).
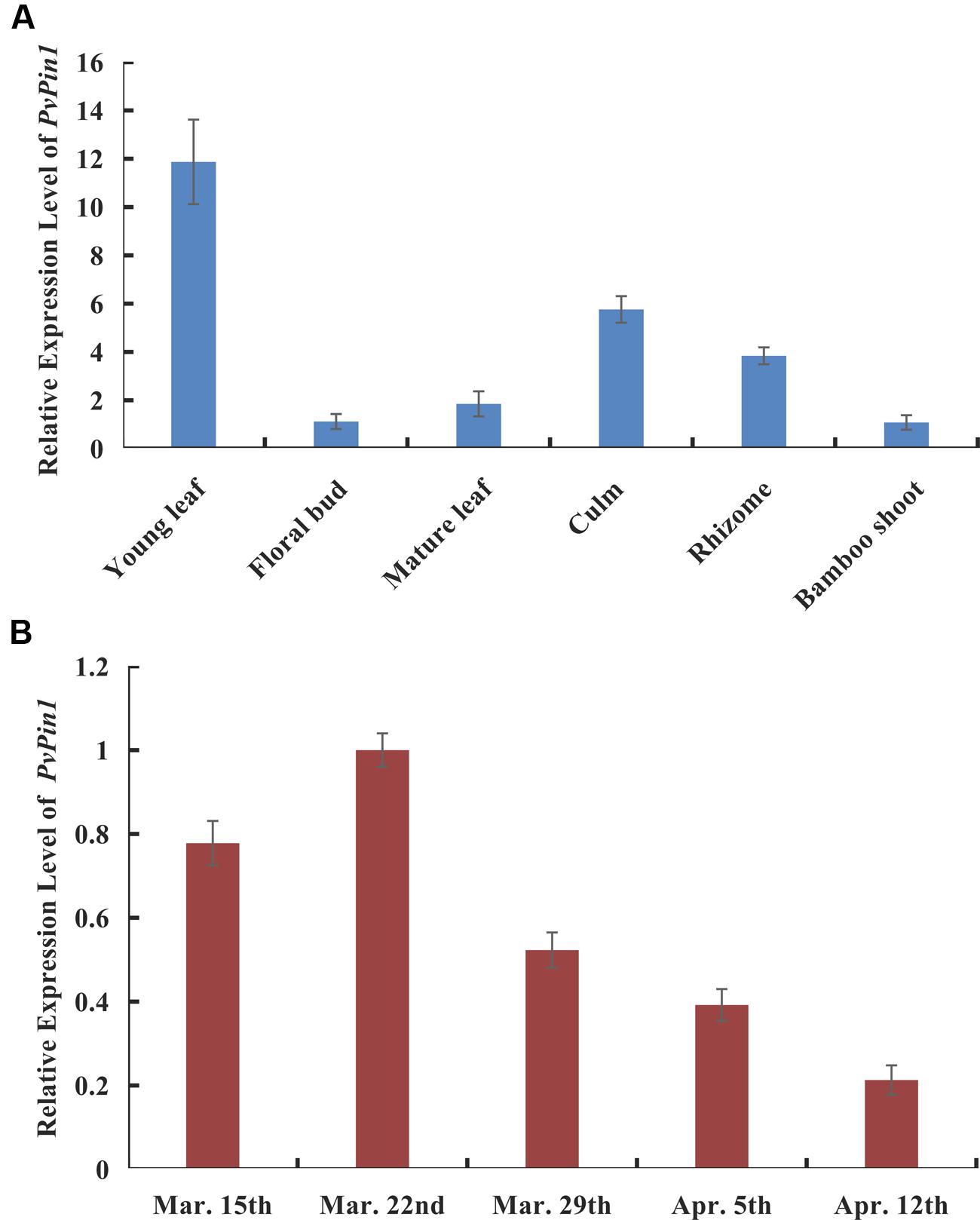
FIGURE 2. Spatial and temporal expression of PvPin1 in P. violascens by RT-qPCR. (A) Relative expression of PvPin1 in different tissues. (B) Relative expression of PvPin1 in young leaves during flower development. In the first stage (before March 29), the meristem morphology of flower buds was similar with vegetative buds; in the second stage (March 29), the inflorescence had presented, and the floral organs started to take shape; in the third stage (April 5), the floral organs continued to develop and gradually become mature; and in the fourth stage (April 12), the bloom stage, the anther was outcropped from palea (Lin et al., 2012). Data are mean ±SD from three replicates.
Also, we used RT-qPCR to detect the temporal expression of PvPin1 in young bamboo leaves at different flowering stages from March 15 to April 12. PvPin1 expression peaked on March 22, before flowering. Although the PvPin1 transcript level increased significantly in young leaves of flowering bamboo plants at the early stage, it declined in leaves during flower development (Figure 2B).
ABRE and TGACG-Motif Elements Exist in Promoters of PvPin1 and PePin1 But Not Pin1At
To determine whether the expression patterns of PvPin1, Pin1At, and PePin1 (Pin1-like in P. edulis) were associated with the regulation of their promoters, we compared and analyzed their promoter sequences. An upstream 1500-bp sequence of PvPin1’s start codon was cloned, and the same length promoter sequences of Pin1At and PePin1 were downloaded from the NCBI database. On sequence alignment, the promoter sequence of PvPin1 shared 39.09% and 60.43% similarity with those of Pin1At and PePin1, respectively. The potential cis-acting regulatory elements were predicted by using PlantCARE. The typical CAAT-box and TATA-box core elements and other elements involving in light-responsive (LTR), MeJA-responsive (CGTCA-motif), endosperm expression (Skn-1_motif), and anaerobic responsive (ARE) were commonly found in these three promoters (Figure 3). However, the ABRE and TGACG-motif cis-acting elements, which are regulated by ABA and MeJA, respectively, were specific to the promoter sequences of bamboo (PvPin1 and PePin1).
ABA and MeJA Treatment Decreased PvPin1 Expression in Leaf ofP. violascens Seedlings
P. violascens plants were treated with ABA and MeJA because the ABRE and TGACG-motif elements were specific to the promoter sequences of bamboo (PvPin1 and PePin1). The mRNA level of PvPin1 in leaf was significantly lower with ABA and MeJA than mock treatment (Figure 4), which suggested that PvPin1 can respond to ABA and MeJA.
