- Department of Plant Sciences and Plant Pathology, Montana State University, Bozeman, MT, United States
Phylogenetic relationships of the Abyssinian pea (Pisum sativum ssp. abyssinicum) to other subspecies and species in the genus were investigated to test between different hypotheses regarding its origin and domestication. An extensive sample of the Pisum sativum ssp. sativum germplasm was investigated, including groups a-1, a-2, b, c, and d as identified by Kwon et al. (2012). A broad sample of P. fulvum but relatively few P. s. ssp. elatius accessions were analyzed. Partial sequences of 18 genes were compared and these results combined with comparisons of additional genes done by others and available in the literature. In total, 54 genes or gene fragment sequences were involved in the study. The observed affinities between alleles in P. ssp. sativum, P. s. ssp. abyssinicum, P. s. ssp. elatius, and P. fulvum clearly demonstrated a close relationship among the three P. sativum subspecies and rejected the hypothesis that the Abyssinian pea was formed by hybridization between one of the P. sativum subspecies and P. fulvum. If hybridization were involved in the generation of the Abyssinian pea, it must have been between P. s. ssp. sativum and P. s. ssp. elatius, although the Abyssinian pea possesses a considerable number of highly unique alleles, implying that the actual P. s. ssp. elatius germplasm involved in such a hybridization has yet to be tested or that the hybridization occurred much longer ago than the postulated 4000 years bp. Analysis of the P. s. ssp. abyssinicum alleles in genomic regions thought to contain genes critical for domestication indicated that the indehiscent pod trait was independently developed in the Abyssinian pea, whereas the loss of seed dormancy was either derived from P. s. ssp. sativum or at least partially developed before the P. s. ssp. abyssinicum lineage diverged from that leading to P. s. ssp. sativum.
Introduction
The Abyssinian pea [Pisum sativum ssp. abyssinicum A. Br.) (alternatively: Lathyrus schaeferi Kosterin) (Kosterin, 2017) is a well-defined taxon of uncertain derivation. It is cultivated in Ethiopia and Yemen along with the typical domesticated pea (Pisum sativum L.) and appears to lack a wild form (see discussion in Kosterin, 2017). Yet it crosses only with difficulty to the cultivated pea, with wild forms of P. sativum (P. s. ssp. elatius (Bieb.) Schmalh.), or with the other species in the genus (P. fulvum Sibeth. & Sm.). The reduced fertility in these crosses appears to be due to both prezygotic and postzygotic barriers (Kosterin, 2017). In flower color and general habit the Abyssinian pea resembles the domesticated pea, although it possesses several traits rarely found in the domesticated pea, such as strongly serrate leaflets and a glossy seed coat. Relevant to the domestication of pea, it shares with P. s. ssp. sativum an indehiscent pod and absence of a seed dormancy mechanism (in Pisum seed dormancy is primarily controlled by a thick, water-impermeable seed coat). These two traits do not segregate in populations derived from crosses between P. s. ssp. abyssinicum and P. s. ssp. sativum), all progeny possessing indehiscent pods and producing seeds that usually germinate immediately after planting (Weeden, 2007, unpublished). Hence, the same loci must have been mutated in each lineage. Finally, genetic diversity within P. s. ssp. abyssinicum is extremely limited, (Weeden and Wolko, 2001; Vershinin et al., 2003), suggesting that the taxon has experienced a recent, severe bottleneck or is a relatively young taxon.
The combination of low genetic diversity, reproductive semi-isolation from close relatives, and numerous unique traits or markers in the Abyssinian pea has led to various alternative hypotheses about its origin. Govorov (1937) suggested that it may have been formed by hybridization between P. sativum and P. fulvum, producing plants that displayed partial sterility with both parental species. All three taxa have the same chromosome number (2n = 14) (Errico et al., 1991) but differ in karyotype or plastid/cytoplasmic compatibility group (Saccardo, 1971; Bogdanova et al., 2015). Studies with various DNA markers have identified a considerable number of markers present in P. s. ssp. abyssinicum that are much more closely related to P. fulvum than to P. s. ssp. sativum (Vershinin et al., 2003), supporting the hybrid hypothesis. Due to the near absence of genetic diversity in the Abyssinian pea, the hybridization event has been postulated to have occurred about 4000 years bp (Kosterin, 2017), and the traits critical for initial domestication must have been obtained from the P. sativum parent.
In contrast, the similarity of P. s. ssp. abyssinicum to P. s. ssp. sativum in flower color, seed size and several other morphological features, as well as the ability to make reasonably fertile crosses between the two, led many students of the genus to designate the Abyssinian pea as a subspecies of the domesticated taxa (Berger, 1928). The marked divergence of this taxon and its limited genetic diversity being attributed to its isolation and cultivation in the Ethiopian highlands. In this latter hypothesis, isolation and intense selection permits the divergence time between the two subspecies to be moved back considerably. In Pisum, the two traits critical for domestication (indehiscent pods and lack of seed dormancy) are influenced by several loci (Weeden et al., 2002), although one locus, Dpo, plays a major role in pod dehiscence (Blixt, 1972). The finding that the same loci were modified in both subspecies led the current author to suggest that the two taxa may have diverged after these mutations had occurred, probably less than 10,000 years bp (Weeden, 2007).
In the present study I attempted to distinguish between the two hypotheses described above by comparing the DNA sequences of portions of 18 genes in accessions of P. fulvum, and the three subspecies of P. sativum. This approach is very similar to that used by Jing et al. (2007), although their study focused on the genetic diversity within the genus rather than testing between the alternate hypotheses above. Hence, I was able to use their data on an additional 34 sequences to greatly expand the coverage of the genome as well as the number of accessions of P. s. ssp. elatius included. In addition, the data from two studies on histone gene diversity (Zaytseva et al., 2012, 2015) were likewise included in my investigation. The 54 genes are distributed across all seven chromosomes of pea, allowing any significant contribution from P. fulvum to the Abyssinian pea to be recognizable as a high similarity between the gene sequences in that region and those in P. fulvum. In contrast, if the Abyssinian pea represents a divergence from P. s. ssp. sativum after initial domestication changes, virtually all Abyssinian alleles should nest within the diversity of P. s. ssp. sativum.
