- Department of Biological Sciences, University of North Carolina at Charlotte, Charlotte, NC, United States
There is a considerable demand for crop improvement, especially considering the increasing growth of world population, continuing climatic fluctuations, and rapidly evolving plant pests and pathogens. Crop wild relatives hold great potential in providing beneficial alleles for crop improvement. Wild soybean, Glycine soja (Siebold & Zucc.), the wild ancestor to the domesticated soybean (Glycine max (L.) Merr.), harbors a high level of genetic variation. Research on G. soja has been largely devoted to understanding the domestication history of the soybean, while little effort has been made to explore its genetic diversity for crop improvement. High genomic diversity and expanded traits make G. soja populations an excellent source for soybean improvement. This review summarizes recent successful research examples of applying wild soybeans in dissecting the genetic basis of various traits, with a focus on abiotic/biotic stress tolerance and resistance. We also discuss the limitations of using G. soja. Perspective future research is proposed, including the application of advanced biotechnology and emerging genomic data to further utilize the wild soybean to counterbalance the rising demand for superior crops. We proposed there is an urgent need for international collaboration on germplasm collection, resource sharing, and conservation. We hope to use the wild soybean as an example to promote the exploration and use of wild resources for crop improvement in order to meet future food requirements.
Soybean and Wild Soybean
Cultivated soybean (Glycine max (L.) Merr.) is an economically important crop grown world-wide with diverse uses in oil and protein consumption for human and livestock, as well as feedstock for biofuel production. Cultivated soybean produces two-thirds of the world's protein meal, and is the leading producer of oilseed (Oecd Food and Nations AOOTU, 2016). With the growing world population, the yield for soybean is at a current deficit of 1.2% annual production per year (Ray et al., 2013). Although soybean yield per acre has increased by approximately 40% in the last quarter century and production has more than doubled (1990–2015) (USDA, 2016), current soybean yield potential is restricted by the narrow genetic variation in G. max, hindering the potential for breeding soybean varieties with high environmental stress tolerance and resistance traits. These concerns regarding the current cultivars are increasing because of the ecological changes caused by climate fluctuations and other factors such as expanded drought or saline environments. Meanwhile, the rapid evolution of pests and pathogen is posing difficult challenges to soybean production. Because of the aforementioned factors, there is an urgent need to use the untapped genetic resources from the wild relatives of cultivated soybean to improve soybean production.
Wild soybean, Glycine soja (Siebold & Zucc.), is the wild ancestor of the domesticated soybean, G. max. Glycine soja is native to East Asia with a broad geographic range, from East Russia to South China, and the species can grow in diverse habitats. Though G. soja and G. max are different in many phenotypic characteristics (Figure 1), they have the same number of chromosomes (2n = 40), exhibit normal meiotic chromosome pairing, and are cross-compatible (Carter et al., 2004). Also, G. soja was found to harbor a high level of genetic diversity, which might have been lost in G. max during their domestication and improvement (Hyten et al., 2006). Considering the higher level of genetic diversity retained in G. soja, as well as its adaptations to harsh environments, G. soja holds great potential to improve its agriculturally important domesticated relative, beyond what is currently known (Stupar, 2010; Qiu et al., 2013; Qi et al., 2014; Zhang et al., 2017d). Research on G. soja has been largely devoted to understanding the domestication history of the soybean, with comparatively little effort made to use it as a genetic reservoir for soybean improvement (Kim et al., 2011; Qi et al., 2014; Zhang et al., 2016). In this review, we will discuss genomic diversity and the current research in G. soja, and we present a potential step forward in our application of G. soja to improve G. max.

Figure 1. Illustration of phenotypic comparison between G. max (A) and G. soja (B) (Photo taken by J. Kofsky).
Domestication History
Domestication of G. soja is reported to have occurred around 6,000-9,000 years ago in regions along the Yellow River or Huang-Huai Valley in Central China (Carter et al., 2004; Li et al., 2010; Han et al., 2016), resulting in landraces of G. max, and with further selection the modern (elite) cultivated material. However, the history of G. soja in relation to G. max is far more complex, with varying contradictory hypotheses. It is likely that the domestication process happened over a long period of time, allowing for frequent introgressions between the wild and cultivated populations during this time (Wang and Li, 2013; Qiu et al., 2014). Although an opposing study of candidate domestication regions in Korean wild soybeans suggests a single selective sweep, detecting no evidence for multiple domestication events in East Asia (Chung et al., 2014). A summation of the contradictory hypotheses of soybean origin and domestication was recently presented in a review of the domestication history (Sedivy et al., 2017), in which three overarching hypotheses were provided: the single origin hypothesis, the multiple origin hypothesis, and the complex hypothesis. The single origin hypothesis states that G. max was diverged from G. soja from a single domestication event in Central China, no earlier than 9,000 years ago, supported by the observation that all the selected domesticated soybeans were clustered together by analyzing whole-genome SNPs of 302 wild, landrace and cultivated soybeans (Zhou et al., 2015b). The multiple origin hypothesis states that G. max was domesticated from G. soja during multiple events between 5,000 and 9,000 years ago. The complex hypothesis incorporates the results from two recent studies (Kim et al., 2010; Li et al., 2014), with a G. soja/G. max complex first diverging before multiple domestication events. The estimated age of the G. soja/G. max complex is 0.27 million years ago (MYA) (Kim et al., 2010) by whole genome comparison of one wild soybean ecotype to one soybean cultivar, or 0.8 MYA (Li et al., 2014) by pan-genome comparison of 7 wild soybean ecotypes. In this last hypothesis, the domestication would have stemmed from an already diverged G. soja/G. max complex (Sedivy et al., 2017). Individuals from either soja or max subpopulation were closely clustered based on their geographic origins (Zhang et al., 2015, 2016). A possible explanation could be that the early-domesticated G. soja or G. soja/G. max complex spread from China to Korean and Japan, and subsequently underwent varying degrees of domestication to meet local needs. Nevertheless, it is widely accepted that G. max was created from G. soja or G. soja/G. max complex through a long, slow, and complex domestication process by countless independent efforts (Sedivy et al., 2017).