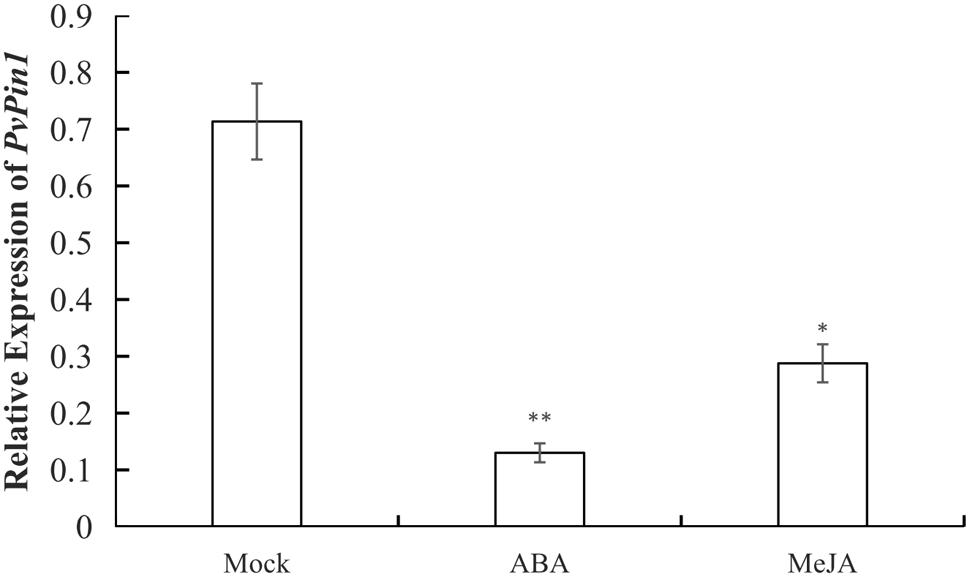
FIGURE 4. Effect of abscisic acid (ABA) and methyl jasmonate (MeJA) treatments on the expression of PvPin1 in leaves of P. violascens seedlings. Data are mean ± SD. ∗p < 0.05; ∗∗p < 0.01 compared to mock, by Student’s t-test.
Ectopic Expression of PvPin1 Delays Flowering Time in Arabidopsis
Establishing a regeneration and genetic transformation system in bamboo is difficult (Zang et al., 2016, 2017). To examine the function of PvPin1 in regulating flowering, we overexpressed PvPin1 under control of the CaMV 35S promoter in the pCAMBIA1301 vector in Arabidopsis. Six independent lines in the homozygous T3 generation grown under greenhouse conditions were chosen for further analysis. 35S::PvPin1 transgenic Arabidopsis showed a significantly late flowering phenotype (Figures 5A,B).
To understand whether the phenotypic alteration of flowering time in transgenic Arabidopsis was related to the expression of PvPin1, we detected the expression of PvPin1 in six homozygous lines of 35S::PvPin1 Arabidopsis. RT-qPCR revealed a positive association between flowering time and the expression of PvPin1 in transgenic Arabidopsis (Figures 5B,C).
In Arabidopsis, SOC1 and AGL24 are important regulatory genes locating at the convergence of the multiple floral induction pathways. We determined the transcript levels of SOC1 and AGL24 in transgenic Arabidopsis by RT-qPCR. The transcript levels of SOC1 and AGL24 in 35S::PvPin1 transgenic plants were greatly decreased (Figure 5D). Hence, PvPin1 delayed the flowering time in Arabidopsis by downregulating the expression of SOC1 and AGL24.
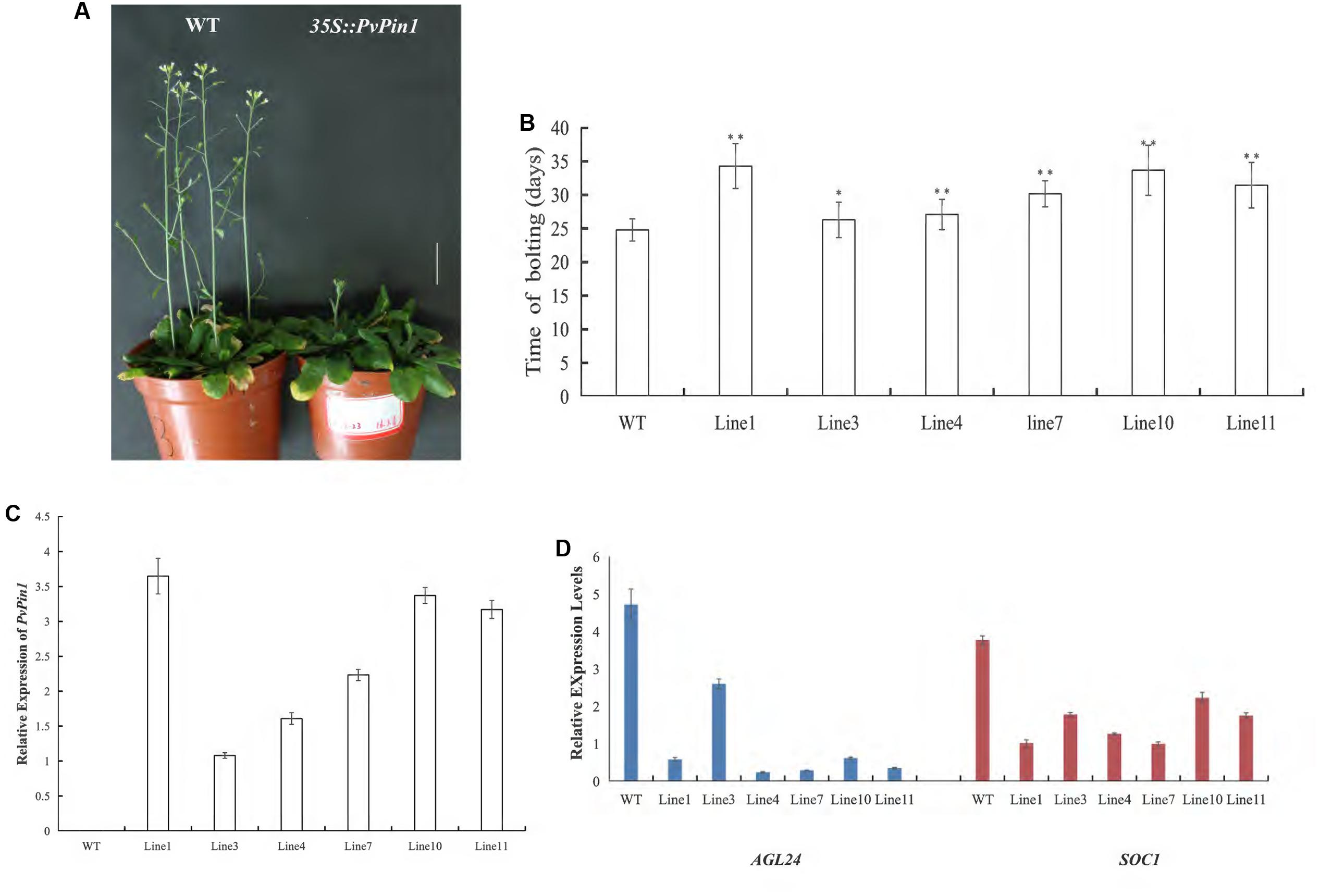
FIGURE 5. Phenotype analysis of 35S::PvPin1 Arabidopsis plants under long-day (LD) conditions. (A) Late flowering phenotype of transgenic Arabidopsis. The scale bar represents 2 cm. (B) Days to flowering of T3 transgenic Arabidopsis (n = 30). (C) RT-qPCR expression analysis of PvPin1. (D) Expression analysis of AGL24 and SOC1. Data are mean ± SD from three replicates. ∗p < 0.05; ∗∗p < 0.01 compared to WT, by Student’s t-test.