Materials and Methods
Plant Material
Ten accessions of P. s. ssp. sativum were obtained from the USDA collection at Pullman, WA, United States (Supplementary Table S1). These accessions represented samples from each of the four major clades (a, b, c, and d) from Kwon et al. (2012). In addition, 66 modern pea varieties and two P. s. ssp. elatius accessions, JI 1794 and JI 261 were included (Supplementary Table S1). JI 1794 is a representative of the ‘northern humile’ group that is considered a likely candidate for the wild ancestor of domesticated pea. Crosses between it and domesticated lines are highly fertile. JI 261 produces partly sterile hybrids in crosses with domesticated lines, but also can be crossed with P. fulvum, again producing a hybrid with reduced fertility (Weeden, personal observation). Nine accessions of P. fulvum, representing a wide sample of the genetic diversity in this species, were generously provided by Dr. S. Abbo, The Hebrew University, Rehovot, Israel. Passport data for these can be obtained in Naim-Feil et al. (2017). Two additional samples of P. fulvum from Israel (VIR 6070 and VIR 6071) were kindly supplied by Dr. F. Gorel, then at the Institute of Cytology & Genetics, Novosibirsk, Russia. All P. fulvum samples were subjected to sequencing of four genes: Pur (PURPLE-PODDED), Rb (ADP-glucose phosphorylase), GlyOH (glucan endo-1,3-beta-glucosidase), and Gpic (cytosolic glucose phosphate isomerase). For the remaining genes, only three P. fulvum sequences were obtained, one from those provided by S. Abbo (usually P. fulvum-19) and the two VIR accessions.
Genes Analyzed
The genes examined, the primers used and the size of the fragment sequenced are presented in Supplementary Table S2. In all cases the primers were designed to match conserved portions of the coding sequence of the gene, but the amplified segment included at least one intron.
PCR Conditions
All PCR reactions (20 μl) contained the following: 4 μl Promega 5X PCR buffer, 2.5 mM MgCl2, 0.3 μM of each dNTP, 0.6 units Promega Taq polymerase, and approximately 25 ng pea genomic DNA. In all cases a touchdown procedure was employed, with annealing temperature starting at 63°C and dropping in 1° steps to the final annealing temperature listed on Supplementary Table S2.
DNA Sequencing
PCR products were purified using QIAquick PCR Purification Kit (QIAGen), and DNA concentrations determined after purification using a Nanodrop 2000 (Thermo Scientific). Frozen DNA samples were placed in microtitre plates and the appropriate primer added before freezing and shipping to Sequetech (Mountain View, CA, United States). Sequence data has been deposited at NCBI (accession numbers are given in Supplementary Table S2).
DNA Sequence Analysis
Forward and Reverse sequences were aligned using MAFFT 7.0.1 Ends were trimmed to generate a common start and end nucleotide for each sequence to be compared. Any ambiguous data were hand checked against the original traces for resolution. All sequences for a particular gene fragment were compared using MAFFT 7.0 and checked for regions of questionable reliability. Those sequences with high background or dubious results (about 2% of the sequences) were discarded or a second sequencing performed. Final sequences were compared using the Neighbor Joining function on MAFFT 7.0 with Bootstrapping set at 500 reiterations. The homologous sequence in Medicago truncatula (Medicago genome 4.0 on the Legume Information System at legumeinfo.org) was used as an outgroup for initial analyses, but as these comparisons rarely changed the branching on the resulting cladograms except to greatly compress the branching within the Pisum accessions, only unrooted cladograms are presented in the Results section. Trees for 34 genes from Jing et al. (2007) were obtained from the online supplementary material (three other gene trees gave very poor resolution or lacked a P. s. ssp. abyssinicum sequence and were not used in the current analysis).
Genetic Mapping
SNP or indel polymorphisms for the 18 genes listed in Supplementary Table S2 and the two histone genes studied by Zaytseva et al. (2012, 2015) were used to locate the genes on the pea linkage map for the JI1794 × Slow RIL population (Weeden et al., 1998). This map now includes over 2000 morphological, isozyme, STS, RAPD and SSR markers, with a resolution of <1 cM in most regions. The location of the genes used by Jing et al. (2007) when not provided by the authors, were approximated by identifying the position of their homologs on the Medicago truncatula genomic sequence (Medicago 4.0) and determining the corresponding position on the pea map using flanking markers.
Results
Allele trees for each of the genes studied generated by the neighbor-joining process are presented in Figures 1–3 and Supplementary Figure S1. In order to make the trees as clear as possible, the bulk of the cultivated accessions have been combined into groups (after the neighbor-joining analysis) and labeled Domes1, Domes2, etc., in order of the size of the group. The alleles within each group are identical or nearly so, and there are no significant branches within a group. The Plant Introduction (PI) accessions representing the four major divisions of genetic diversity in P. s. ssp. sativum (Kwon et al., 2012) are included as separate entities on all trees, as are the P. s. ssp. elatius accessions JI 1794 and JI 261, a P. s. ssp. abyssinicum accession, and three or more P. fulvum samples.
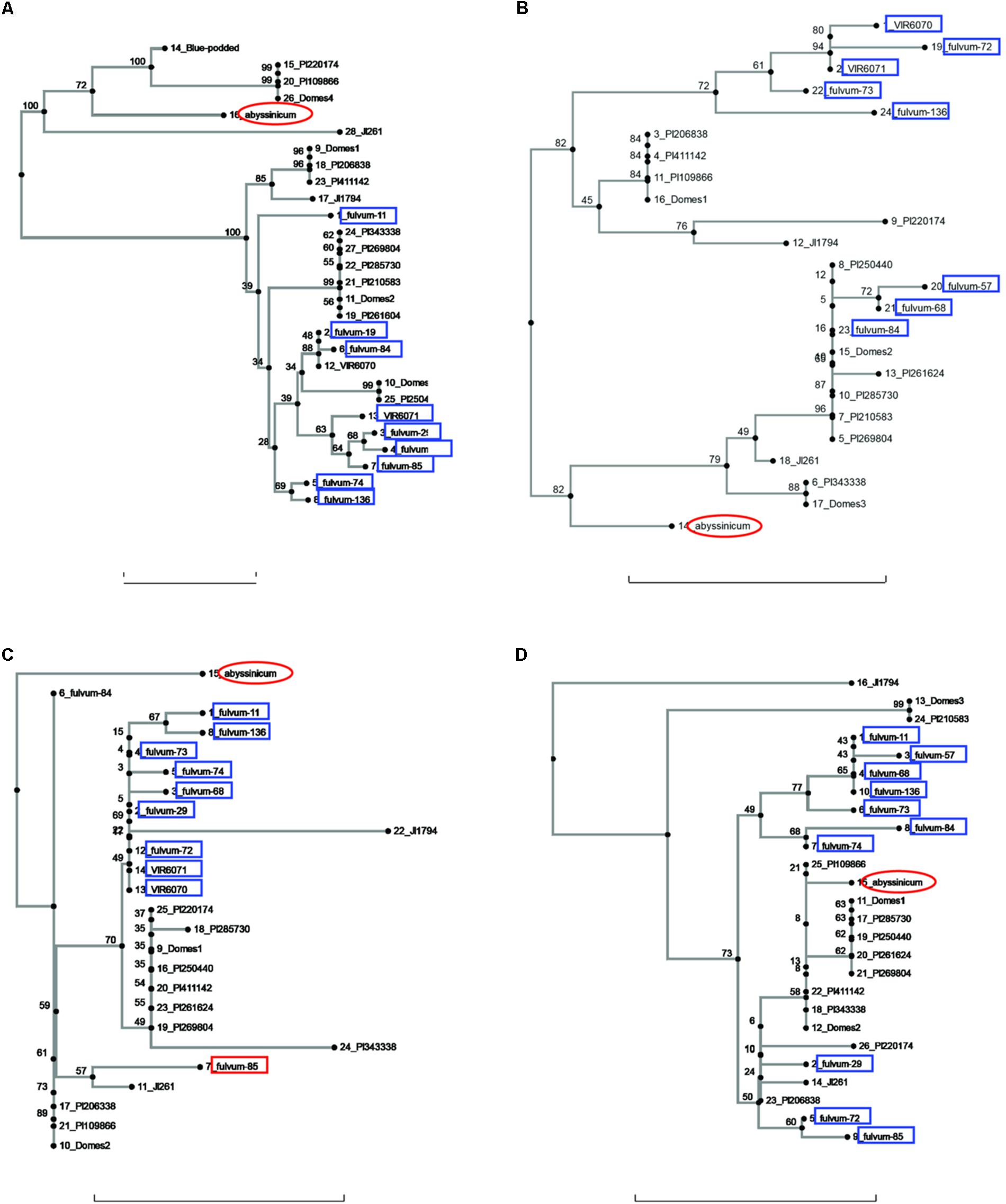
FIGURE 1. Phylogeny trees of the alleles for (A) Pur, (B) Rb, (C) GlyOH, and (D) Gpic. The position of the allele from P. sativum ssp. abyssinicum is circled in red, those for P. fulvum accessions are boxed in blue. The position of each PI accession is presented individually, but those for the modern varieties (except Blue-podded Shelling in (A) are lumped into groups labeled Domes1, Domes2, etc). Bar at base of each cladogram represents a nucleotide substitution rate of 0.01.
