- Department of Cell and Metabolic Biology, Leibniz-Institut für Pflanzenbiochemie, Halle, Germany
Glandular trichomes contribute to the high resistance of wild tomato species against insect pests not only thanks to the metabolites they produce but also because of morphological and developmental features which support the high production of these defense compounds. In Solanum habrochaites, type VI trichomes have a distinct spherical shape and a large intercellular storage cavity where metabolites can accumulate and are released upon breaking off of the glandular cells. In contrast, the type VI trichomes of S. lycopersicum have a four-leaf clover shape corresponding to the four glandular cells and a small internal cavity with limited capacity for storage of compounds. To better characterize the genetic factors underlying these trichome morphological differences we created a back-cross population of 116 individuals between S. habrochaites LA1777 and S. lycopersicum var. cerasiforme WVa106. A trichome score that reflects the shape of the type VI trichomes allowing the quantification of this trait was designed. The scores were distributed normally across the population, which was mapped with a total of 192 markers. This resulted in the identification of six quantitative trait locus (QTLs) on chromosomes I, VII, VII, and XI. The QTL on chromosome I with the highest LOD score was confirmed and narrowed down to a 500 gene interval in an advanced population derived from one of the back-cross lines. Our results provide the foundation for the genetic dissection of type VI trichome morphology and the introgression of these trichome traits into cultivated tomato lines for increased insect resistance.
Key Message: This work shows that the shape of type VI glandular trichomes in tomato is a genetically defined trait controlled by multiple QTLs with one on chromosome I being the major contributor.
Introduction
Trichomes are differentiations of the epidermis that occur at the surface of the aerial parts of many higher plants. Although they come in a great variety of shapes, they can be classified as either non-glandular or glandular. Glandular (or secreting) trichomes typically have one to a few secretory cells located at the tip of a uni- or multicellular stalk. Glandular trichomes are present in about 30% of all vascular plants (Fahn, 2000). These structures accumulate large quantities of secondary metabolites involved in the protection against herbivores and pathogens (Glas et al., 2012; Tissier, 2012), but also are the source of industrially important ingredients in the flavor, fragrance and pharmaceutical areas (Schilmiller et al., 2008). In most cases, biosynthesis of the compounds takes place in the glandular cells, a feature that is typically correlated with high and specific expression of the pathway enzymes. This has greatly facilitated the identification of the genes involved in the biosynthesis pathways, in particular with the help of trichome specific Express Sequence Tag (EST) and more recently next generation sequencing transcriptome databases. Thus, the biosynthesis of menthol in mint (Croteau et al., 2005), of artemisin in Artemisia annua (Brown, 2010), of various terpenoids (Sallaud et al., 2009; Schilmiller et al., 2009) as well as acyl-sugars in tomato (Fan et al., 2016; Schilmiller et al., 2016) or diterpenes in tobacco or rosemary (Sallaud et al., 2012; Scheler et al., 2016) and monoterpenes in thyme (Majdi et al., 2017) could be completely or partially elucidated. In contrast to the progress made in understanding trichome-specific biosynthesis pathways, very little is known about the genes controlling the development and differentiation of glandular trichomes. Much work has been done on the development of non-glandular trichomes in Arabidopsis thaliana with numerous transcription factors and regulators identified [reviewed by (Pattanaik et al., 2014)]. However, there is indication that the regulatory mechanisms controlling glandular trichome development may be different for non-glandular trichomes (Serna and Martin, 2006), although initiation mechanisms are likely to be shared (Maes et al., 2011).