PvPin1 Overexpression Delays Flowering in Rice
To further examine its function, PvPin1 was transformed into O. sativa (Dongjing), a member of the same grass family as bamboo. We analyzed the flowering time in six independent lines in the homozygous T3 generation that were grown under field conditions. 35S::PvPin1 transgenic rice plants showed a significantly late-flowering phenotype (Figures 6A,B). Moreover, days to heading were positively associated with the expression of PvPin1 in transgenic rice (Figures 6B,C).
OsMADS50 and OsMADS56 are two SOC1 homolog genes in rice. Because SOC1 expression was markedly decreased in 35S::PvPin1 transgenic Arabidopsis plants, we determined the expression of OsMADS50 and OsMADS56 in transgenic rice lines 4, 5, and 6. OsMADS56 expression was greatly increased in these lines, with no significant difference in expression of OsMADS50 in comparison with wild-type rice (Figure 6D), so overexpression of PvPin1 inhibited flowering in transgenic rice by upregulating the expression of OsMADS56.
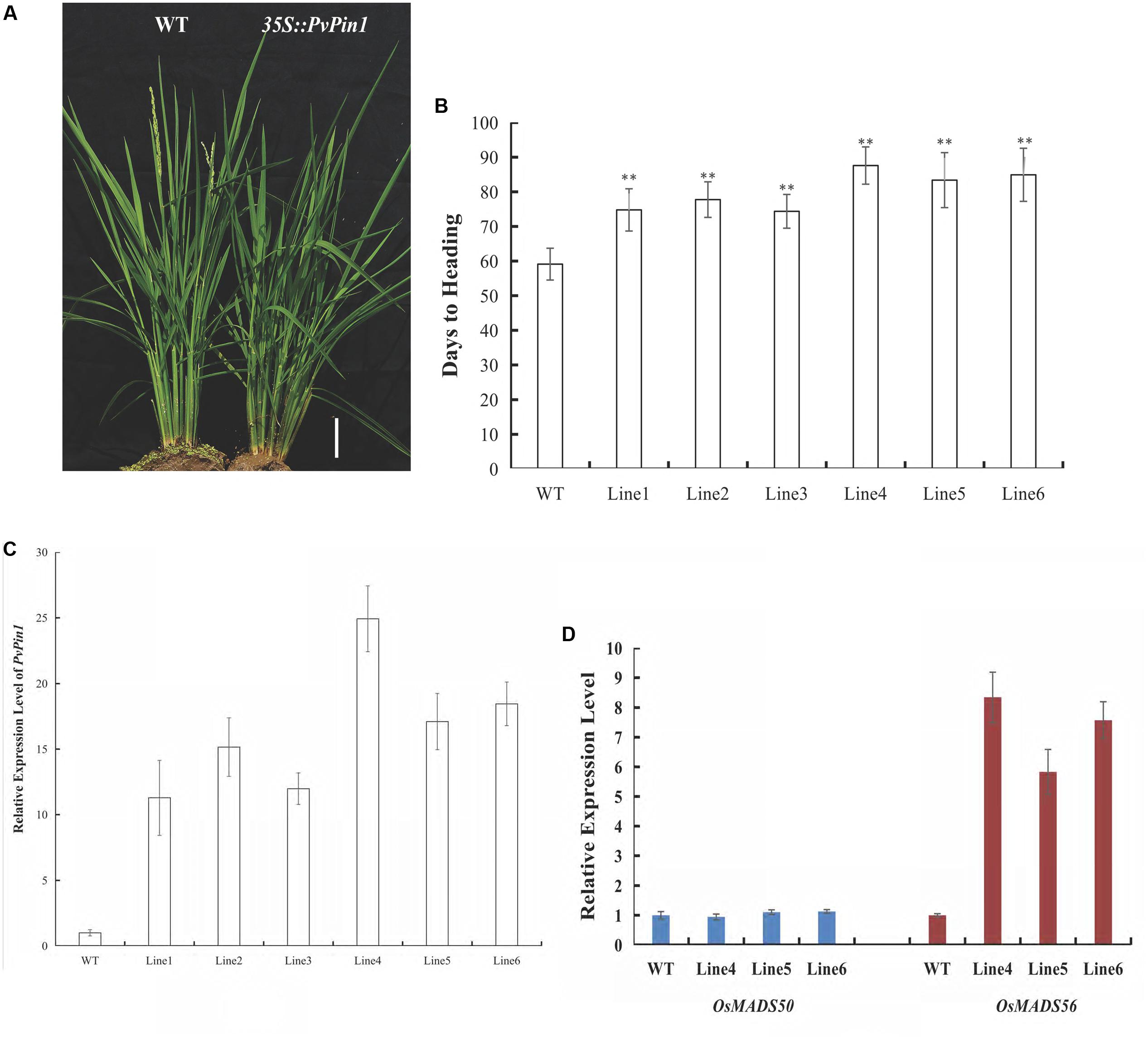
FIGURE 6. Phenotypic differences between 35S::PvPin1 and WT rice grown in the field. (A) Late-flowering phenotype of transgenic rice. The scale bar represents 5 cm. (B) Days to heading of T3 homozygous transgenic plants (n = 30). (C) RT-qPCR expression analysis of PvPin1. (D) Expression analysis of OsMADS50 and OsMADS56. Data are mean ± SD from three replicates. ∗∗p < 0.01 compared to WT, by Student’s t-test.
PvPin1 Was Localized in the Nucleus and Cytolemma
Pin1At localizes in both the nucleus and cytoplasm (Wang et al., 2010). We used the infection method (Escobar et al., 2003) to determine the subcellular localization of PvPin1 protein. The fusion protein PvPin1-GFP located in the nucleus and cytolemma of epidermal cells of tobacco (Nicotiana benthamiana), whereas as a control, the GFP protein distributed in the whole tobacco cells (Figure 7). The different localization between Pin1At and PvPin1 implies that they might have different function.