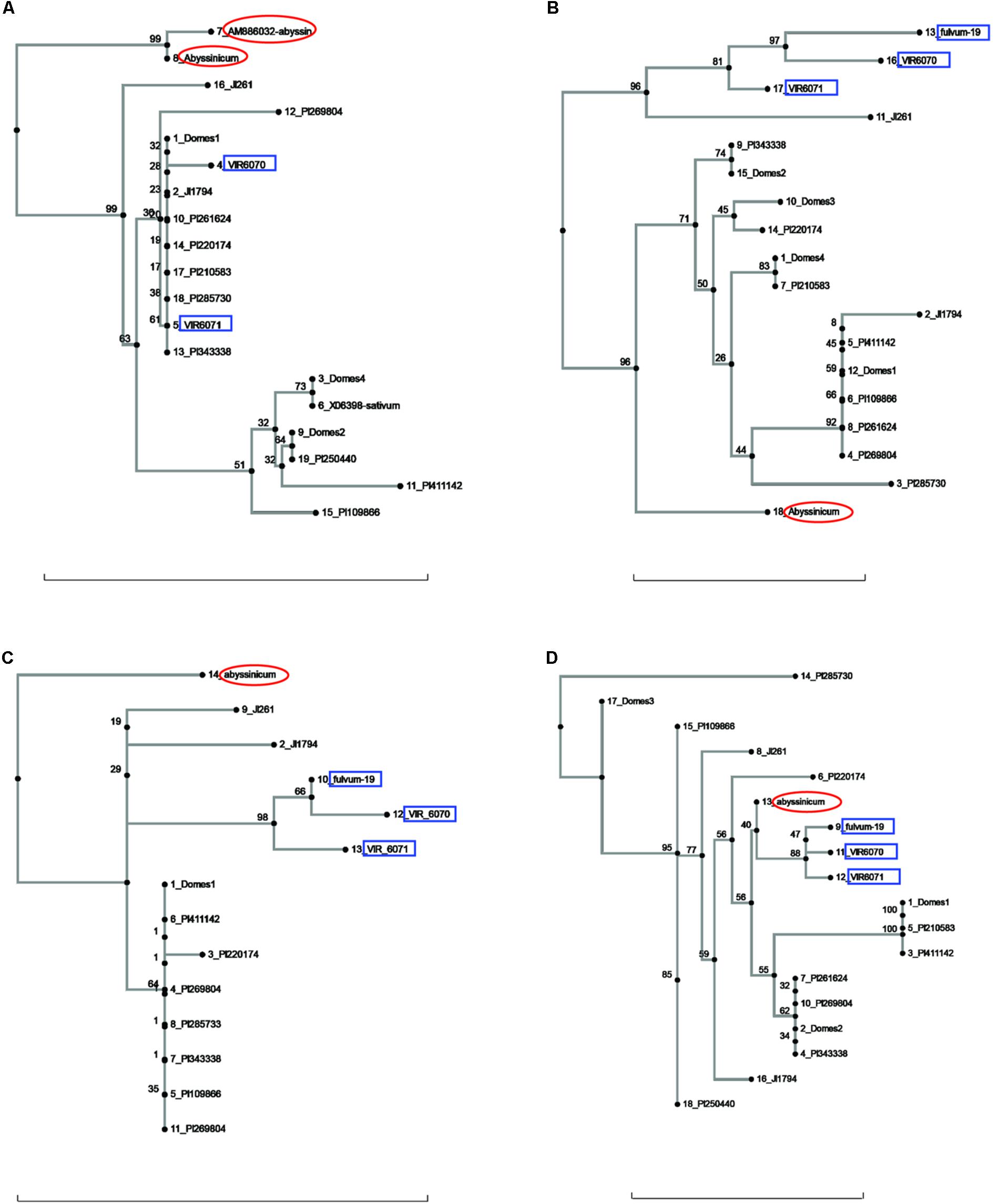
FIGURE 2. Phylogeny trees of the alleles for (A) Cvc, (B) Gib2ox, (C) Er1, and (D) Skdh. The position of the allele from P. sativum ssp. abyssinicum is circled in red, those for P. fulvum accessions are boxed in blue. The position of each PI accession is presented individually, but those for the modern varieties are lumped into groups labeled Domes1, Domes2, etc. Bar at base of each cladogram represents a nucleotide substitution rate of 0.01.