Tomato is a major vegetable crop species with over 170 million tons harvested worldwide in 2014, doubling the yield of 1994 (FAOSTAT, 2017). Plants of the Solanum genus possess different types of both glandular and non-glandular trichomes but the abundance and composition of these organs is very diverse (Luckwill, 1943; Channarayappa et al., 1992). The type I and type IV glandular trichomes typically have a single gland cell on a stalk and they are the major site of biosynthesis for acyl sugars (McDowell et al., 2011). Type VII trichomes have a single-celled stalk and a multicellular berry-shaped head and do not seem to be involved in the biosynthesis of secondary metabolites. The type VI is the most prevalent type of glandular trichomes and is found in almost all tomato species (reviewed by Glas et al., 2012). This specific type of trichomes has a four-celled head producing a variety of metabolites including terpenoids or methylketones (in S. habrochaites ssp. glabratum) but the amount and composition of these metabolites is highly variable in different species and subspecies. The main components in S. habrochaites LA1777 are the sesquiterpenes santalene and bergamotene and their carboxylic acids (Coates et al., 1988), both of which are absent in the cultivated tomato Solanum lycopersicum. In addition to these distinct chemical contents, there are obvious morphological differences between the type VI trichomes of S. habrochaites and S. lycopersicum. While the head of the wild tomato species forms a round sphere, the cultivated tomato head looks like a four-leaf clover (Ben-Israel et al., 2009). Furthermore, observations of the type VI trichomes under the microscope show that there is a large intercellular cavity in the trichome head of the wild tomato, whereas this cavity is absent or much smaller in the cultivated tomato (Bergau et al., 2015). This morphological difference is important because it allows large quantities of secondary metabolites to accumulate in the wild species, and thereby contribute to an increased resistance against pests. In a previous study, Ben-Israel et al. (2009) showed a correlation between the expression of secondary metabolite pathway genes and the shape of the trichomes, suggesting that this intercellular storage compartment could arise due to the high flux of metabolites. Recently we could show that the total amount of terpenoids per trichome is 97 fold lower in the S. lycopersicum accession LA4024 compared to LA1777 which can be explained at least in part by the near absence of the intercellular cavity in the cultivated tomato (Balcke et al., 2017). However, the genes responsible for this morphological character are still unknown. Because this compartment is extracellular, potential candidates are cell wall modifying genes such as xyloglucan endotransglycosylases/hydrolases (Van Sandt et al., 2007) or pectin methylesterases (Micheli, 2001), components of the cytoskeleton such as microtubules (Lloyd, 2011) or regulating transcription factors like the Arabidopsis thaliana MYB5 (Li et al., 2009). Three mutants affecting type VI trichome development in tomato have been described so far, hairless (hl), chalcone isomerase (chi) and odorless-2 (od-2), but these do not seem to affect the development of the storage cavity (Kang et al., 2010, 2014, 2016). The hl mutant is affected in a component of the WAVE complex controlling actin nucleation, and thereby supports a link of the cytoskeleton with trichome morphology and terpenoid production. The chi mutant has no major morphological defects except a smaller size of type VI trichomes that produce severely reduced levels of terpenes, indicating a metabolic connection between flavonoid and terpenoid metabolisms. The gene responsible for the od-2 mutation has not been identified to date.
A quantitative trait locus (QTL) is a region on chromosome that can be linked to a particular phenotype (Miles and Wayne, 2008). QTL analyses have been successfully used to map various trichome traits such as trichome specific acyl-sugars (Leckie et al., 2012) as well as type IV trichome density in S. habrochaites (Momotaz et al., 2010) and the presence of type IV trichomes in S. galapagense (Firdaus et al., 2013).
To investigate the genetic loci underlying the morphological differences in the type VI trichomes between S. habrochaites and S. lycopersicum, we created a backcross population using S. lycopersicum as the recurrent parent. This population was genetically mapped using High-Resolution Melt Analysis (HRM) (Wittwer et al., 2003) and other types of molecular markers. A score was designed to quantify the shape of the type VI trichome heads and used to perform a QTL analysis. Our results indicate that there are multiple loci, with one on chromosome I being the major factor, contributing to this distinct trichome morphology.
Materials and Methods
Plant Material and Growth Conditions
Seeds of the tomato accessions were obtained from the Tomato Genetics Resource Center, UC Davis, United States. Seeds of S. lycopersicum var. cerasiforme ‘West Virginia 106’ (WVa 106) and S. lycopersicum var. Ferum were provided by Mathilde Causse (INRA, France). Seeds of the other tomato accessions were obtained from the Tomato Genetics Resource Center, UC Davis, United States. Plants were grown under 16:8 h long day conditions in a climate-controlled greenhouse at 21–24°C and 65% humidity.
Backcross Population
Solanum habrochaites LA1777 and S. lycopersicum var. cerasiforme ‘West Virginia 106’ (WVa 106) were crossed by using the pollen of LA1777 to manually fertilize emasculated flowers of WVa106. The backcross population was then created by taking the pollen of F1 plants to manually fertilize an emasculated flower of WVa106. The backcross plants were numbered according to the F1 plant from which they originated. For example, plant 4-06 is the sixth backcross plant bred from F1 plant number 4.
Successful backcrosses were confirmed by determining the trichome score (see below), analyzing leaf surface GC-MS profiles and preliminary genotyping for six independent loci.
Two advanced backcross lines were created by self-pollination of the BC1 plants 4-06 and 2-11.