Advantages of Using Wild Soybean
G. soja gene pool is indisputably more diverse than G. max due to artificial selection during domestication and continued loss due to modern breeding practices. A comparison of 102 gene sequences from 26 G. soja, 52 landraces, 17 North American ancestors, and 25 elite cultivar isolates suggested that the most significant loss in genetic diversity occurred during the domestication bottleneck, and a secondary loss of diversity during modern breeding. The domestication bottleneck has resulted in an 81% loss of rare alleles, 60% gene allele frequency change, and almost halving the nucleotide diversity (π) from G. soja (π = 2.47 × 10−3) to landrace (π = 1.47 × 10−3). After domestication, intense selection toward the elite cultivars (π = 1.17 x 10−3) resulted in an additional loss of 23% nucleotide diversity (π), and a 21% loss of rare alleles (Hyten et al., 2006). These artificial selection processes resulted in morphological differences of many agriculturally-important traits between G. max and G. soja, such as pod shattering resistance (Dong et al., 2014), determinate growth habit (Tian et al., 2010), and seed-related traits (Zhou et al., 2015a). Directional selection during modern soybean breeding practice also reduced the genetic diversity surrounding the regions conferring these agriculturally important traits, known as selective sweep. A recent genome-wide sequencing analysis showed that half of the resistance-related genes/loci in G. soja were not found in landraces or the domesticated soybean (Zhou et al., 2015b). Thus, modern breeding practices have further shrunk the gene pool by selecting from a small fraction of landraces to produce elite cultivars (Song et al., 2015). These observations suggest that the morphological characters of the elite cultivars are genetically controlled by a combined effect of these genomic regions that were selectively swept or lost during domestication and artificial selection. It is possible that reduced genetic diversity in elite cultivars can be significantly increased by introgression of the favorable variation in G. soja in modern breeding program, while additional efforts are needed to balance the selection of agriculturally and adaptively important traits.
Natural populations of G. soja were strongly influenced by climatic fluctuations. The Quaternary glaciation around 2 MYA resulted in the shrinking or distinction of many plant populations (Qiu et al., 2011). Bottlenecks of most natural populations resulted in loss of genetic diversity within populations and differentiation among populations by genetic drift and environmental selection (Guo et al., 2012; Li et al., 2014; Leamy et al., 2016). It is likely that G. soja populations then expanded rapidly through Asia (Figure 2A) after glaciation due to its high adaptability (Sakai et al., 2003; Wang et al., 2016). This evolutionary history accounts for the wide range of genetic and phenotypic diversity found among G. soja populations. The population size of G. soja is expanding, while G. max has been relatively constant (Wang et al., 2015), with correlated disparity in genetic diversity seen in Figure 2B. Adaptations to local environments has enabled G. soja to develop sophisticated mechanisms to tolerate many biotic and abiotic stressors. Many of the resistance-causal genes have been found to exist in manners of copy number variations (CNVs), such as rhg1 conferring soybean cyst nematode (SCN) resistance (Cook et al., 2012). Environmental/niche isolation played a stronger role than isolation by geographic distance in the genetic differentiation of G. soja, suggesting the presence of many environmentally tailored adaptations in natural populations (He et al., 2016; Leamy et al., 2016). Lee et al. (2015) found that the origin of the tandem duplication of the 31.2-kb segment at the rhg1 locus (Cook et al., 2012), a major QTL conferring SCN resistance, occurred prior to the divergence of G. max and G. soja or the formation of the ancestor of G. max/G. soja complex, implying that the copy number variation in rhg1 evolved from a G. soja population in East Asia, where the SCN populations most likely originated (Tylka and Marett, 2014).
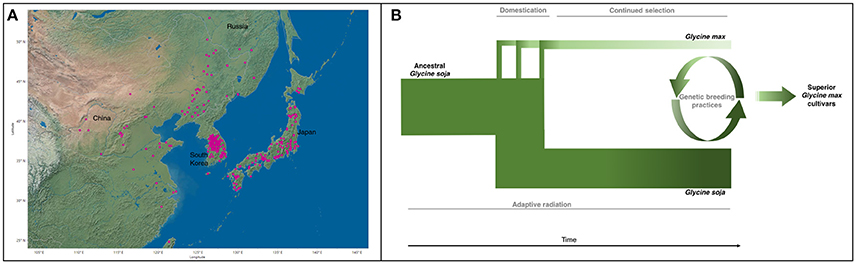
Figure 2. (A) Geographic distribution of all G. soja ecotypes with identified locations (Geographic location data were retrieved from URL: https://training.ars-grin.gov/gringlobal/search.aspx). (B) G. max diverged from ancestral G. soja as a result of multiple domestication events. G. max then underwent continued artificial selection for traits of agronomic importance, further reducing the genetic diversity found in G. max. During this time, G. soja continued to adapt to its various environments, maintaining and potentially increasing genetic variability. Genetic breeding practices incorporate select components from the G. soja gene pool to improve modern cultivars, developing superior G. max cultivars. Width and color represent genetic diversity (Sedivy et al., 2017).
The concept of revitalizing the cultivated gene pool with a wild progenitor species has been well applied in many crops. Plant domestication limits the range of obtainable phenotypes by artificial selection, often removing unforeseen traits of interest down the line. For example, during the 1970s, commercial corn crops (Zea mays L.) were devastated by blight affecting as much as 50% of the yield in the United States, until blight-resistant alleles from the wild relative (Mexican maize, Tripascum dactyloides L.) were introduced into the domesticated population (Maxted and Kell, 2009). Examples of the use of crop wild relatives to improve abiotic stress tolerance, biotic stress resistance, and agronomic traits of cultivated crops has been seen in major crops: rice, barley, wheat, tomato, potato, and peanut (Zhang et al., 2017c). The development of superior soybean cultivars by incorporating genes/alleles from G. soja (Figures 2B, 3) is a promising and environmentally friendly solution for soybean improvement, moderating or negating the need for pesticides and fertilizers.
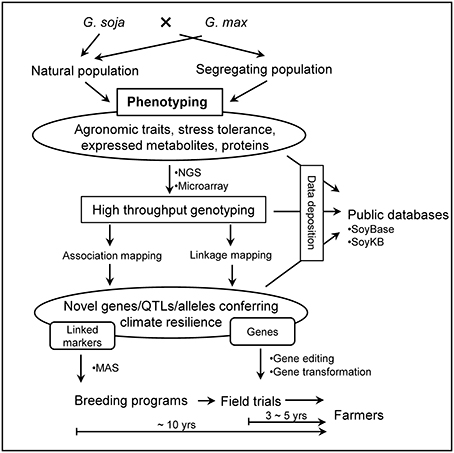
Figure 3. Research pipeline of crop improvement of G. max using G. soja (Fernie and Schauer, 2009).
Genomic Resources and Discoveries from G. SOJA
In 2010, a G. soja ecotype (IT182932) and 17 additional ecotypes were resequenced and assembled using G. max as a reference, serving as the first reported whole-genome sequences of G. soja (Kim et al., 2010; Lam et al., 2010). Structural characteristics of the genome were reported 2 years later, creating a physical map, useful for investigating genome architectural differences between G. max and G. soja (Ha et al., 2012). The development of the SoySNP50K iSelect Illumina BeadChip and the release of up to 50,000 genome-wide single nucleotide polymorphism (SNPs) in 2013 further promoted the studies and applications of wild soybean (Song et al., 2013). Information regarding soybean research, including all SNPs for over 20,000 of USDA soybean accessions, is publically available at the SoyBase database (https://soybase.org/), serving as an excellent resource for various studies, such as genomic diversity and gene discovery in wild soybeans.