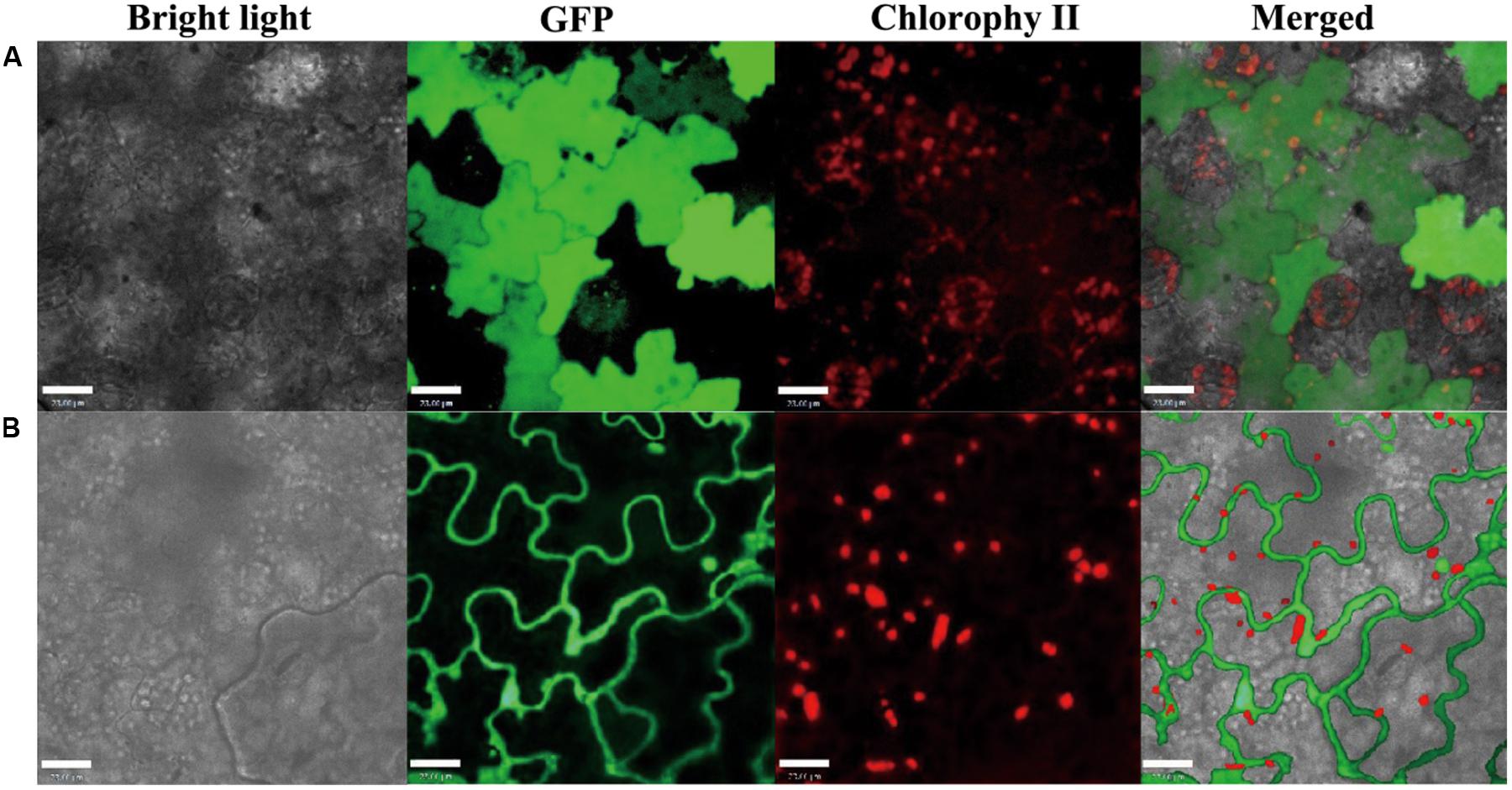
FIGURE 7. Subcellular localization of PvPin1 in leaf cells of N. benthamiana. (A) Fluorescence of the control green fluorescent protein (GFP) distributed throughout the cell. (B) PvPin1::GFP localized in both the cytolemma and nucleus. Bar = 23 μm.
Discussion
Bamboo usually experiences a long vegetative phase before flowering, so some floral suppressors may be working during this long phase to inhibit bamboo flowering. The inhibiting effect of these flower suppressors could be relieved when bamboo is under stress or undergoing a lengthy vegetative growth. Flowering locus C (FLC) is an important flower repressor in Arabidopsis; however, no FLC homologs were determined in monocot plants until now (Doi et al., 2004; Helliwell et al., 2006). Many flowering promoters in bamboo have been reported (Tian et al., 2005; Lin et al., 2009, 2010; Guo et al., 2016; Liu et al., 2016a). We previously showed that BoTFL1-like and PvFRIL might be possible floral suppressors of bamboo (Zeng et al., 2015; Liu et al., 2016c). In this study, we identified and characterized another possible floral suppressor, a Pin1-like gene from P. violascens named PvPin1. PvPin1 was expressed in all tested tissues in bamboo, but its expression was highest in young leaf and lowest in flower bud (Figure 2A). PvPin1 expression peaked before flowering then gradually decreased (Figure 2B). Overexpression of PvPin1 conferred a significantly late flowering phenotype in both greenhouse-grown Arabidopsis and field-grown rice. Hence, PvPin1 might be a flowering repressor in bamboo.
Genome sequence analysis showed that PvPin1, Pin1At, and Pin1 homologs in rice and corn have only one intron. Amino acid sequence alignment showed that PvPin1 contains only a C-terminal PPIase catalytic domain with four additional amino acids (Figure 1B) like Pin1At (Wang et al., 2010) and other Pin1-like proteins in plant. Therefore, the gene and protein structure of Pin1-like in plants are conserved. However, PvPin1 delaying flowering time in transgenic Arabidopsis grown under greenhouse conditions and transgenic rice grown under field conditions, which differs from Pin1At, known as a flowering promoter. Furthermore, PvPin1 also has different protein localization and promoter cis-elements from Pin1At. These differences might be caused by evolutionary diversity of the genes and result in the unique flowering characteristics of bamboo.
A protein’s location is mainly determined by its amino acid sequence (Olson et al., 2002). The nuclear localization of hPin1 from human is responsibly directed by the Pin1-WW domain (Rippmann et al., 2000). Recent studies showed that Pin1At from Arabidopsis and DlPar13 from Digitalis lanata localized in the nucleus and cytoplasm (Metzner et al., 2001; Wang et al., 2010), and we found that PvPin1 localized in nuclear and cytomembrane. These three plant proteins have no WW domain but have four additional amino acids (Figure 8). Whether the four additional amino acids are associated with nuclear localization is unknown. In addition, comparison of protein sequences showed that Pin1At and DlPar13 have the same 10 amino acids, which differs from PvPin1 (Figure 8) and may result in their different protein localization and further lead to different functions in flowering regulation.