Trichome Score
Type VI glandular trichome heads were imaged with a Nikon AZ100 stereoscope and the trichome diameters were measured using Adobe Photoshop. The trichome score is defined as the ratio of the sum of the diameters through the constriction sites to the sum of the diameters through the wide edges (Figure 1). Six to twelve trichomes were scored for each line. A score of 1 would imply a perfectly circular shape.
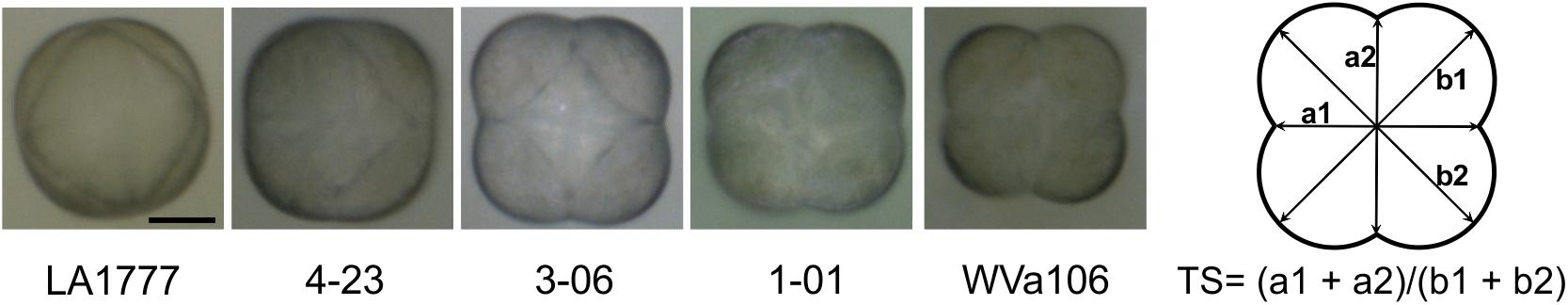
FIGURE 1. The type VI trichome heads of the parental lines LA1777 and WVa 106 as well as representative backcross lines with intermediate scores as observed with a bright field microscope. Scale bar is 20 μm. The right panel shows the formula to calculate the trichome score (TS).
Confocal Microscopy of Type VI Trichomes
Microscopy analysis of type VI trichomes was performed on intact leaves with a LSM 780 (Zeiss, Jena, Germany) confocal microscope. Autofluorescence was excited at 405 nm and recorded at 420–545 nm (to visualize the cytoplasm and cell wall) and 645–735 nm (for chlorophyll). The trichomes were analyzed from the top and the images were taken from the middle part of the trichome. The areas of the secretory cavities and the entire trichomes as well as the different diameters were measured by ImageJ1 (Fiji, 32 bit).
GC–MS Analysis of Hexane Surface Extracts
Surface extracts were collected from 6 leaflets of circa 5 cm length by adding 2 ml n-hexane and shaking for 1 min. The hexane was taken off and centrifuged at 16,000 g for 90 s to remove debris. 1 μl of the supernatant was injected directly into GC–MS for analysis on a Trace GC Ultra gas chromatograph coupled to an ISQ mass spectrometer (Thermo Scientific) as previously described (Brückner et al., 2014).
Mapping
Genomic DNA was isolated from 50 to 100 mg fresh leaf material using the Qiagen DNeasy Plant kit and 10 ng DNA was used for each mapping point.
The backcross lines were genotyped for 192 markers by a combination of different techniques. Several markers were taken from the literature (Dal Cin et al., 2009) but most markers were designed utilizing single nucleotide polymorphisms (SNPs) between the parental lines using the tomato genome (Tomato-Genome-Consortium, 2012) for the cultivated tomato and the Solanum trichome EST library (McDowell et al., 2011) as well as our own unpublished RNA sequencing data for LA1777. The markers were chosen to cover the whole tomato genome and aligned to the published tomato genome version SL2.40 annotation ITAG2.3 by José M Jiménez-Gómez, Max Planck Institute for Plant Breeding Research, Cologne, Germany.
Oligonucleotide primers were designed using primer3 (Rozen and Skaletsky, 2000) based tools and synthesized by Eurofins Genomics (Ebersberg, Germany).
High-Resolution-Melt Analysis
High-resolution melt curves were obtained for 119 markers in a 96 well format using my-Budget 5x EvaGreen® QPCR Mix II (Bio&Sell, Feucht, Germany) on a Bio-Rad Connect 96x Real-Time PCR system. Data analysis was performed using the Bio-Rad Precision Melt Analysis Software V1.1.
Sequenom iPlex SNP Genotyping
For 66 markers an iPlex Gold multiplex genotyping assay was performed by ATLAS Biolabs (Berlin, Germany).