Research on G. soja thus far has mostly focused on the understanding of domestication and evolutionary history of G. soja transition into cultivated crop. Whereas, few studies have made full use of this wild genetic reservoir. Although the utilization of the genetic diversity contained in G. soja is lacking, the potential of G. soja in soybean improvement has been gradually recognized. Table 1 summarizes the recent research and findings using G. soja toward soybean improvement. We discuss some representative examples in the following paragraphs and provide an in-depth consideration of these findings. G. soja has also been used in many studies as a negative control to identify genes for traits present in G. max, for example in the study of seed weight or protein content (Maughan et al., 1996; Zhou et al., 2015b). In these cases, G. soja gene pool that was not used as a direct source of resistance, tolerance, or improvement is not presented here as such.
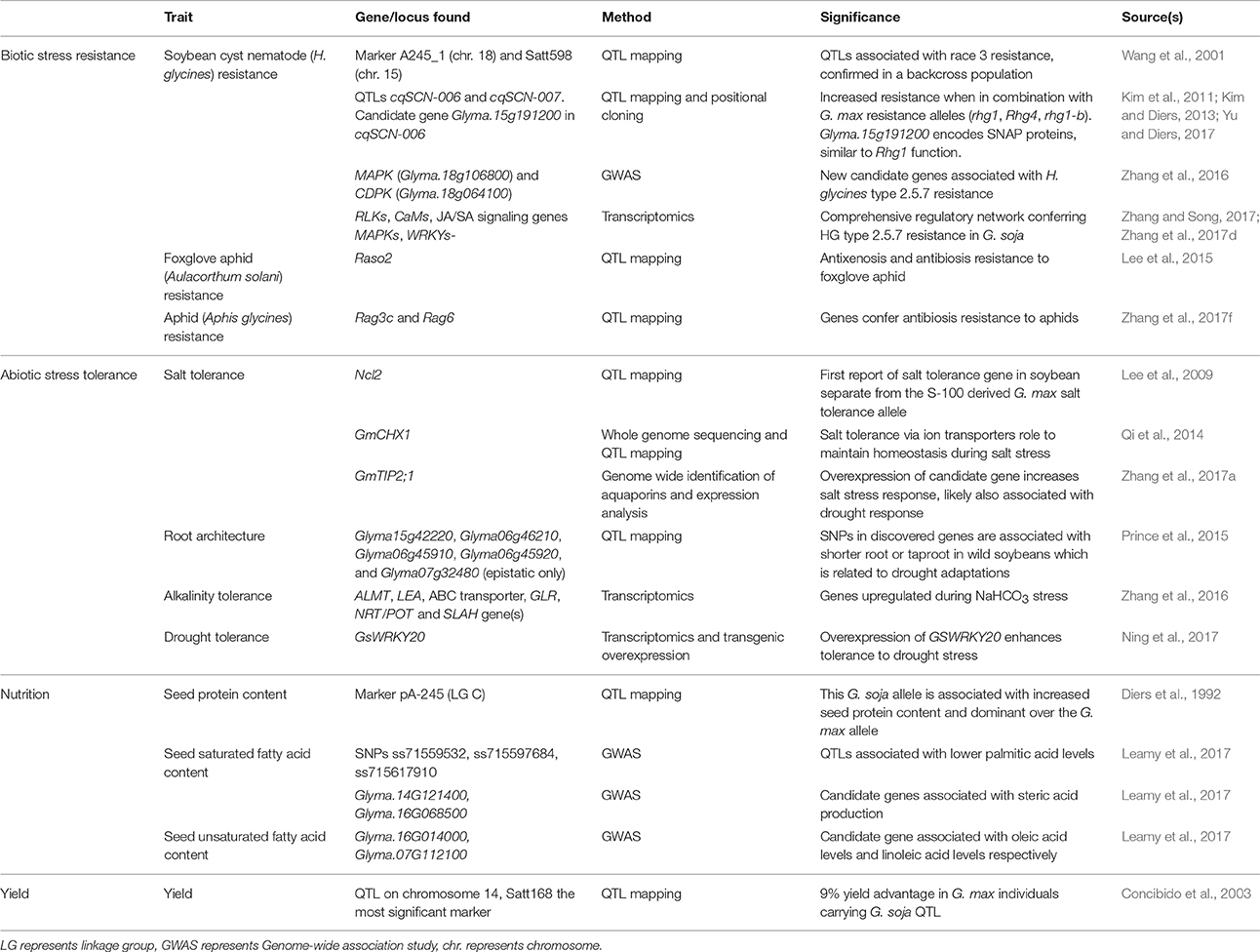
Table 1. Findings from G. soja including biotic stress resistance, abiotic stress tolerance, nutrition, and yield related traits.
Biotic Stress Resistance
The soybean aphid (Aphis glycines) is native to Asia and was introduced to the United States in 2000. Infestations by A. glycines can directly affect soybean biomass and yield, and indirectly affect yield with the transmission of the soybean mosaic virus (SMV). The screening of G. soja for soybean aphid resistance has revealed that three G. soja genotypes showed resistance to the soybean aphid (Hesler, 2013). The follow-up studies identified two new QTLs, Rag3c and Rag6, related to aphid resistance using linkage mapping (Zhang et al., 2017e,f). Rag3c explains 12.5–22.9%, while Rag6 explains 19.5–46.4%, of the phenotypic variance in different trials. These novel aphid-resistance gene(s) derived from G. soja are proven valuable in the development of aphid-resistant soybean cultivars (Zhang et al., 2017e). Map-based cloning of the two QTLs and the functional verification of candidate genes are needed to uncover the underlying mechanism. It is likely that the candidate genes within the QTL regions encode canonical nucleotide-binding site leucine-rich repeat (NBS-LRR)-containing proteins, which belong to a resistance (R) protein that has been found to be resistant to potato aphid (Rossi et al., 1998).
The first report of the genetic identification of foxglove aphid (Aulacorthum solani) resistance gene, Raso2, in G. soja was in 2015 (Lee et al., 2015). Up to this point, five other aphid resistance genes had been identified, with all being mapped from G. max cultivars. Raso2 differs from those isolated from G. max, with strong antixenosis and antibiosis responses to the foxglove aphid (Lee et al., 2015), which suggests a novel mechanism of A. solani resistance exists in G. soja and merits further functional dissection.