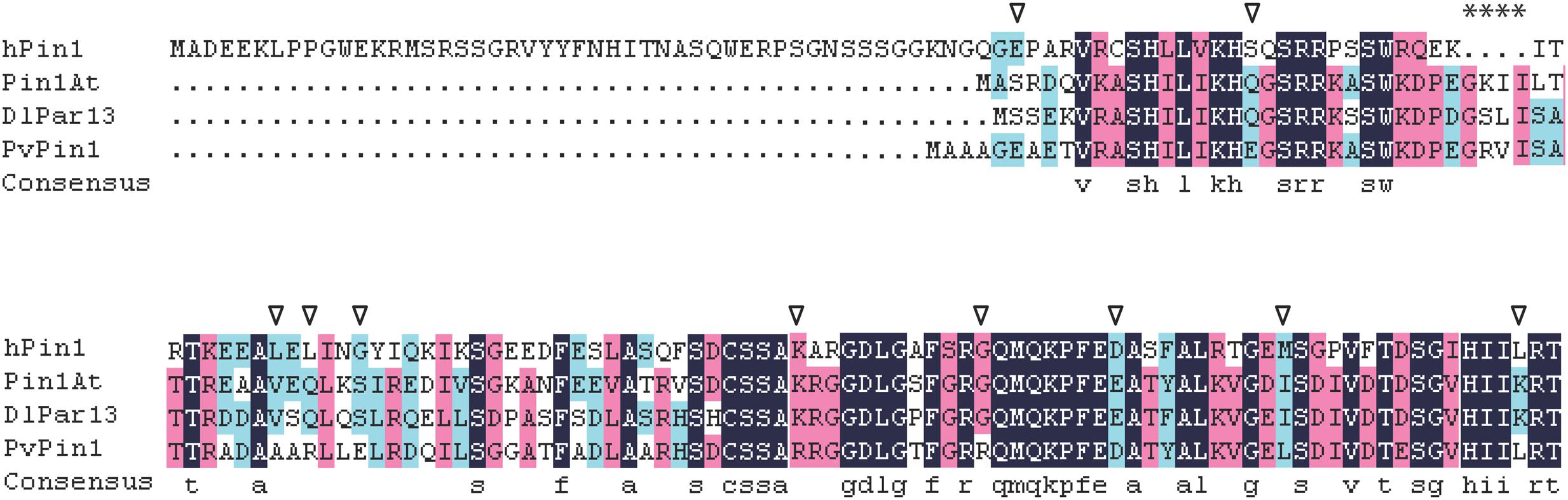
FIGURE 8. Alignment of amino acid sequences for hPin1 in human, Pin1At, DlPar13 in Digitalis lanata, and PvPin1. Asterisks indicate four unique amino acids in plant Pin1 homologs, triangle indicates special amino acid of PvPin1 in comparison with Pin1At and DlPar13.
Multiple genetic pathways coordinately control floral transition in Arabidopsis (Koornneef et al., 1998; Levy and Dean, 1998; Mouradov et al., 2002). SOC1 and AGL24 are essential regulatory genes involved in multiple floral induction pathways (Lee et al., 2000; Samach et al., 2000; Yu et al., 2002; Michaels et al., 2003; Liu et al., 2007). OsMADS50 and OsMADS56 are SOC1 homologous genes in rice (Nam et al., 2005). OsMADS50 acts as a flowering activator whose overexpression could promote flowering in transgenic Arabidopsis (Tadege et al., 2003). OsMADS50 and OsMADS56 may form a complex to delay rice heading time (Ryu et al., 2009). In this study, we found that overexpression of PvPin1 could downregulate the expression of AGL24 and SOC1 in Arabidopsis and upregulate OsMADS56 in rice to delay the flowering time. However, whether the regulation of SOC1 and AGL24 in Arabidopsis and OsMADS56 in rice is indirect or direct requires further experiments.
Plant hormones are related to flower development. Lu et al. (2012) showed that flower bud differentiation could be promoted with a high ABA level in P. violascens. Abscisic acid could promote flowering by activating the key floral gene flowering locus T (FT) in Arabidopsis (Conti et al., 2014). Methyl jasmonate could also affect flowering time in some other species (Diallo et al., 2014). In this study, ABRE (responding to ABA) and TGACG-motif (responding to MeJA) were found as specific cis-acting elements in the promoter of PvPin1 and PePin1 (bamboo) in comparison with Pin1At (Arabidopsis). ABA and MeJA treatments could reduce the expression of PvPin1 (possible flowering repressor) in P. violascens. In addition, ABA and MeJA might promote flowering by upregulating the expression of PvMADS56 (flower promoter) in P. violascens (Liu et al., 2016b). Thus, ABA and MeJA might promote flowering by affecting multiple genes such as PvMADS56 and PvPin1 via a complicated regulatory network in bamboo, and their inhibitor may be used for inhibiting bamboo flowering for bamboo forest management.
We found that PvPin1 is evolutionarily conserved in gene and protein structure in comparison with Pin1-like homologs from other plants, especially monocots; however, unlike Pin1At, PvPin1 might be a flowering repressor that can delay bamboo flowering. In addition, our results indicate that ABA and MeJA can significantly reduce the expression of PvPin1 to promote bamboo flowering. Our results are helpful to disclose the bamboo flowering mechanism and could be used for developing new technologies to inhibit bamboo flowering.
Materials and Methods
Plant Materials and Growth Conditions
P. violascens samples were collected from the campus of Zhejiang Agriculture and Forestry University. Wild-type (ecotype Columbia) and transgenic plants of A. thaliana were cultivated in a controlled temperature room under 22°C with 16-h light/8-h dark. N. benthamiana was grown in a controlled temperature room under 28°C with 10-h light/14-h dark. Rice (O. sativa cv. Dongjing) plants are cultivated in the field of Lin’an (Zhejiang, China, north latitude 30°14′ and east longitude 119°42′).
Isolation of PvPin1 cDNA and Its Intron Sequence from P. violascens
Total RNA from P. violascens was isolated by using RNAiso Plus (Takara, Shiga, Japan), then Reverse Transcriptase M-MLV (Takara, Japan) was used to synthesize first-strand cDNA. A specific Pin1-like cDNA fragment (approximately 300 bp) was amplified by using the pair of primers (Pin1-1 and Pin1-2, Table 1), which were designed by comparing the amino acid sequences of Pin1 homologs from grass family plants including P. edulis (FP099633.1), O. rufipogon (CU406178.1), O. sativa (AK243434.1), Triticum aestivum (AK333419.1), and Zea mays (NM001157033.1). The 3′ end and 5′ partial cDNA of Pin1-like were isolated with the RACE kit (Invitrogen) by using gene-specific primers (3′-1 and 3′-2; 5′-1 and 5′-2, Table 1). Finally, the full-length ORF sequence was obtained by using the primers (ORF-F and ORF-R, Table 1) based on the known 5′ and 3′ sequences.
Genomic DNA was isolated by the modified CTAB method (Reichardt and Rogers, 1993) from leaves. Then a 2709-bp intron sequence of PvPin1 was obtained by using the primers ORF-F and ORF-R.
Isolation of PvPin1 Promoter from P. violascens
The sequence of PvPin1 ORF was used for a BLAST search in the transcript online database for P. edulis (affinis species of P. violascens) (Peng et al., 2013)1. A sequence (ID: FP099633.1) that exists between PH01001300G0520 and PH01001300G0540 with the highest identity to PvPin1 was identified. Then the correlative genomic sequence in PH01001300 was extracted from the genome database of P. edulis (Peng et al., 2013) and used to design the primers (S1-F, S1-R; S2-F, S2-R, Table 1) for amplifying the promoter of PvPin1. A promoter sequence of 1500 bp was obtained from the DNA by using Nested PCR (Gundersen and Lee, 1996).