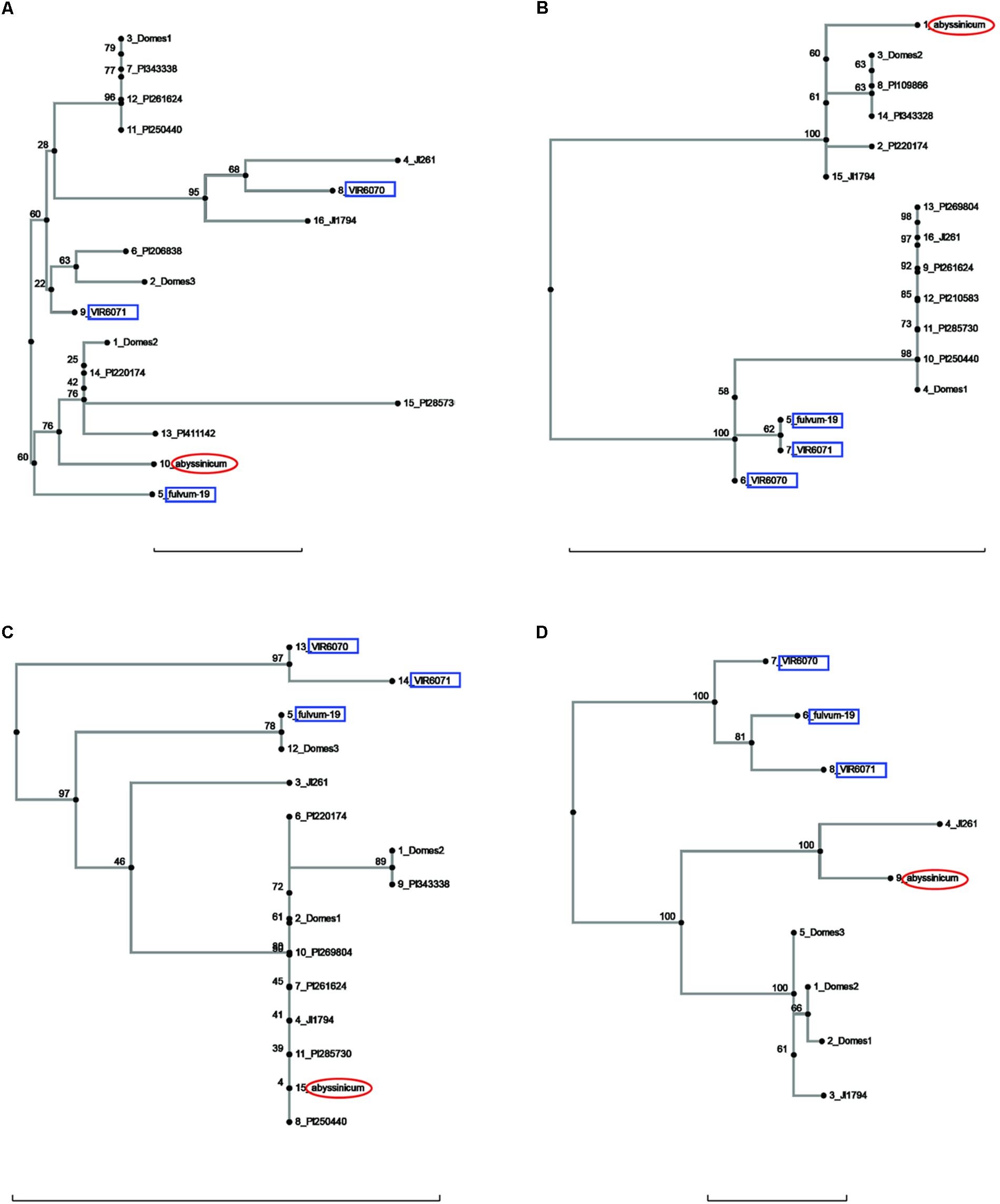
FIGURE 3. Phylogeny trees of the alleles for (A) Peam5, (B) Ntfy, (C) Alat, and (D) Sbm1. The position of the allele from P. sativum ssp. abyssinicum is circled in red, those for P. fulvum accessions are boxed in blue. The position of each PI accession is presented individually, but those for the modern varieties are lumped into groups labeled Domes1, Domes2, etc. Bar at base of each cladogram represents a nucleotide substitution rate of 0.01.
Sequences for the bulk of the P. fulvum samples were obtained only for four genes (Pur, Rb, GlyOH, and Gpic). Trees for each of these genes are shown in Figure 1. In each tree most of the P. fulvum sequences clustered in one region, but in each case there were 2 or 3 sequences that fell outside the cluster. The diverging P. fulvum accessions were different for each gene, although fulvum-84 and -85 were divergent in two cases. For Pur, Rb, and Gpic, the sequence for P. s. ssp. abyssinicum was located in a different part of the tree from any of the P. fulvum sequences, indicating that P. fulvum was not the source of the allele in the Abyssinian pea. For GlyOH, the P. s. ssp. abyssinicum allele was highly divergent, but branched from the tree between fulvum-84 and a mix of other accessions (Figure 1C). In this case the source of the P. s. ssp. abyssinicum allele is ambiguous, with no close affinities among the germplasm tested.
Several other genes gave ambiguous affinities for the P. s. ssp. abyssinicum allele (Figure 2). In the cases of Cvc, Gib2ox, and Er1, the P. s. ssp. abyssinicum allele displayed considerable divergence from all other alleles identified. For convicilin, several additional sequences, including one from P. s. ssp. abyssinicum, were available in the literature. As expected, the P. s. ssp. abyssinicum sequence from NCBI (AM8886032) was very similar to that obtained in the current study, both possessing relatively large unique insertions as well as several SNPs. A P. s. ssp. sativum sequence (X06398) was identical to that found in the Domes4 group (Figure 2A). However, several other sequences in the literature, one for P. fulvum (AM886036), two for P. s. ssp. elatius (AM66034 and AM66035) and one for P. s. ssp. sativum (AM66033) all contained a large unique insertion and formed a different branch on the tree with 100% bootstrap support. These sequences were not included on the final tree because they are most likely from an ortholog. The final gene, Skdh (Figure 2D), branched off the tree between the P. fulvum cluster and several P. s. ssp. sativum groups. However, the tree is compressed in this region, with only 40% bootstrap support for the placement of the P. s. ssp. abyssinicum sequence. Thus, assessment of phylogenetic relationships is difficult. The P. s. ssp. abyssinicum Peam5 allele also was placed between a P. fulvum allele and a P. s. ssp. sativum cluster (Figure 3A), but in this case the three P. fulvum accessions were dispersed on the tree, and it would be difficult to assign much significance to the similarity between the P. s. ssp. abyssinicum and P. fulvum alleles.
The sequences of three genes exhibited a similarity between the P. s. ssp. abyssinicum allele and those found in P. s. ssp. elatius. If the tree for Ntfy is oriented so that the P. fulvum cluster is basal, the branches lead to two clusters of accessions, both of which contain P. s. ssp. elatius and P. s. ssp. sativum taxa (Figure 3B). However, the branch to which the P. s. ssp. abyssinicum sequence belongs contains the ‘northern humile’ P. s. ssp. elatius (JI 1794), the sequence from a landrace from Afghanistan (PI 220174) and several other primitive landraces, suggesting that it might represent a group more closely aligned to the original ancestor of the cultivated pea. The tree for Alat (Figure 3C) shows a similar pattern, although in this case the P. s. ssp. abyssinicum allele groups with nearly all the P. s. ssp. sativum, as well as with JI 1794 and PI 220174. On the other hand, the Sbm1 allele from P. s. ssp. abyssinicum clearly groups with the sequence from the ‘wilder’ P. s. ssp. elatius accession, the northern humile accession grouping with P. s. ssp. sativum (Figure 3D).