Soybean cyst nematode (SCN) is the most devastating pest in cultivated soybeans. Breeding SCN-resistant soybean cultivars in the United States are dependent on very limited genetic sources. In 2005, 94% of the SCN-resistant cultivars grown in Illinois were sourced from a G. max accession (PI88788) that has shown reduced resistance to the majority of SCN populations found in the soil (Kim et al., 2011). Nematodes have demonstrated the ability to adapt and overcome resistance in G. max, and therefore new sources of SCN resistance are needed. G. soja has been shown to exhibit varying resistance to different SCN populations currently affecting crops in the United States (Kim et al., 2011; Zhang et al., 2016). A large portion of G. soja accessions is yet to be screened for resistance, and the investigation into the genetic mechanisms conferring resistance is still underway. So far, several studies have reported loci and candidate genes that might function differently from those thus far detected in G. max conferring SCN resistance using linkage mapping (Kim et al., 2011) and genome-wide association study (Zhang et al., 2016, 2017d), suggesting species-specific resistance mechanisms in G. soja. For example, two SCN-resistance QTLs (cqSCN-006 on chr.15 and cqSCN-007 on chr.18) identified in G. soja genotype PI468916 differ from the two major SCN resistance QTLs (rhg1 on chr.18 or Rhg4 on chr.8) identified in G. max (Kim et al., 2011), providing an alternative or additional sources of SCN resistance other than PI88788. On-going efforts in fine mapping of the underlying QTLs have identified SCN resistant candidate genes including SNAP gene Glyma.15g191200 (Yu and Diers, 2017). Further molecular cloning experiments are expected to be made to increase our understanding of defense mechansims in G. soja.
On the other hand, extensive research on different races of SCN may increase our understanding of SCN resistance. Currently, the majority of studies has been mostly focused on resistance mechanisms to SCN HG type 0 (race3), which is prevalent in the central United States. However, few studies focused on other less-studied races, such as SCN HG type 2.5.7 (known as race 5), which is prevalent in the southeast United States. A recent study identified G. soja genotypes resistant to SCN HG type 2.5.7 (Zhang et al., 2016) and has identified novel QTLs in G. soja relating to HG type 2.5.7 resistance using genome-wide association study (Zhang et al., 2016). Further analysis of one of the resistant genotypes with RNA-seq data revealed a biologically-sound defense regulatory network involved in SCN resistance (Zhang and Song, 2017; Zhang et al., 2017b). Given the variability in virulence and rapid evolution of SCN populations, it is critical to develop soybean cultivars with broad-spectrum resistance to multiple SCN types. This can be achieved by identifying new sources of resistance and applying genome editing or gene stacking to develop soybean lines with broad resistance to SCN.
Abiotic Stress Tolerance
Soil salinity is a growing challenge for today's crops. As the most important protein-providing crop, cultivated soybean is salt sensitive and has been cultured in well-plowed soil. The discovery of salt tolerance in G. soja was especially significant and has led to the first investigation of this trait in 1997, but only weak correlation has been found between the genetic markers chosen and the salt tolerance traits (Hu and Wang, 1997). Further investigation using randomly amplified polymorphism DNA markers revealed associations between 6 markers (OPF05-213, OPF19-4361, OPF19-1727, OPF19-14000-, OPF19-700, OPH02-1350) and salt tolerance in G. soja, but with no certainty of the genomic location of these markers (Zhang et al., 1999). The first discovered salt tolerance gene in G. soja was Ncl2, which has not been identified in G. max (Lee et al., 2009). Screening and sequencing of another wild soybean ecotype W05 have led to the discovery of a different salt-tolerant gene, GmCHX1 (Qi et al., 2014), suggesting genotype-specific salt tolerance mechanisms. The mechanism of salt tolerance in a wild soybean genotype, Tongyu06311, was found to be regulated by amino acid and organic acid metabolism where compatible solutes were accumulated instead of relying on the consumption of ATP (Yang et al., 2017). Most recently, aquaporin gene GmTIP2;1, was found associated with salt tolerance in G. soja (Zhang et al., 2017a). Given the role of aquaporins in water transport, it is likely that more aquaporin alleles found in G. soja might be associated with salt stress tolerance.
The drought tolerance gene GsWRKY20 from G. soja was recently identified and functionally validated by overexpressing it in soybean. The transgenic soybean exhibits increased yield, plant height, and root length over the non-transgenic plant in the same drought conditions. The mechanisms governing the drought tolerance is related to stomatal density and closure speed (Ning et al., 2017). This study provides promising solutions to farming in arid and semi-air environments.
A landscape genomics study of G. soja revealed candidate SNPs associated with varying environmental factors, including monthly precipitation, substrate sand percentage and substrate silt percentage (Anderson et al., 2016). These findings indirectly suggest these loci might play roles in certain abiotic stress tolerance, functional evaluation is needed for each of these genes before they can be considered for soybean crop improvement.
Nutrition
Reduction of saturated fatty acid content of soybeans is highly desired due to its association with cardiovascular disease (Hu et al., 1997). A GWAS study on seed composition in G. soja revealed three new markers associated with palmitic acid levels, a saturated fatty acid, although increased resolution is needed to identify candidate genes. This result has led to the identification of a putative candidate genes, Glyma.07G112100, associated with biosynthesis of linoleic acid in G. soja (Leamy et al., 2017). Linoleic acid is a polyunsaturated fatty acid that is often partially hydrogenated and attributed to cardiovascular health. This recent discovery of a gene controlling both unwanted fatty acids has the potential to make marked improvements on the nutritional profile of the cultivated soybean. The same study of G. soja identified two candidate genes, Glyma.14G121400 and Glyma.16G068500, which are associated with a saturated fatty acid, steric acid, and revealed a candidate gene associated with the “good” fatty acid, unsaturated oleic acid (Leamy et al., 2017). These candidate genes were reported with G. max gene IDs, although they were identified in G. soja. In addition, seed protein content of G. soja has been shown to be higher than G. max on average, likely due to selection on increased yield and oil content (Diers et al., 1992; Chen and Nelson, 2004; Leamy et al., 2017).
Yield Related Traits
An investigation into the variation of early plant height in wild soybeans revealed significant differences among G. soja accessions (Chen and Nelson, 2006). This variation suggests that the trait has been under selection in the natural environment, and may have underlying genetic controls that can be introduced to cultivated soybean. However, characterizing G. soja height beyond 30 days is especially challenging due to the fragile vining nature of the species, limiting growth rate phenotyping to early stages. Nevertheless, early growth rates are important indicators of a plant's eventual success and yield (Chen and Wiatrak, 2011; Filho, 2015).
Yield has been strongly selected in cultivars, often making G. max superior in this trait. However, one study has identified a QTL in G. soja correlated with improved yield. QTL mapping of a cross population of G. soja with G. max lead to the discovery of a yield related QTL in G. soja on chromosome 14. This QTL is responsible for a 9.4% yield advantage and has been validated in two elite genetic backgrounds (Concibido et al., 2003).
Perspectives and Limitations
G. soja holds great potential to provide novel genes/alleles soybean and other legume species for crop improvement. A prerequisite for using G. soja to improve the agricultural potential of G. max is to dissect the genetic architecture underlying the traits of interest and uncover the molecular, physiological, and biochemical mechanisms involved. Some studies using GWAS and linkage mapping have facilitated the dissection of potentially useful traits, and an integration of these strategies coupled with investigation of gene expression (transcriptomes), protein expression (proteomics), and metabolite profiling (metabolomes) can be helpful to understand the mechanisms involved in phenotypic differences. High throughput sequencing technology and biotechnology (such as genome editing) can significantly facilitate novel gene discovery and transfer useful genes to soybean.