Expression Pattern of PvPin1
The RT-qPCR primers (qPCR-F and qPCR-R, Table 1) were designed by using the full-length ORF sequence of PvPin1. Here, PeUBC18 was used as the internal control gene (Qi et al., 2013; Liu et al., 2016b; Table 1) because of the close relationship between P. edulis and P. violascens. CFX96TM Real-Time PCR Detection System (Bio-Rad) and the SYBR Premix ExTaq II mix (Takara) were used for PCR amplification. The program was 95°C for 3 min, followed by 40 cycles of amplification (95°C for 15 s, 60°C for 30 s). Reactions were performed in 20-μl mixtures consisting of 10 μl 2× SYBR Premix Ex Taq II Mix, 0.5 μl each of forward or reverse primer, 1 μl cDNA template (50 ng/μl), and 8 μl double distilled H2O (Liu et al., 2016b). The data were analyzed by the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Binary Plasmid Construction and Analysis of Transgenic Plants
The full-length ORF for PvPin1 was cloned into the binary vector pCAMBIA1301 under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. Recombinant vector was transferred into A. tumefaciens strain GV3101, then into Arabidopsis by the floral dip method (Clough and Bent, 1998). Transformants were screened in media with 50 μg/ml kanamycin. The same construct was also transformed into rice plants (Dongjing) mediated by A. tumefaciens strain EHA105 as described (Xu et al., 2017). Positive transgenic rice lines were confirmed by genomic PCR. The expression of SOC1 and AGL24 genes in transgenic Arabidopsis in six T3 lines and WT Arabidopsis, and the expression of OsMADS50 and OsMADS56 in transgenic rice in three T3 lines and WT rice were analyzed by real-time qPCR with gene-specific primers (Table 1) following the protocol in expression pattern of PvPin1 section “Expression Pattern of PvPin1.”
Subcellular Localization of PvPin1
The full-length coding sequence without terminator codon (TAA) of PvPin1 was cloned into the CaMV 35S-GFP vector that allowed the system to generate a PvPin1-GFP fusion protein for investigating subcellular location in epidermal cells from tobacco (N. benthamiana) and the transient expression assay method (Escobar et al., 2003) was adopted. The tobacco epidermal cells were visualized on confocal laser scanning microscopy (LSM510, Zeiss, Germany).
Bioinformatics Analysis
A BLAST search in the NCBI database was used to obtain the protein sequences of Pin1-like. The phylogenetic tree was constructed by the neighbor-joining method with the parameter bootstrap (10,000 replicates) in MEGA 5.0. The software ProtParam from ExPASy2 was used to obtain the molecular weights (MW) and theoretical isoelectric point (pI) of PvPin1 protein. PlantCARE (Lescot et al., 2002) was used to analyze cis-acting regulatory elements in the PvPin1 promoter.
ABA and MeJA Treatment
Leaves of P. violascens seedlings were sprayed with ABA (100 μM), MeJA (100 μM), and water as a blank control once a day for 9 days. Every treatment was performed with three biological replicates. The PvPin1 transcript level was detected after treatment.
Author Contributions
XL, ZZ, and XY conceived and designed the experiments; YF contributed materials of transgenic rice; XL and LZ monitored the experimental work; ZZ, XY, and HW performed the experiments; ZZ analyzed the data; and ZZ and XL wrote the paper.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 31270715 and 31000295), the Ministry of Science and Technology of China (No. 2012CB723008), and the Top Key Discipline of Forestry of Zhejiang Province (KF201304).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Brambilla, V., and Fornara, F. (2013). Molecular control of flowering in response to day length in rice. J. Integr. Plant Biol. 55, 410–418. doi: 10.1111/jipb.12033
Chen, X. (2003). Promotion of bamboo for poverty alleviation and economic development. J. Bamboo Rattan 2, 345–350. doi: 10.1163/156915903322700386
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Conti, L., Galbiati, M., and Tonelli, C. (2014). “ABA and the floral transition,” in Abscisic Acid: Metabolism, Transport and Signaling, ed. D. P. Zhang (Beijing: Tsinghua University), 365–384.
Diallo, A. O., Agharbaoui, Z., Badawi, M. A., Ali-Benali, M. A., Moheb, A., Houde, M., et al. (2014). Transcriptome analysis of an mvp mutant reveals important changes in global gene expression and a role for methyl jasmonate in vernalization and flowering in wheat. J. Exp. Bot. 65, 2271–2286. doi: 10.1093/jxb/eru102
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. doi: 10.1101/gad.1189604
Escobar, N. M., Haupt, S., Thow, G., Boevink, P., Chapman, S., and Oparka, K. (2003). High-throughput viral expression of cDNA–green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell 15, 1507–1523. doi: 10.1105/tpc.013284
Fischer, G., Bang, H., and Mech, C. (1983). Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides. Biomed. Biochim. Acta 43, 1101–1111.
Franklin, D. C. (2004). Synchrony and asynchrony: observations and hypotheses for the flowering wave in a long lived semelparous bamboo. J. Biogeogr. 31, 773–786. doi: 10.1111/j.1365-2699.2003.01057.x
Gao, J., Ge, W., Zhang, Y., Cheng, Z., Li, L., Hou, D., et al. (2015). Identification and characterization of microRNAs at different flowering developmental stages in moso bamboo (Phyllostachys edulis) by high-throughput sequencing. Mol. Genet. Genomics 290, 2335–2353. doi: 10.1007/s00438-015-1069-8
Gao, J., Zhang, Y., Zhang, C., Qi, F., Li, X., Mu, S., et al. (2014). Characterization of the floral transcriptome of Moso bamboo (Phyllostachys edulis) at different flowering developmental stages by transcriptome sequencing and RNA-seq analysis. PLOS ONE 9:e98910. doi: 10.1371/journal.pone.0098910
Gundersen, D., and Lee, I.-M. (1996). Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol. Mediterr. 35, 144–151.