The analyses on the remaining six genes all indicate a strong relationship between the P. s. ssp. abyssinicum allele and alleles in P. s. ssp. sativum (Supplementary Figure S1). The trees all have the same general pattern, with the P. fulvum sequences forming a cluster, the P. s. ssp. elatius sequences usually intermediate between P. fulvum and P. s. ssp. sativum, the P. s. ssp. sativum in one or more groups, occasionally with one or two associated with a P. s. ssp. elatius allele, and the P. s. ssp. abyssinicum allele included within or derived from one of the P. s. ssp. sativum clusters. In some cases, e.g., Pgmc and Pao, the P. s. ssp. abyssinicum allele is highly divergent from the P. s. ssp. sativum cluster, but it is also distant from any P. s. ssp. elatius or P. fulvum allele.
In order to try to summarize the affinities of the P. s. ssp. abyssinicum alleles for the various genes, the most closely related sequence(s) to the P. s. ssp. abyssinicum allele is given for each gene in Table 1. Included in this table are the results of 36 comparisons performed previously by others (Jing et al., 2007; Zaytseva et al., 2012, 2015). Of the 54 P. s. ssp. abyssinicum sequences compared, 12 (11) were closest to a P. s. ssp. sativum sequence, 6 (5) were scored as basal to P. s. ssp. sativum (only P. s. ssp. sativum on branches above), 8 (6) as between P. s. ssp. sativum and P. s. ssp. elatius, 20 (16) closest to P. s. ssp. elatius, 5(3) between P. s. ssp. elatius and P. fulvum, 2 (2) between P. s. ssp. sativum and P. fulvum, and 1 (1) P. fulvum (but highly divergent). Numbers in parentheses indicate the total if all those with ‘poor resolution’ marked in the comments column of Table 1 are eliminated from consideration, thereby avoiding most of the ambiguous data. Either set of results indicate that very little of the nuclear genome of P. s. ssp. abyssinicum is derived from P. fulvum.
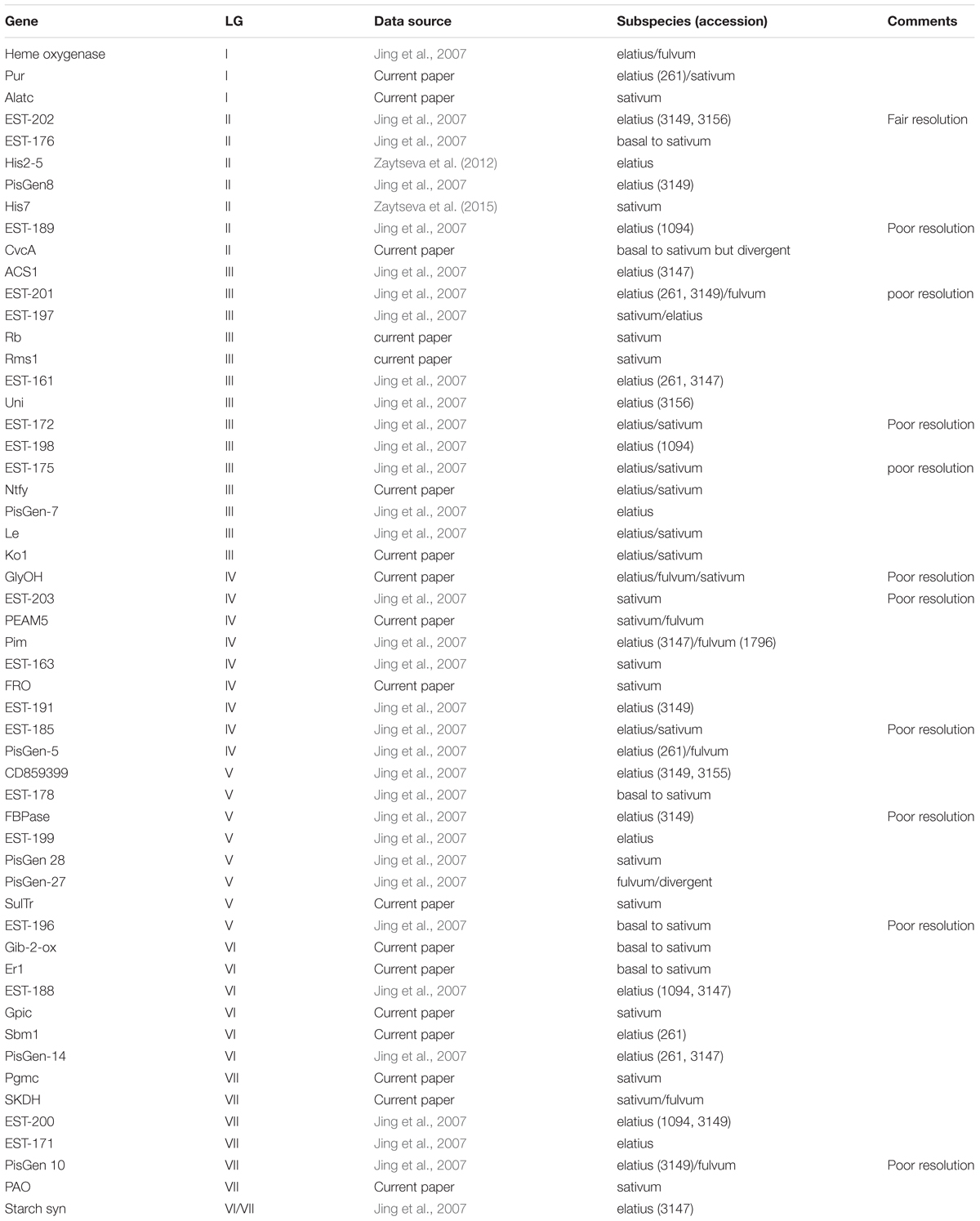
TABLE 1. Pisum taxon (taxa) with the allele displaying the greatest similarity to the allele in the Abyssinian pea.