Cleaved Amplified Polymorphic Sequences (CAPS)
To analyze the 7 CAPS marker a PCR with specific primers was performed using DreamTaq DNA polymerase (Thermo Scientific) followed by a digest with the respective restriction enzyme (NEB) and separation on an agarose gel.
QTL Mapping
QTL analysis was performed by composite interval mapping (CIM) using the Windows QTL Cartographer software V2.5 (Wang et al., 2012), at 1 cM walk speed and with a 10 cM window size. Following the guidelines by (Lander and Kruglyak, 1995) a logarithm of odds (LOD) threshold of 2.5, calculated from 1000 permutations at a probability of 0.05, was used for declaring significant QTLs.
Epistatic interactions were determined by using the QTLNetwork software version 2.1 (Yang et al., 2008). Window size, working speed and filtration window for the genome scan were set at 10, 1, and 10 cM, respectively. The F-test using Henderson method III was used to determine significance with 1,000 permutations.
Results
Designing a Score for Type VI Trichome Shape
When seen from above, the shape of type VI trichomes in the cultivated tomato (S. lycopersicum) and in the wild species S. habrochaites can be clearly differentiated (Figure 1). The type VI trichomes of S. habrochaites have the shape of a ball, whereas in S. lycopersicum the four glandular cells can be clearly distinguished giving the head of the trichome the appearance of a four-leaf clover. Observations of harvested type VI trichomes under a fluorescence microscope confirm the presence of four cells and of a large extracellular cavity between the four glandular cells in S. habrochaites (Supplementary Figure S1). To quantify this difference in the shape of the trichomes, we designed a score TS = (a1 + a2)/(b1 + b2) where a1 and a2 represent the two shortest diameters and b1 and b2 the two longest diameters of the trichome viewed from above. For S. habrochaites this score approaches 1 (0.972 ± 0.004), reflecting its almost perfect circular shape, whereas the S. lycopersicum score is 0.780 ± 0.006. We calculated this score for a number of different tomato species and cultivars (Figure 2). This revealed that all three different S. habrochaites lines have the highest score, whereas cultivars of S. lycopersicum and S. pimpinellifolium have the lowest score. Other wild species like S. pennellii, S. chmielewskii or S. peruvianum have scores that are intermediate and are statistically significantly distinguishable from both S. habrochaites and S. lycopersicum correlating with their slightly rounder shape (Figure 2 and Supplementary Figure S2).
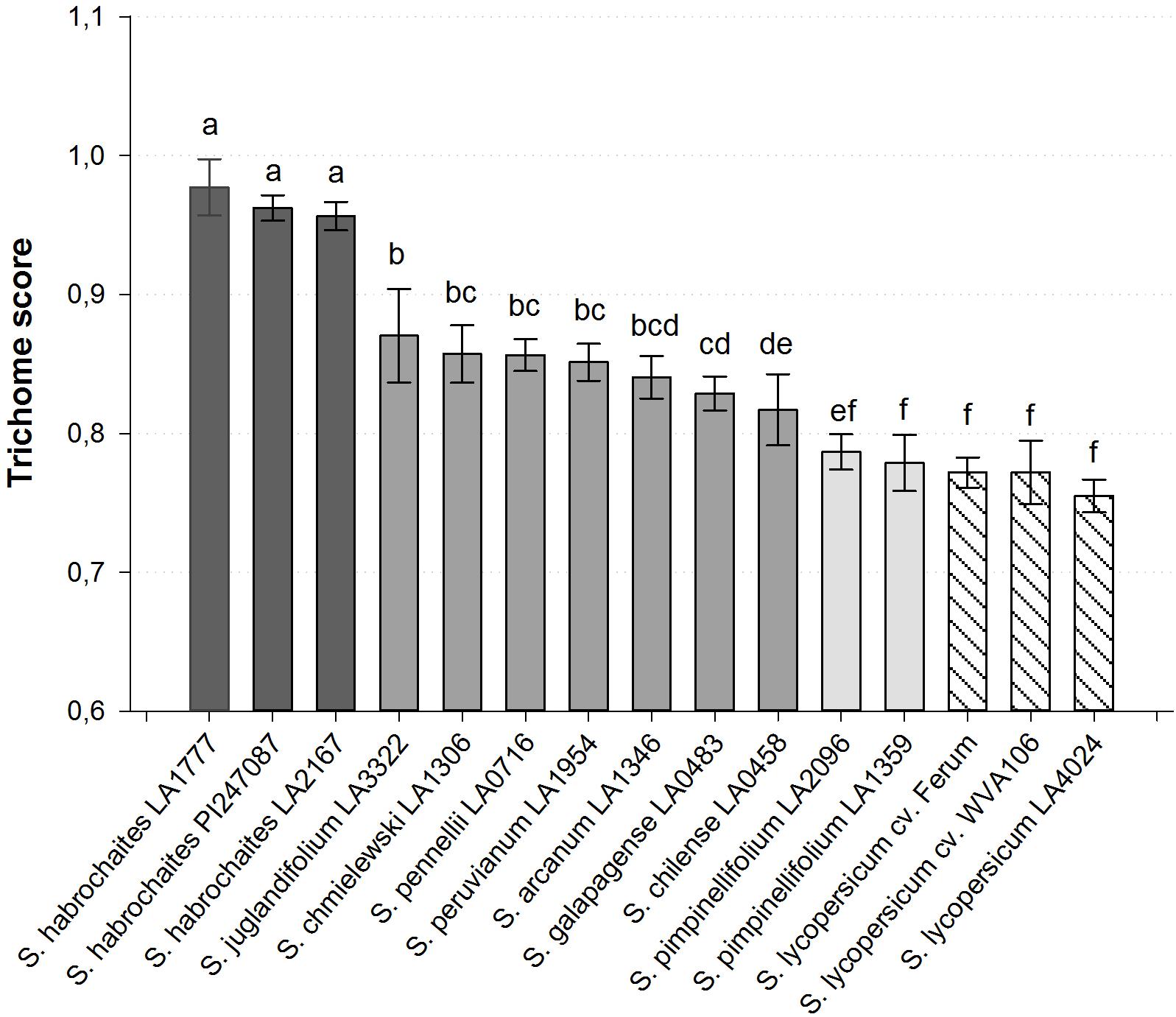