Guo, X. Q., Wang, Y., Wang, Q., and Xu, Z. E. (2016). Molecular characterization of FLOWERING LOCUS T(FT)genes from bamboo (Phyllostachys violascens). J. Plant Biochem. Biotechnol. 25, 168–178. doi: 10.1007/s13562-015-0322-x
Hanes, S. D., Shank, P. R., and Bostian, K. A. (1989). Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast 5, 55–72. doi: 10.1002/yea.320050108
Hani, J., Stumpf, G., and Domdey, H. (1995). PTF1 encodes an essential protein in Saccharomyces cerevisiae, which shows strong homology with a new putative family of PPIases. FEBS Lett. 365, 198–202. doi: 10.1016/0014-5793(95)00471-K
He, Z., Li, L., and Luan, S. (2004). Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 134, 1248–1267. doi: 10.1104/pp.103.031005
Helliwell, C. A., Wood, C. C., Robertson, M., James, P. W., and Dennis, E. S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular weight protein complex. Plant J. 46, 183–192. doi: 10.1111/j.1365-313X.2006.02686.x
Hsu, T., McRackan, D., Vincent, T. S., and De Couet, H. G. (2001). Drosophila Pin1 prolyl isomerase Dodo is a MAP kinase signal responder during oogenesis. Nat. Cell Biol. 3, 538–543. doi: 10.1038/35078508
Hunter, T. (1998). Prolyl isomerases and nuclear function. Cell 92, 141–143. doi: 10.1016/S0092-8674(00)80906-X
Izawa, T. (2007). Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58, 3091–3097. doi: 10.1093/jxb/erm159
Jack, T. (2004). Molecular and genetic mechanisms of floral control. Plant Cell 16(Suppl. 1), S1–S17. doi: 10.1105/tpc.017038
Jarillo, J. A., and Piñeiro, M. (2011). Timing is everything in plant development. The central role of floral repressors. Plant Sci. 181, 364–378. doi: 10.1016/j.plantsci.2011.06.011
Kiefhaber, T., Quaas, R., Hahn, U., and Schmid, F. X. (1990). Folding of ribonuclease T1. 2. Kinetic models for the folding and unfolding reactions. Biochemistry 29, 3061–3070. doi: 10.1021/bi00464a024
Koornneef, M., Alonso-Blanco, C., Peeters, A. J., and Soppe, W. (1998). Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Biol. 49, 345–370. doi: 10.1146/annurev.arplant.49.1.345
Landrieu, I., De Veylder, L., Fruchart, J. S., Odaert, B. T., Casteels, P., Portetelle, D., et al. (2000). The Arabidopsis thaliana PIN1At gene encodes a single-domain phosphorylation-dependent peptidyl prolyl cis/trans isomerase. J. Biol. Chem. 275, 10577–10581. doi: 10.1074/jbc.275.14.10577
Lee, H., Suh, S. S., Park, E., Cho, E., Ahn, J. H., Kim, S. G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. doi: 10.1101/gad.813600
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Levy, Y. Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10, 1973–1989. doi: 10.1105/tpc.10.12.1973
Lin, E. P., Peng, H. Z., Jin, Q. Y., Deng, M. J., Li, T., Xiao, X. C., et al. (2009). Identification and characterization of two Bamboo (Phyllostachys praecox) APl/SQUA-like MADS-box genes during floral transition. Planta 231, 109–120. doi: 10.1007/s00425-009-1033-0
Lin, X., Chow, T., Chen, H., Liu, C., Chou, S., Huang, B., et al. (2010). Understanding bamboo flowering based on large-scale analysis of expressed sequence tags. Genet. Mol. Res. 9, 1085–1093. doi: 10.4238/vol9-2gmr804
Lin, X. C., Yuan, X. L., Lin, R., Lou, Y. F., and Fang, W. (2012). Morphogenesis of indefinite inflorescence of Phyllostachys violascens (Carr.) A. et Riv. J. Fujian College Forestr. 32, 141–145.
Liu, C., Zhou, J., Bracha-Drori, K., Yalovsky, S., Ito, T., and Yu, H. (2007). Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134, 1901–1910. doi: 10.1242/dev.003103
Liu, S., Ma, T., Ma, L., and Lin, X. (2016a). Ectopic expression of PvSOC1, a homolog of SOC1 from Phyllostachys violascens, promotes flowering in Arabidopsis and rice. Acta Physiol. Plant. 38, 1–9. doi: 10.1007/s11738-016-2186-7
Liu, S., Qi, T., Ma, J., Ma, T., Ma, L., and Lin, X. (2016c). Ectopic expression of a SOC1 homolog from Phyllostachys violascens alters flowering time and identity of floral organs in Arabidopsis thaliana. Trees 30, 2203–2215. doi: 10.1007/s00468-016-1445-y
Liu, S., Zhu, L., Lin, X., and Ma, L. (2016b). Overexpression of the repressor gene PvFRI-L from Phyllostachys violascens delays flowering time in transgenic Arabidopsis thaliana. Biol. Plant. 3, 401–409. doi: 10.1007/s10535-016-0614-6
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Louis, B., Waikhom, S. D., Goyari, S., Jose, R. C., Roy, P., and Talukdar, N. C. (2015). First proteome study of sporadic flowering in bamboo species (Bambusa vulgaris and Dendrocalamus manipureanus) reveal the boom is associated with stress and mobile genetic elements. Gene 574, 255–264. doi: 10.1016/j.gene.2015.08.010
Lu, K. P., Hanes, S. D., and Hunter, T. (1996). A human peptidyl–prolyl isomerase essential for regulation of mitosis. Nature 380, 544–547. doi: 10.1038/380544a0
Lu, K. P., and Zhou, X. Z. (2007). The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat. Rev. Mol. Cell Biol. 8, 904–916. doi: 10.1038/nrm2261
Lu, P. J., Zhou, X. Z., Shen, M., and Lu, K. P. (1999). Function of WW domains as phosphoserine-or phosphothreonine-binding modules. Science 283, 1325–1328. doi: 10.1126/science.283.5406.1325
Lu, Y., Yuan, X., Lin, X., and Fang, W. (2012). Endogenous hormone changes during floral bud morphological differentiation of Phyllostachys violascens. J. Zhejiang A F Univ. 29, 161–165.
Metzner, M., Stoller, G., Rücknagel, K. P., Lu, K. P., Fischer, G., Luckner, M., et al. (2001). Functional replacement of the essential ESS1 in yeast by the plant parvulin DlPar13. J. Biol. Chem. 276, 13524–13529. doi: 10.1074/jbc.M007005200
Michaels, S. D., Ditta, G., Gustafson Brown, C., Pelaz, S., Yanofsky, M., and Amasino, R. M. (2003). AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 33, 867–874. doi: 10.1046/j.1365-313X.2003.01671.x
Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time interacting pathways as a basis for diversity. Plant Cell 14(Suppl. 1), S111–S130.
Nam, J., Kaufmann, K., Theißen, G., and Nei, M. (2005). A simple method for predicting the functional differentiation of duplicate genes and its application to MIKC-type MADS-box genes. Nucleic Acids Res. 33:e12. doi: 10.1093/nar/gni003
Olson, V. A., Wetter, J. A., and Friesen, P. D. (2002). Baculovirus transregulator IE1 requires a dimeric nuclear localization element for nuclear import and promoter activation. J. Virol. 76, 9505–9515. doi: 10.1128/JVI.76.18.9505-9515.2002
Pastorino, L., Sun, A., Lu, P. J., Zhou, X. Z., Balastik, M., Finn, G., et al. (2006). The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-β production. Nature 440, 528–534. doi: 10.1038/nature04543
Peng, Z., Lu, Y., Li, L., Zhao, Q., Feng, Q., Gao, Z., et al. (2013). The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 45, 456–461. doi: 10.1038/ng.2569
Putterill, J., Laurie, R., and Macknight, R. (2004). It’s time to flower: the genetic control of flowering time. Bioessays 26, 363–373. doi: 10.1002/bies.20021
Qi, F., Hu, T., Peng, Z., and Gao, J. (2013). Screening of reference genes used in qRT-PCR and expression analysis of PheTFL1 gene in Moso Bamboo. Acta Bot. Boreali Occident. Sin. 1, 011.