G. soja and G. max have the same chromosome number and can be easily crossed to create fertile hybrids, which make it possible to transfer useful genes from G. soja to G. max by traditional breeding practice. However, introgression of G. soja into G. max may result in linkage drag by bringing in unwanted parts of the genome together with the selected genes due to linkage disequilibrium. Linkage drag usually results in drags of unfavorable traits, such as reduced yield, apt to shatter, and lodging etc. This limitation can be resolved by the rapid progress of biotechnology, such as improved genetic transformation and cutting edge genome editing (e.g., Kim et al., 2017). The advanced breeding technologies, such as marker-assisted backcrossing (MABC) (e.g., Iftekharuddaula et al., 2011; Hasan et al., 2015) and genomic selection (e.g., Duhnen et al., 2017) can also improve the efficiency.
Thus far, G. soja has been used to determine the nature of domesticated traits in G. max (Li et al., 2014; Zhou et al., 2015b), while the associated genetic variation in G. soja has not been fully investigated. Knowledge of the genomic regions with selective sweeps in G. soja is helpful to understand the evolutionary mechanisms that produced and altered traits in G. max resulting from domestication, and can facilitate the use of G. soja-derived variations to improve soybean. Investigations of phenotypic variation should shift toward the screening for biotic and abiotic resistance under different environmental conditions and stressor regimes. Such a shift in research focus would produce results (and future directions) that are more relevant to current needs as they pertain to growing food needs and climate change. Although some studies have identified SCN-resistant G. soja accession by screening a small portion of G. soja collection at USDA germplasm (Kim et al., 2011; Zhang et al., 2016), a systematic screening is needed by collecting more ecotypes from many different natural populations, taking into account the observed association between the SCN resistance and G. soja origins. Meanwhile, it is critical to find out the genes/alleles associated with important traits in G. soja are species-specific by comparing them with those from cultivated soybean collections.
On the other hand, it is still time-consuming and labor intensive to measure agriculturally-important traits in G. soja because of its extensive lateral branching phenotype and indeterminate growth habit. The lack of high-throughput phenotyping technology poses challenges for research of genotype-phenotype interactions, especially those requiring large datasets (Houle et al., 2010). For example, phenotyping of root traits infected with root pathogens requires additional efforts and time. The measurement of SCN resistance in soybean roots requires at least 25 days of nematode inoculation (a life cycle of SCN), making it one of the most difficult traits to screen for.
Crop improvement by using wild soybean has been progressing, but is still lacking in some promising areas. Currently, very little work on the Soybean Mosaic Virus (SMV) has been reported in G. soja, a native host to the virus. G. soja plants infected by SMV have been observed in South Korea (Seo et al., 2009). More work is needed to develop SMV-resistant soybean varieties. In addition, the observed variation in early vigor traits in wild soybeans is also an understudied area with potential to improve the early success and eventual yield of soybean crop.
The chloroplast genome has been used in only a few studies with Glycine, and in those cases only to discuss diversity and domestication. Plastids regulate photosynthesis and the production of many metabolites; it would not be a great leap to assume that some of those attributes are (a) important to cultivation and (b) phenotypically found to vary in the wild population of soybean. The chloroplast genome of soybeans underwent multiple selection events, and diverged into two major haplotypes early on in domestication (Fang et al., 2016). Whole chloroplast genome assembly of G. soja compared to nine other Glycine species reported high conservation with no major rearrangements within Glycine. Phylogenetic topology is consistent between nuclear and plastid genomes, and an in-depth investigation into plastid genomic differences between G. soja and G. max may reveal attributes worth studying for crop improvement (Asaf et al., 2017).
Efforts should also be made to conserve the wild soybean populations, along with other crop wild relatives (Khoury et al., 2010; Castañeda-Álvarez et al., 2016). It has been shown that these diverse populations exhibit desired phenotypes and diverse genotypes that can be exploited for crop improvement. Conservation of these populations is required in order to continue benefiting from them. A recent study in Japan reported low risk of gene flow between genetically modified and wild soybeans (Goto et al., 2017). Soybeans self-fertilize, and therefore gene flow can easily be maintained by reducing seed transport and spillage and increasing the distance between crops and wild populations.
The natural populations of wild soybean are distributed in East Asia and Russia. Current research mainly uses a little over 1,000 genotypes from USDA collections. To fully investigate the natural variation of this species and identify useful resources, international collaborations between scientists from diverse disciplines are needed to collect and share germplasm, as well as share genomic resources. More findings from wild soybean are expected in the near future.
Conclusion
The wild soybean has been the source of much advancement in crop improvement and aided in research related to the evolution of the soybean. The domestication history of soybean is now widely understood. Continued research and screening of more ecotypes for potentially useful traits in G. soja will help to reveal additional valuable genetic sources for genetic improvement of legumes. Collaborations and effort should be put toward finishing what has already begun—to link the discovery of candidate genes with studies on relevant gene regulatory networks and ultimately, the molecular and practical procedures for crop improvement. Lastly, the conservation of natural soybean populations needs to be promoted to ensure the continued existence of adaptable, wild traits that could later be used for crop improvement. International collaboration is needed to carry out goal-directed, comprehensive research of G. soja to make full use of this untapped genetic reservoir.
Author Contributions
JK, HZ, and B-HS wrote the manuscript and approved the final manuscript.
Funding
B-HS was supported by the National Institute of General Medical Sciences of the National Institutes of Health, Award Number: R15GM122029; North Carolina Biotechnology Center, Award Number: 2014-CFG-8005; the North Carolina Soybean Producers Association, and University of North Carolina at Charlotte.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Anderson, J. E., Kono, T. J., Stupar, R. M., Kantar, M. B., and Morrell, P. L. (2016). Environmental association analyses identify candidates for abiotic stress tolerance in Glycine soja, the wild progenitor of cultivated soybeans. G3 (Bethesda) 6, 835–843. doi: 10.1534/g3.116.026914
Asaf, S., Khan, A. L., Aaqil Khan, M., Muhammad Imran, Q., Kang, S. M., Al-Hosni, K., et al. (2017). Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PLoS ONE 12:182281. doi: 10.1371/journal.pone.0182281
Hymowitz, T., and Nelson, R. L. (2004). “Biogeography, local adaptation, vavilov, and genetic diversity in soybean,” in Biological Resources and Migration, ed D. Werner (Berlin; Heidelberg: Springer), 47–59.