The genes in Table 1 are listed in the approximate order that they have on the linkage map for P. sativum (there are translocations known in Pisum, but most of the domesticated germplasm possesses the same karyotype). By examining the changes in the P. s. ssp. abyssinicum allele affinities as one moves down the column, we can determine if there are any regions of the P. s. ssp. abyssinicum genome that are primarily derived from P. fulvum or from one of the P. sativum subspecies. Such an exercise reveals a very low association with P. fulvum alleles but indicates approximately even distribution of P. s. ssp. sativum and P. s. ssp. elatius affinities. For LG I, the genes investigated cover only the upper half of the linkage group. No clear pattern is evident, all markers being somewhat ambiguous. LG II is reasonably saturated with markers, and most of the alleles indicate a P. s. ssp. elatius pedigree. The one exception, His7, is near the center of the linkage group, a location not known for any genes critical for domestication. The coverage of LG III is very good, and most of the alleles favor a P. s. ssp. elatius heritage or are ambiguous between P. s. ssp. elatius and P. s. ssp. sativum, although a region on the upper portion (around Rb) has a P. s. ssp. sativum preference. The P. s. ssp. abyssinicum alleles from genes on LG IV appear to favor P. s. ssp. sativum sequences except on its lower end. Affinites on LG V and LG VII are about equal, and those on LG VI slightly favor P. s. ssp. elatius. Such results are consistent with a P. s. ssp. sativum × P. s. ssp. elatius hybrid origin of the Abyssinian pea. The P. s. ssp. elatius accessions that often appear in Table 1 as having the most similar allele to P. s. ssp. abyssinicum are from Turkey. The region from Turkey north into Georgia was identified as potentially important in the domestication of pea (Zaytseva et al., 2017). However, much of the P. s. ssp. elatius germplasm tested by Jing et al. (2007) was originally collected in this country, and the significance of the Turkish connection to the development of the Abyssinian pea is questionable.
Discussion
General Comments Regarding the Study
Sequence analysis of 18 gene fragments confirmed the finding of previous studies that there is considerable genetic diversity present in Pisum and that the alleles are distributed across taxa, presumably by outcrossing and hybridization. The sequences from the accessions of P. fulvum examined here generally formed a cluster, occasionally with one or more accessions outside this cluster and sometimes lying within a P. s. ssp. sativum or P. s. ssp. elatius cluster. It is evident that a sample of 11 P. fulvum accessions is inadequate to reveal all the sequence diversity within that species for the gene segments used in this study, and a sample size of three certainly limits the conclusions that can be drawn, particularly if the sequences do not form a cluster on the tree such as was the case for Peam5 (Figure 3A). The three accessions of P. fulvum chosen as a minimum sample of the species represent those found in preliminary studies to be the most genetically divergent. The conclusion that P. fulvum did not make a significant contribution to the P. s. ssp. abyssinicum nuclear genome is based on the observation that the allele found in P. s. ssp. abyssinicum never clusters with the common allele of P. fulvum whether one considers any of the 18 genes examined in this study or any of the 36 genes that had been subjected to similar analyses previously.
A similar criticism of small sample size could be raised regarding having only two representatives of P. s. ssp. elatius sequences in the current study. Indeed, no conclusion regarding the relationship between P. s. ssp. abyssinicum and P. s. ssp. elatius could have been made had it not been for the availability of the excellent data set of Jing et al. (2007). They included a wide selection of P. s. ssp. elatius accessions in their study, and it is their data that reveal the close relationship between P. s. ssp. abyssinicum alleles and P. s. ssp. elatius alleles in over half the fragments studied. The two studies by Zaytseva et al. (2012, 2015) also included numerous P. fulvum and P. s. ssp. elatius sequences, providing a strong basis for concluding that the P. s. ssp. abyssinicum His2-5 allele is close to those of P. s. ssp. elatius and the His7 allele is close to those of P. s. ssp. sativum.
It may be useful to compare the positions of the P. s. ssp. abyssinicum allele in each cladogram relative to those for PI 220174. This latter accession represents the subgroup a-2 of Kwon et al. (2012) and more significantly is a sample of a large group of landraces grown in the region between Iran and Nepal. This group is clearly a domesticated pea but appears to represent one of the earliest major branches of Pisum germplasm post domestication (Jing et al., 2007). In most of the cladograms, the allele of PI 220174 is identical to an allele found in one of the modern cultivars. In a number of these cases (e.g., Figures 1A, 3A–C) the P. s. ssp. abyssinicum allele is also identical or very similar. In most other cladograms the allele in P. s. ssp. abyssinicum is more distant from the alleles in the modern cultivars than is that of PI 220174, as expected if P. s. ssp. abyssinicum diverged from the modern domesticated germplasm earlier (or was derived from a hybridization with a lineage that had diverged earlier than group a-2). However, in one case, Gpic (Figure 1D), the allele in PI 220174 is placed between the Abyssinian pea allele and those in P. fulvum, contradicting the general pattern. For both the position of PI 220174 and the phylogenetic relationship of the Abyssinian pea with other members of the genus, the general trend appears to be more reliable than results for any particular gene.
The current study contained a large number of accessions of known cultivars, and it is of interest to examine if these cultivars added much to the various gene trees or whether the selected PI accessions would have covered the genetic diversity. A total of six cases can be seen in diagrams (excluding those for Sbm1 and FRO1, for which a complete data set for the PI accessions was not available) where one or more of the pea accessions contained an allele not found in the selected PIs. The unique Pur allele found in Blue Podded Shelling (Figure 1B) is understandable, because none of the PIs had purple pods. In Alat (Figure 3C), Domes4 clustered with a P. fulvum accession, and the nearest PI was several nodes away. This Domes4 group consisted of one winter pea variety, ‘Walechia,’ and it must have had some P. fulvum in its pedigree. The other four cases (Pao, Peam5, Cvc, and Skdh) are less easily explained and indicate how difficult it is to capture all the genetic diversity present in modern cultivars in a small set of PIs. Alternatively, there are many instances on the phylogeny trees where at least one PI accession was present, but none of the modern cultivars possessed a similar allele, reflecting the more limited genetic diversity of modern pea varieties.
Associations With Genes Involved in Domestication
There are two very important changes required for the domestication of a pulse crop: elimination of seed dormancy and elimination of the seed dispersal mechanism (the dehiscent pod). Genetic studies on the dehiscent pod character identified a locus, Dpo, on LG III that has a major influence on pod dehiscence (Blixt, 1972). A recessive allele at this locus is primarily responsible for the indehiscent character of most cultivated pea varieties. If the indehiscent pod trait in the Abyssinian pea was derived from an early P. s. ssp. sativum lineage, one would expect to observe P. s. ssp. sativum alleles in this region of LG III. In Table 1, Dpo should be positioned between Ntfy and PisGen 7. The P. s. ssp. abyssinicum allele for PisGen 7 appears to have come from P. s. ssp. elatius, while the ancestry of the Ntfy allele is ambiguous. Should this region be confirmed to be derived from P. s. ssp. elatius stock, it would imply that Dpo mutated independently in the lineage leading to the Abyssinian pea after the divergence of this lineage from that leading to P. s. ssp. sativum.