Ranganathan, R., Lu, K. P., Hunter, T., and Noel, J. P. (1997). Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 89, 875–886. doi: 10.1016/S0092-8674(00)80273-1
Reichardt, M., and Rogers, S. (1993). “Plant DNA isolation using CTAB,” in Current Protocols in Molecular Biology, eds F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, et al. (Hoboken, NJ: John Wiley and Sons).
Rippmann, J. F., Hobbie, S., Daiber, C., Guilliard, B., Bauer, M., Birk, J., et al. (2000). Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell Growth Differ. 11, 409–416.
Ryu, C. H., Lee, S., Cho, L. H., Kim, S. L., Lee, Y. S., Choi, S. C., et al. (2009). OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD) dependent flowering in rice. Plant Cell Environ. 32, 1412–1427. doi: 10.1111/j.1365-3040.2009.02008.x
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. doi: 10.1126/science.288.5471.1613
Schiene-Fischer, C. (2015). Multidomain peptidyl prolyl cis/trans Isomerases. Biochim. Biophys. Acta 1850, 2005–2016. doi: 10.1016/j.bbagen.2014.11.012
Shih, M. C., Chou, M. L., Yue, J. J., Hsu, C. T., Chang, W. J., Ko, S. S., et al. (2014). BeMADS1 is a key to delivery MADSs into nucleus in reproductive tissues-De novo characterization of Bambusa edulis transcriptome and study of MADS genes in bamboo floral development. BMC Plant Biol. 14:179. doi: 10.1186/1471-2229-14-179
Simpson, G. G., and Dean, C. (2002). Arabidopsis, the rosetta stone of flowering time. Science 296, 285–289. doi: 10.1126/science.296.5566.285
Song, X. Z., Zhou, G. M., Jiang, H., Yu, S. Q., Fu, J. H., Li, W. Z., et al. (2011). Carbon sequestration by chinese bamboo forests and their ecological benefits: assessment of potential, problems, and future challenges. Environ. Rev. 19, 418–428. doi: 10.1139/a11-015
Tadege, M., Sheldon, C. C., Helliwell, C. A., Upadhyaya, N. M., Dennis, E. S., and Peacock, W. J. (2003). Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 1, 361–369. doi: 10.1046/j.1467-7652.2003.00034.x
Tian, B., Chen, Y., Yan, Y., and Li, D. (2005). Isolation and ectopic expression of a bamboo MADS-box gene. Chin. Sci. Bull. 50, 217–224. doi: 10.1007/BF02897530
Wang, Y., Liu, C., Yang, D., Yu, H., and Liou, Y. C. (2010). Pin1At encoding a peptidyl-prolyl cis/trans isomerase regulates flowering time in Arabidopsis. Mol. Cell. 37, 112–122. doi: 10.1016/j.molcel.2009.12.020
Wu, C. Y., You, C. J., Li, C. S., Long, T., Chen, G. X., Byrne, M. E., et al. (2008). RID1 encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl. Acad. Sci. U.S.A 105, 12915–12920. doi: 10.1073/pnas.0806019105
Xu, Y., Julien, S., Wu, Y., Fu, Y., Zhu, L., Li, J., et al. (2017). Rice sucrose partitioning mediated by a putative pectin methyltransferase and homogalacturonan methylesterification. Plant Physiol. 3, 1595–1608. doi: 10.1104/pp.16.01555
Yaffe, M. B., Schutkowski, M., Shen, M., Zhou, X. Z., Stukenberg, P. T., Rahfeld, J. U., et al. (1997). Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278, 1957–1960. doi: 10.1126/science.278.5345.1957
Yao, J. L., Kops, O., Lu, P. J., and Lu, K. P. (2001). Functional conservation of phosphorylation-specific prolyl isomerases in plants. J. Biol. Chem. 276, 13517–13523. doi: 10.1074/jbc.M007006200
Yu, H., Xu, Y., Tan, E. L., and Kumar, P. P. (2002). AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. U.S.A. 99, 16336–16341. doi: 10.1073/pnas.212624599
Zang, Q., Jiao, Y., Guo, X., Zhuge, F., Yeh, K., and Lin, X. (2017). Callus induction and plant regeneration from lateral shoots of herbaceous bamboo Mniochloa abersend. J. Hortic. Sci. Biotechnol. 92, 168–174. doi: 10.1080/14620316.2016.1232610
Zang, Q., Zhou, L., Zhuge, F., Yang, H., Wang, X., and Lin, X. (2016). Callus induction and regeneration via shoot tips of Dendrocalamus hamiltonii. SpringerPlus. 5:1799. doi: 10.1186/s40064-016-3520-7
Zeng, H., Lu, Y., Yang, X., Xu, Y., and Lin, X. (2015). Ectopic expression of the BoTFL1-like gene of Bambusa oldhamii delays blossoming in Arabidopsis thaliana and rescues the tfl1 mutant phenotype. Genet. Mol. Res. 14, 9306–9317. doi: 10.4238/2015.August.10.11
Zhang, X. M., Zhao, L., Larson-Rabin, Z., Li, D. Z., and Guo, Z. H. (2012). De novo sequencing and characterization of the floral transcriptome of Dendrocalamus latiflorus (Poaceae: Bambusoideae). PLOS ONE 7:e42082. doi: 10.1371/journal.pone.0042082
Zhao, H., Peng, Z., Fei, B., Li, L., Hu, T., Gao, Z., et al. (2014). BambooGDB: a bamboo genome database with functional annotation and an analysis platform. Database 2014:bau006. doi: 10.1093/database/bau006
Keywords: Phyllostachys violascens, peptidylprolyl cis/trans isomerases, flowering, repressor, ecotopic expression, abscisic acid, methyl jasmonate
Citation: Zheng Z, Yang X, Fu Y, Zhu L, Wei H and Lin X (2017) Overexpression of PvPin1, a Bamboo Homolog of PIN1-Type Parvulin 1, Delays Flowering Time in Transgenic Arabidopsis and Rice. Front. Plant Sci. 8:1526. doi: 10.3389/fpls.2017.01526
Received: 06 June 2017; Accepted: 21 August 2017;
Published: 08 September 2017.
Edited by:
Hanjo A. Hellmann, Washington State University, United StatesReviewed by:
Yin Tongming, Nanjing Forestry University, ChinaRen Wang, Institute of Botany, Jiangsu Province and Chinese Academy of Sciences, China
Copyright © 2017 Zheng, Yang, Fu, Zhu, Wei and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinchun Lin, linxcx@163.com
 Zhigang Zheng1
Zhigang Zheng1 Xinchun Lin
Xinchun Lin