Castañeda-Álvarez, N. P., Khoury, C. K., Achicanoy, H. A., Bernau, V., Dempewolf, H., Eastwood, R. J., et al. (2016). Global conservation priorities for crop wild relatives. Nature Plants 2:16022. doi: 10.1038/nplants.2016.22
Chen, G. H., and Wiatrak, P. (2011). Seeding rate effects on soybean height, yield, and economic return. Agron. J. 103, 1301–1307. doi: 10.2134/agronj2010.0427
Chen, Y. W., and Nelson, R. L. (2004). Genetic variation and relationships among cultivated, wild, and semiwild soybean. Crop Sci. 44, 316–325. doi: 10.2135/cropsci2004.3160
Chen, Y. W., and Nelson, R. L. (2006). Variation in early plant height in wild soybean. Crop Sci. 46, 865–869. doi: 10.2135/cropsci2005.07-0202
Chung, W. H., Jeong, N., Kim, J., Lee, W. K., Lee, Y. G., Lee, S. H., et al. (2014). Population structure and domestication revealed by high-depth resequencing of Korean cultivated and wild soybean genomes. DNA Res. 21, 153–167. doi: 10.1093/dnares/dst047
Concibido, V. C., La Vallee, B., Mclaird, P., Pineda, N., Meyer, J., Hummel, L., et al. (2003). Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theor. Appl. Genet. 106, 575–582. doi: 10.1007/s00122-002-1071-5
Cook, D. E., Lee, T. G., Guo, X., Melito, S., Wang, K., Bayless, A. M., et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–1209. doi: 10.1126/science.1228746
Diers, B. W., Keim, P., Fehr, W. R., and Shoemaker, R. C. (1992). Rflp analysis of soybean seed protein and oil content. Theor. Appl. Genet. 83, 608–612. doi: 10.1007/BF00226905
Dong, Y., Yang, X., Liu, J., Wang, B. H., Liu, B. L., and Wang, Y. Z. (2014). Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat. Commun. 5:3352. doi: 10.1038/ncomms4352
Duhnen, A., Gras, A., Teyssèdre, S., Romestant, M., Claustres, B., Daydé, J., et al. (2017). Genomic selection for yield and seed protein content in soybean: a study of breeding program data and assessment of prediction accuracy. Crop Sci. 57, 1325–1337. doi: 10.2135/cropsci2016.06.0496
Fang, C., Ma, Y., Yuan, L., Wang, Z., Yang, R., Zhou, Z., et al. (2016). Chloroplast DNA underwent independent selection from nuclear genes during soybean domestication and improvement. J. Genet. Genomics 43, 217–221. doi: 10.1016/j.jgg.2016.01.005
Fernie, A. R., and Schauer, N. (2009). Metabolomics-assisted breeding: a viable option for crop improvement. Trends Genet. 25, 39–48 doi: 10.1016/j.tig.2008.10.010
Filho, J. M. (2015). Seed vigor testing: an overview of the past, present and future perspective. Sci. Agric. 72, 363–374. doi: 10.1590/0103-9016-2015-0007
Goto, H., Mcpherson, M. A., Comstock, B. A., Stojšin, D., and Ohsawa, R. (2017). Likelihood assessment for gene flow of transgenes from imported genetically modified soybean (Glycine max (L.) Merr.) to wild soybean (Glycine soja Seib. et Zucc.) in Japan as a component of environmental risk assessment. Breed Sci. 67, 348–356. doi: 10.1270/jsbbs.16134
Guo, J., Liu, Y., Wang, Y., Chen, J., Li, Y., Huang, H., et al. (2012). Population structure of the wild soybean (Glycine soja) in China: implications from microsatellite analyses. Ann. Bot. 110, 777–785. doi: 10.1093/aob/mcs142
Ha, J., Abernathy, B., Nelson, W., Grant, D., Wu, X., Nguyen, H. T., et al. (2012). Integration of the draft sequence and physical map as a framework for genomic research in soybean (Glycine max (L.) Merr.) and wild soybean (Glycine soja Sieb. and Zucc.). G3 (Bethesda) 2, 321–329. doi: 10.1534/g3.111.001834
Han, Y., Zhao, X., Liu, D., Li, Y., Lightfoot, D. A., Yang, Z., et al. (2016). Domestication footprints anchor genomic regions of agronomic importance in soybeans. New Phytol. 209, 871–884. doi: 10.1111/nph.13626
Hasan, M. M., Rafii, M. Y., Ismail, M. R., Mahmood, M., Rahim, H. A., Alam, M. A., et al. (2015). Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol. Biotechnol. Equip. 29, 237–254. doi: 10.1080/13102818.2014.995920
He, S. L., Wang, Y. S., Li, D. Z., and Yi, T. S. (2016). Environmental and historical determinants of patterns of genetic differentiation in wild soybean (Glycine soja Sieb. et Zucc). Sci. Rep. 6:22795. doi: 10.1038/srep22795
Hesler, L. S. (2013). Resistance to soybean aphid among wild soybean lines under controlled conditions. Crop Prot. 53, 139–146. doi: 10.1016/j.cropro.2013.06.016
Houle, D., Govindaraju, D. R., and Omholt, S. (2010). Phenomics: the next challenge. Nat. Rev. Genet. 11, 855–866. doi: 10.1038/nrg2897
Hu, F. B., Stampfer, M. J., Manson, J. E., Rimm, E., Colditz, G. A., Rosner, B. A., et al. (1997). Dietary fat intake and the risk of coronary heart disease in women. N. Eng.J. Med. 337, 1491–1499. doi: 10.1056/NEJM199711203372102
Hu, Z. A., and Wang, H. X. (1997). Salt tolerance of wild soybean (Glycine soja) in populations evaluated by a new method. Soybean Genet. Newsl. 24, 79 - 80.