The genetic basis of seed dormancy has also been studied in Pisum, but with less success. The primary mechanism for dormancy is known to be a thick, impervious testa, however, few studies have reported specific genes influencing this trait, and those genes identified do not appear to have been involved in the domestication process. Unpublished results from a study of the JI 1794 × Slow RIL in my laboratory have suggested that regions near the upper end of LG III and lower end of LG IV may contain genes influencing this character (Brauner et al., unpublished). As the hard-seeded trait is present in JI 1794 and absent in Slow, these could be the genes modified during the domestication of pea. Examination of these regions in P. s. ssp. abyssinicum (Table 1) reveals the P. s. ssp. sativum allele is present in both the Rb, Rms1 region of LG III and the EST-163, FRO1 region on LG IV. The Rb/Rms1 region precisely corresponds with the QTL for seed dormancy on LG III, whereas that on LG IV would have centered on the FRO1/EST-191 segment. However, should either or both QTL represent a gene from JI 1794 producing a thick testa, the presence of P. s. ssp. sativum-related alleles at these locations in the Abyssinian pea would be consistent with the thinning of the testa occurring before the P. s. ssp. sativum and P. s. ssp. abyssinicum lineages split.
Does the Abyssinian Pea Represent an Independent Domestication of Pisum sativum?
To address this question, one must first define what is meant by “to domesticate.” Some have defined it as the acquisition of a considerable number of traits that we observed in modern day cultivars (Hammer, 1984; Harlan, 1992), while others see it as the modification of one or two characters that allow man to harvest sufficiently more seed than what was initially planted, thereby allowing a society to feed itself while still maintaining a seed stock for planting the next season (Abbo et al., 2012). Another way of defining the process is the loss of natural adaptations of an organism until it becomes dependent on man for survival. For many crops (e.g., maize, dates, cereal grains, and pulses) domestication has involved changes in reproductive structures and seed dispersal mechanisms (Hammer, 1984). Pulse crops become dependent on man for survival when they can no longer disperse their seeds (possess an indehiscent pod) and cannot maintain a seed bank in the soil to allow survival through years of drought or other factors that prevent seed production during 1 or 2 years (loss of seed dormancy).
Using this last definition, the Abyssinian pea would represent an independent domestication only if seed dormancy and pod dehiscence were independently lost in the two lineages, one leading to P. s. ssp sativum and one leading to P. s. ssp. abyssinicum. Convincing evidence pertaining this question remains lacking, but the knowledge that there are several ways to obtain indehiscent pods in Pisum (the dpo mutation, loss of schlerenchyma in pod wall, making the pod wall thicker with callus growths such as seen in some neoplasm {Np} lines), and several avenues to loss of seed dormancy (causing the testa to crack by wrinkling the testa, having the seeds stick to each other, thinning the testa by interfering with phenylpropanoid synthesis) it appears the onus is on those claiming an independent domestication for the Abyssinian pea to demonstrate that the Dpo gene and those genes controlling seed dormancy actually did mutate independently in the P. s. ssp. abyssinicum lineage. The results of the present study provide some evidence for an independent mutation at Dpo, but indicate that the lack of seed dormancy may have preceded the divergence of the two subspecies.
It is of interest to ask why P. s. ssp. sativum has become such a widespread crop while P. s. ssp. abyssinicum has a very restricted range. The answer may lie in the considerable difference in genetic diversity present in the two taxa. The former has a genetic diversity that rivals that present in maize, whereas the Abyssinian pea is nearly monomorphic at all loci that have been tested. The much greater genetic diversity present in P. s. spp. sativum undoubtedly facilitates adaptation of the crop to different environments and modification of the actual crop (not just the dry seeds, but also leaves, tendrils, pods, and flowers are known to be eaten). The difficulty in crossing the two subspecies probably prevented breeder/growers in Ethiopia and Yemen from moving traits available in the more widespread domesticate into the Abyssinian pea. Hence, it is now much more appropriate to consider this latter domesticate as a potential, albeit limited source of novel traits for pea improvement.
The Problem of Highly Unique Alleles Versus Narrow Genetic Diversity in P. s. ssp. abyssinicum
The Abyssinian pea presents an interesting dilemma when trying to explain its genetic characteristics. Many of its alleles are highly diverged from any allele yet identified in the other Pisum taxa, suggesting a very ancient divergence from these other groups, yet the extremely low genetic diversity of P. s. ssp. abyssinicum, particularly relative to the other Pisum taxa, suggests a very recent origin or genetic bottleneck. The unique alleles present in Abyssinian pea are strong evidence against a recent origin (<5000 years bp), whether hybrid or not, because we should still find those alleles in other accessions at the numerous germplasm collections throughout the world. We are left with the genetic bottleneck explanation, with isolation and strong selection being the obvious factors. However, if P. s. ssp. abyssinicum was widespread at one point, what was its distribution and why, based on how alleles seem to be freely exchanged in the rest of the genus, don’t we see some of the P. s. ssp. abyssinicum alleles appearing rarely in other accessions?
Taxonomic Considerations
The taxonomic treatment of a related group of organisms can change as new information is obtained and ideas of what constitutes a particular taxonomic level are modified. Whether only two (Smartt, 1984) or up to six species (Govorov, 1937) should be recognized within the genus Pisum has been a moot point since the defining of the genus. Most of the problem exists in trying to further divide the ‘wild’ material referred to as P. s. ssp. elatius (sensu latu) into species. This germplasm is a diverse assemblage of types, yet the variation appears continuous within this complex (Vershinin et al., 2003; Jing et al., 2007; Kosterin and Bogdanova, 2008; Smykal et al., 2012; Zaytseva et al., 2012, 2015, 2017) and certainly intergrades with P. s. ssp. sativum. The analysis presented in this paper is consistent with treating ‘sativum’ and ‘elatius’ as a panmictic complex and demonstrates that the P. s. ssp. abyssinicum germplasm was derived from this complex. If we are not willing or able to define a P. elatius at the species level, then what justification is there to define a P. abyssinicum?
Conclusion
The comparison of alleles at 54 genes distributed throughout the pea nuclear genome demonstrated that Pisum sativum ssp. abyssinicum alleles displayed a much closer relationship to alleles in either P. s. ssp. sativum or P. s. ssp. elatius than to those in P. fulvum. The possibility that the Abyssinian pea was derived from a P. sativum × P. fulvum hybridization within the last 10,000 years is thus rejected. The possibility remains that the Abyssinian pea is a product of a P. s. ssp. sativum × P. s. ssp. elatius hybridization, with the most likely source of P. s. ssp. elatius germplasm being Asia Minor. The question whether the Abyssinian pea represents an independent domestication event remains debatable. The indehiscent pod trait required for domestication may have arisen independently in this lineage based on the P. s. ssp. elatius-related alleles in the Dpo region of the P. s. ssp. abyssinicum genome. However, little evidence is available to determine if the lack of seed dormancy in the Abyssinian pea evolved after this lineage separated from that leading to P. s. ssp. sativum.
Author Contributions
The author performed the research and wrote the manuscript.