Hyten, D. L., Song, Q., Zhu, Y., Choi, I. Y., Nelson, R. L., Costa, J. M., et al. (2006). Impacts of genetic bottlenecks on soybean genome diversity. Proc. Nat.l Acad. Sci. U.S.A. 103, 16666–16671. doi: 10.1073/pnas.0604379103
Iftekharuddaula, K. M., Newaz, M. A., Salam, M. A., Ahmed, H. U., Mahbub, M. A. A., Septiningsih, E. M., et al. (2011). Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into BR11, the rainfed lowland rice mega variety of Bangladesh. Euphytica 178, 83–97. doi: 10.1007/s10681-010-0272-2
Khoury, C., Laliberte, B., and Guarino, L. (2010). Trends in ex situ conservation of plant genetic resources: a review of global crop and regional conservation strategies. Genet. Resour. Crop Evol. 57, 625–639. doi: 10.1007/s10722-010-9534-z
Kim, H., Kim, S.-T., Ryu, J., Kang, B.-C., Kim, J.-S., and Kim, S.-G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8:14406 doi: 10.1038/ncomms14406
Kim, M., and Diers, B. W. (2013). Fine mapping of the SCN Resistance QTL cqSCN-006 and cqSCN-007 from Glycine soja PI 468916. Crop Sci. 53, 775–785. doi: 10.2135/cropsci2012.07.0425
Kim, M., Hyten, D. L., Niblack, T. L., and Diers, B. W. (2011). Stacking resistance alleles from wild and domestic soybean sources improves soybean cyst nematode resistance (vol 51, pg 934, 2011). Crop Sci. 51, 2301–2301. doi: 10.2135/cropsci2010.08.0459
Kim, M. Y., Lee, S., Van, K., Kim, T. H., Jeong, S. C., Choi, I. Y., et al. (2010). Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc. Natl. Acad. Sci. U.S.A. 107, 22032–22037. doi: 10.1073/pnas.1009526107
Lam, H. M., Xu, X., Liu, X., Chen, W., Yang, G., Wong, F. L., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–U1041. doi: 10.1038/ng.715
Leamy, L. J., Lee, C. R., Song, Q., Mujacic, I., Luo, Y., Chen, C. Y., et al. (2016). Environmental versus geographical effects on genomic variation in wild soybean (Glycine soja) across its native range in northeast Asia. Ecol. Evol. 6, 6332–6344. doi: 10.1002/ece3.2351
Leamy, L. J., Zhang, H., Li, C., Chen, C. Y., and Song, B. H. (2017). A genome-wide association study of seed composition traits in wild soybean (Glycine soja). BMC Genomics 18:18. doi: 10.1186/s12864-016-3397-4
Lee, J. D., Shannon, J. G., Vuong, T. D., and Nguyen, H. T. (2009). Inheritance of salt tolerance in wild soybean (Glycine soja Sieb. and Zucc.) accession PI483463. J. Heredity 100, 798–801. doi: 10.1093/jhered/esp027
Lee, J. S., Yoo, M. H., Jung, J. K., Bilyeu, K. D., Lee, J. D., and Kang, S. (2015). Detection of novel QTLs for foxglove aphid resistance in soybean. Theor. Appl. Genet. 128, 1481–1488. doi: 10.1007/s00122-015-2519-8
Li, Y. H., Li, W., Zhang, C., Yang, L., Chang, R. Z., Gaut, B. S., et al. (2010). Genetic diversity in domesticated soybean (Glycine max) and its wild progenitor (Glycine soja) for simple sequence repeat and single-nucleotide polymorphism loci. New Phytol. 188, 242–253. doi: 10.1111/j.1469-8137.2010.03344.x
Li, Y. H., Zhou, G., Ma, J., Jiang, W., Jin, L. G., Zhang, Z., et al. (2014). De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32, 1045–1052. doi: 10.1038/nbt.2979
Maughan, P. J., Maroof, M. A.S., and Buss, G. R. (1996). Molecular-marker analysis of seed weight: genomic locations, gene action, and evidence for orthologous evolution among three legume species. Theor.Appl. Genet. 93, 574–579. doi: 10.1007/BF00417950
Maxted, N., and Kell, S. P. (2009). Establishment of a Global Network for the In Situ Conservation of Crop Wild Relatives: Status and Needs. (Rome: Food and Agriculture Organization of the United Nations Commission on Genetic Resources for Food and Agriculture), 1–265.
Ning, W., Zhai, H., Yu, J., Liang, S., Yang, X., Xing, X., et al. (2017). Overexpression of Glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol. Breed. 37:19. doi: 10.1007/s11032-016-0614-4
Oecd Food and Nations AOOTU (2016). OECD-FAO Agricultural Outlook 2016–2025. Paris: OECD Food and Nations AOOTU.
Prince, S. J., Song, L., Qiu, D., Maldonado Dos Santos, J. V., Chai, C., Joshi, T., et al. (2015). Genetic variants in root architecture-related genes in a Glycine soja accession, a potential resource to improve cultivated soybean. BMC Genomics 16:132. doi: 10.1186/s12864-015-1334-6
Qi, X., Li, M. W., Xie, M., Liu, X., Ni, M., Shao, G., et al. (2014). Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 5:4340. doi: 10.1038/ncomms5340
Qiu, J., Wang, Y., Wu, S., Wang, Y. Y., Ye, C. Y., Bai, X., et al. (2014). Genome re-sequencing of semi-wild soybean reveals a complex Soja population structure and deep introgression. PLoS ONE 9:e108479. doi: 10.1371/journal.pone.0108479
Qiu, L. J., Xing, L. L., Guo, Y., Wang, J., Jackson, S. A., and Chang, R. Z. (2013). A platform for soybean molecular breeding: the utilization of core collections for food security. Plant Mol. Biol. 83, 41–50. doi: 10.1007/s11103-013-0076-6
Qiu, Y. X., Fu, C. X., and Comes, H. P. (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world's most diverse temperate flora. Mol. Phylogenet. Evol. 59, 225–244. doi: 10.1016/j.ympev.2011.01.012
Ray, D. K., Mueller, N. D., West, P. C., and Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428. doi: 10.1371/journal.pone.0066428
Rossi, M., Goggin, F. L., Milligan, S. B., Kaloshian, I., Ullman, D. E., and Williamson, V. M. (1998). The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. U.S.A. 95, 9750–9754. doi: 10.1073/pnas.95.17.9750
Sakai, M., Kanazawa, A., Fujii, A., Thseng, F. S., Abe, J., and Shimamoto, Y. (2003). Phylogenetic relationships of the chloroplast genomes in the genus Glycine inferred from four intergenic spacer sequences. Plant Syst. Evol. 239, 29–54. doi: 10.1007/s00606-002-0226-9
Sedivy, E. J., Wu, F., and Hanzawa, Y. (2017). Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol. 214, 539–553. doi: 10.1111/nph.14418
Seo, J. K., Ohshima, K., Lee, H. G., Son, M., Choi, H. S., Lee, S. H., et al. (2009). Molecular variability and genetic structure of the population of Soybean mosaic virus based on the analysis of complete genome sequences. Virology 393, 91–103. doi: 10.1016/j.virol.2009.07.007
Song, Q., Hyten, D. L., Jia, G., Quigley, C. V., Fickus, E. W., Nelson, R. L., et al. (2013). Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLOS ONE 8:e54985. doi: 10.1371/journal.pone.0054985