Funding
This research was funded in part by a grant from the USA Dry Pea and Lentil Commission.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author would like to express his sincere appreciation to Clare Coyne and the staff at the USDA Western Regional Plant Introduction Station for supplying Pea Plant Introduction lines, to Shahal Abbo for the P. fulvum lines and stimulating discussions regarding the domestication of pea, and to T. H. N. Ellis and Mike Ambrose for pea germplasm and their leadership in investigations of Pisum genetic diversity. Thanks are also extended to Aish Kothari and Wendy Rivero for their excellent technical assistance in obtaining the data and to the USA Pea and Lentil Commission for their extended support of my research and specifically their contribution to this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00515/full#supplementary-material
FIGURE S1 | Phylogeny trees for the six genes that indicate a close relationship between the allele from the Abyssinian pea (circled in red) and alleles found in P. sativum ssp. sativum. Alleles from P. fulvum accessions are boxed in blue. The position of each PI accession is presented individually, but those for the modern varieties are lumped into groups labeled Domes1, Domes2, etc. Bar at base of each cladogram represents a nucleotide substitution rate of 0.01.
Footnotes
References
Abbo, S., Lev-Yadun, S., and Gopher, A. (2012). Plant domestication and crop evolution in the Near East: on events and Process. Crit. Rev. Plant Sci. 31, 241–257. doi: 10.1080/07352689.2011.645428
Berger, A. (1928). “Systematic botany of peas and their allies. Peas of New York,” in The Vegetables of New York, ed. U Hedrick (Albany, NY: J.B. Lyon Company), 1–132.
Bogdanova, V., Zasytsva, O., Mglinets, A., Shatskaya, N., Kosterin, O., and Vasiliev, G. (2015). Nuclear-cytoplasmic conflict in pea (Pisum sativum L.) is associated with nuclear and plastidic candidate genes encoding acetyl-CVoA carboxylase subunits. PLoS One 10:e0119835. doi: 10.1371/journal.pone.0119835
Errico, A., Conicella, C., and Venora, G. (1991). Karyotype studies on Pisum fulvum and Pisum sativum, using a chromosome image analysis system. Genome 34, 105–108. doi: 10.1139/g91-017
Govorov, L. (1937). “Pisum,” in Flora of Cultivated Plants. IV. Grain Leguminosae, eds N. Vavilov and E. Wulff (Moscow: State Agricultural Publishing Company), 231–336.
Jing, R., Johnson, R., Seres, A., Kiss, G., Ambrose, M., Knox, M., et al. (2007). Gene-based sequence diversity analysis of field pea (Pisum). Genetics 177, 2263–2275. doi: 10.1534/genetics.107.081323
Kosterin, O. (2017). Abyssinian pea (Lathyrus schaeferi Kosterin nom. Nov. pro Pisum abyssinicum A. Br.) is a problematic taxon. Vavilov J. Genet. Breed. 21, 158–169. doi: 10.18699/VJ17.234
Kosterin, O., and Bogdanova, V. (2008). Relationship of wild and cultivated forms of Pisum L. as inferred from an analysis of three markers, of the plastid, mitochondrial and nuclear genomes. Genet. Resour. Crop Evol. 55, 735–755. doi: 10.1007/s10722-007-9281-y
Kwon, S.-J., Brown, A., Hu, J., McGee, R., Watt, C., Kisha, T., et al. (2012). Genetic diversity, population structure and genome-wide marker-trait association analysis emphasizing seed nutrients of the USDCA pea (Pisum sativum L.) core collection. Genes Genomics 34, 305–320. doi: 10.1007/s13258-011-0213-z
Naim-Feil, E., Toran, M., Aubert, G., Rubinstein, M., Rosen, A., Eshed, R., et al. (2017). Drought response and genetic diversity in Pisum fulvum, a wild relative of domesticated pea. Crop Sci. 57, 1145–1159. doi: 10.2135/cropsci2016.10.0880
Smartt, B. (1984). Evolution of grain legumes. I. Mediterranean pulses. Exp. Agric. 20, 275–296. doi: 10.1017/S0014479700017968
Smykal, P., Kenicer, G., Flavell, A., Corander, J., Kosterin, O., Redden, R., et al. (2012). Phylogeny, phylogeography and genetic diversity of the Pisum genus. Plant Genet. Resour. 9, 4–18. doi: 10.1017/thg.2012.108
Vershinin, A., Allnutt, T., Knox, M., Ambrose, M., and Ellis, T. (2003). Transposable elements reveal the impact of introgression, rather than transposition, in Pisum diversity, evolution, and domestication. Mol. Biol. Evol. 20, 2067–2075. doi: 10.1093/molbev/msg220
Weeden, N. (2007). Genetic changes accompanying the domestication of Pisum sativum: Is there a common genetic basis to the ‘domestication syndrome’ for legumes? Ann. Bot. 100, 1017–1025. doi: 10.1093/aob/mcm122
Weeden, N., and Wolko, B. (2001). Allozyme analysis of Pisum sativum ssp. abyssinicum and the development of a genotype definition for this subspecies. Pisum Genet. 33, 21–25.
Weeden, N. F., Brauner, S., and Przyborowski, J. A. (2002). Genetic analysis of pod dehiscence in pea (Pisum sativum L.). Cell. Mol. Biol. Lett. 7, 657–663.
Weeden, N., Ellis, N., Timmerman-Vaughan, G., Swiecicki, W., Rozov, S., and Berdnikov, V. (1998). A consensus linkage map for Pisum sativum. Pisum Genet. 30, 1–4.
Zaytseva, O., Bogdanova, V., and Kosterin, O. (2012). Phylogenetic reconstruction at the species and intraspecies levels in the genus Pisum (L.) (peas) using a histone H1 gene. Gene 504, 192–202. doi: 10.1016/j.gene.2012.05.026
Zaytseva, O., Bogdanova, V., Mglinets, A., and Kosterin, O. (2017). Refinement of the collection of wild peas (Pisum L.) and search for the area of pea domestication with a deletion in the plastidic psbA-trnH spacer. Genet. Resour. Crop Evol. 64, 1417–1430. doi: 10.1007/s10722-016-0446-4
Keywords: Pisum sativum, domestication, gene trees, genetic diversity, Pisum fulvum
Citation: Weeden NF (2018) Domestication of Pea (Pisum sativum L.): The Case of the Abyssinian Pea. Front. Plant Sci. 9:515. doi: 10.3389/fpls.2018.00515
Received: 17 January 2018; Accepted: 04 April 2018;
Published: 18 April 2018.
Edited by:
Shahal Abbo, The Hebrew University of Jerusalem, IsraelReviewed by:
Matthew Nicholas Nelson, Royal Botanic Gardens, Kew, United KingdomTom Warkentin, University of Saskatchewan, Canada
Judith Lichtenzveig, Curtin University, Australia
Copyright © 2018 Weeden. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Norman F. Weeden, nweeden@montana.edu
 Norman F. Weeden
Norman F. Weeden