Song, Q., Hyten, D. L., Jia, G., Quigley, C. V., Fickus, E. W., Nelson, R. L., et al. (2015). Fingerprinting soybean germplasm and its utility in genomic research. G3 (Bethesda). 5, 1999–2006. doi: 10.1534/g3.115.019000
Stupar, R. M. (2010). Into the wild: the soybean genome meets its undomesticated relative. Proc. Natl. Acad. Sci. U.S.A.107, 21947–21948. doi: 10.1073/pnas.1016809108
Tian, Z. X., Wang, X. B., Lee, R., Li, Y. H., Specht, J. E., Nelson, R. L., et al. (2010). Artificial selection for determinate growth habit in soybean. Proc. Natl. Acad. Sci. U.S.A. 107, 8563–8568. doi: 10.1073/pnas.1000088107
Tylka, G. L., and Marett, C. C. (2014). Distribution of the soybean cyst nematode, Heterodera glycines, in the United States and Canada: 1954 to 2014. Plant Health Prog. 15, 13–15. doi: 10.1094/PHP-BR-14-0006
USDA (2016). “Soybeans: Acreage planted, harvested, yield, production, value, and loan rate, U.S., 1960-2015,” in Oil Crops Yearbook 2016, ed ERSUSDO (Agriculture United States Department of Agriculture; Economic Research Service, USDA). Available online at: https://www.ers.usda.gov/data-products/oil-crops-yearbook/
Wang, D., Diers, B. W., Arelli, P. R., and Shoemaker, R. C. (2001). Loci underlying resistance to Race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theoret. Appl. Genet. 103, 561–566. doi: 10.1007/PL00002910
Wang, K. J., and Li, X. H. (2013). Genetic diversity and gene flow dynamics revealed in the rare mixed populations of wild soybean (Glycine soja) and semi-wild type (Glycine gracilis) in China. Genet. Resour. Crop Evol. 60, 2303–2318. doi: 10.1007/s10722-013-9998-8
Wang, Y., Shahid, M. Q., and Baloch, F. S. (2016). Phylogeographical studies of Glycine soja: implicating the refugium during the Quaternary glacial period and large-scale expansion after the Last Glacial Maximum. Turk.J. Agric. Forestry 40, 825–838. doi: 10.3906/tar-1607-30
Wang, Y., Shahid, M. Q., Ghouri, F., Baloch, F. S., Wang, Y., and Huang, H. (2015). Evaluation of the geographical pattern of genetic diversity of Glycine soja and Glycine max based on four single copy nuclear gene loci: for conservation of soybean germplasm. Biochem. Syst. Ecol. 62, 229–235. doi: 10.1016/j.bse.2015.09.006
Yang, D.-S., Zhang, J., Li, M.-X., and Shi, L.-X. (2017). Metabolomics analysis reveals the salt-tolerant mechanism in Glycine soja. J. Plant Growth Regul. 36, 460–471. doi: 10.1007/s00344-016-9654-6
Yu, N., and Diers, B. W. (2017). Fine mapping of the SCN resistance QTL cqSCN-006 and cqSCN-007 from Glycine soja PI 468916. Euphytica 213:54. doi: 10.1007/s10681-016-1791-2
Zhang, D. Y., Kumar, M., Xu, L., Wan, Q., Huang, Y. H., Xu, Z. L., et al. (2017a). Genome-wide identification of major intrinsic proteins in Glycine soja and characterization of GmTIP2;1 function under salt and water stress. Sci. Rep. 7:4106. doi: 10.1038/s41598-017-04253-z
Zhang, H., Kjemtrup-Lovelace, S., Li, C., Luo, Y., Chen, L. P., and Song, B. H. (2017b). Comparative RNA-seq analysis uncovers a complex regulatory network for soybean cyst nematode resistance in wild soybean (Glycine soja). Sci. Rep. 7:9699. doi: 10.1038/s41598-017-09945-0
Zhang, H., Li, C., Davis, E. L., Wang, J., Griffin, J. D., Kofsky, J., et al. (2016). Genome-wide association study of resistance to soybean cyst nematode (Heterodera glycines) HG Type 2.5.7 in wild soybean (Glycine soja). Front. Plant Sci. 7:1214. doi: 10.3389/fpls.2016.01214
Zhang, H., Mittal, N., Leamy, L. J., Barazani, O., and Song, B. H. (2017c). Back into the wild—Apply untapped genetic diversity of wild relatives for crop improvement. Evol. Appl. 10, 5–24. doi: 10.1111/eva.12434
Zhang, H. Y., and Song, B. H. (2017). RNA-seq data comparisons of wild soybean genotypes in response to soybean cyst nematode (Heterodera glycines). Genomics Data 14, 36–39. doi: 10.1016/j.gdata.2017.08.001
Zhang, H. Y., Song, Q. J., Griffin, J. D., and Song, B. H. (2017d). Genetic architecture of wild soybean (Glycine soja) response to soybean cyst nematode (Heterodera glycines). Mol. Genet. Genom. 292, 1257–1265. doi: 10.1007/s00438-017-1345-x
Zhang, J. P., Singh, A., Mueller, D. S., and Singh, A. K. (2015). Genome-wide association and epistasis studies unravel the genetic architecture of sudden death syndrome resistance in soybean. Plant J. 84, 1124–1136. doi: 10.1111/tpj.13069
Zhang, Q., Wang, H., and Hu, Z. (1999). RAPD Markers Associated With Salt Tolerance in Wild Soybean Populations. Soybean Genet. Newsl. 26. Available online at: https://www.soybase.org/sgn/article.php?issue_id=2&autoID=6
Zhang, S., Zhang, Z., Bales, C., Gu, C., Difonzo, C., Li, M., et al. (2017e). Mapping novel aphid resistance QTL from wild soybean, Glycine soja 85-32. Theor. Appl. Genet. 130, 1941–1952. doi: 10.1007/s00122-017-2935-z
Zhang, S., Zhang, Z., Wen, Z., Gu, C., An, Y. C., Bales, C., et al. (2017f). Fine mapping of the soybean aphid-resistance genes Rag6 and Rag3c from Glycine soja 85-32. Theor. Appl. Genet. 130, 2601–2615. doi: 10.1007/s00122-017-2979-0
Zhou, L., Wang, S. B., Jian, J., Geng, Q. C., Wen, J., Song, Q., et al. (2015a). Identification of domestication-related loci associated with flowering time and seed size in soybean with the RAD-seq genotyping method. Sci. Rep. 5:9350. doi: 10.1038/srep09350
Keywords: wild soybean, Glycine soja, crop wild relative, genomics and genetics, biotechnology, crop improvement, stress response and tolerance
Citation: Kofsky J, Zhang H and Song B-H (2018) The Untapped Genetic Reservoir: The Past, Current, and Future Applications of the Wild Soybean (Glycine soja). Front. Plant Sci. 9:949. doi: 10.3389/fpls.2018.00949
Received: 15 March 2018; Accepted: 12 June 2018;
Published: 09 July 2018.
Edited by:
Roberto Papa, Università Politecnica delle Marche, ItalyReviewed by:
Francesca Sparvoli, Istituto di Biologia e Biotecnologia Agraria (IBBA), ItalyMichael Benjamin Kantar, University osf Hawaii, United States
Copyright © 2018 Kofsky, Zhang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Hua Song, bsong5@uncc.edu
 Janice Kofsky
Janice Kofsky Hengyou Zhang
Hengyou Zhang Bao-Hua Song
Bao-Hua Song