FIGURE 2. Quantified type VI trichome shape of various wild and cultivated tomato accessions. The trichome score was calculated with standard deviation for n ≥ 10 measured trichomes. All Solanum habrochaites accessions show a score greater 0.95, S. lycopersicum and S. pimpinellifolium accession accessions have a score of 0.76–0.79 and all other wild species score between 0.82 and 0.87. Bars with no common letters above them indicate a significantly different score (p < 0.001; one-way ANOVA, Tukey pairwise test).
Phenotyping a S. habrochaites × S. lycopersicum Backcross Population for Trichome Traits
In order to investigate the shape of type VI glandular trichomes in wild and cultivated tomatoes we created a backcross population (BC1) by crossing the wild S. habrochaites species LA1777 to the cultivated S. lycopersicum var. cerasiforme WVa106 tomato line, which was used as the recurrent parent. A total of 116 independent backcross lines were identified and confirmed by determining the trichome score, analyzing GC-MS profiles and preliminary genotyping for six independent loci.
The population shows a great diversity in the shape of the type VI glandular trichomes, which we could quantify by calculating the trichome score. The wild tomato LA1777 reaches a score of 0.972 with a standard derivation of 0.018 and the cultivated tomato scores 0.780 ± 0.022. While no backcross line reached scores as high as the wild parent, the highest ones scored about 0.92, around the same as the F1 crosses (0.912 ± 0.019), indicating that the morphology of S. habrochaites trichomes is a dominant trait. A few lines showed a phenotype comparable to the cultivated line but the great majority falls in-between the parents, forming a homogeneous gradient (Figure 3). According to the Anderson–Darling test the scores followed a normal distribution with a p-value of 0.594. To test whether the trichome score reflects the size of the internal cavity we selected four backcross lines with distinct trichome scores and estimated the size of their storage cavity, along with those of the two parental lines and an additional cultivar of tomato (S. lycopersicum LA4024). This was done by fluorescence microscopy, which allows us to clearly distinguish the cavity from the cells, by taking pictures where the area of the cavity is the largest. The area of the cavity expressed as percentage of the total area of the trichome head correlates well with the trichome score using a polynomial regression of the second degree (r2 = 0.93) (Supplementary Figure S3). Thus, the trichome score can be considered as a proxy for the size of the intercellular storage cavity of type VI trichomes. The backcross population was screened for the volatile composition. 42 of the 116 lines accumulated the LA1777 specific sesquiterpenes santalene and bergamotene (Sallaud et al., 2009), while the remaining 74 did not. When determining the trichome score of the two subsets, the population lines accumulating santalene scored slightly higher (0.872 ± 0.032) than the other lines (0.853 ± 0.032). A t-test shows that the difference is significant with p < 0.01.
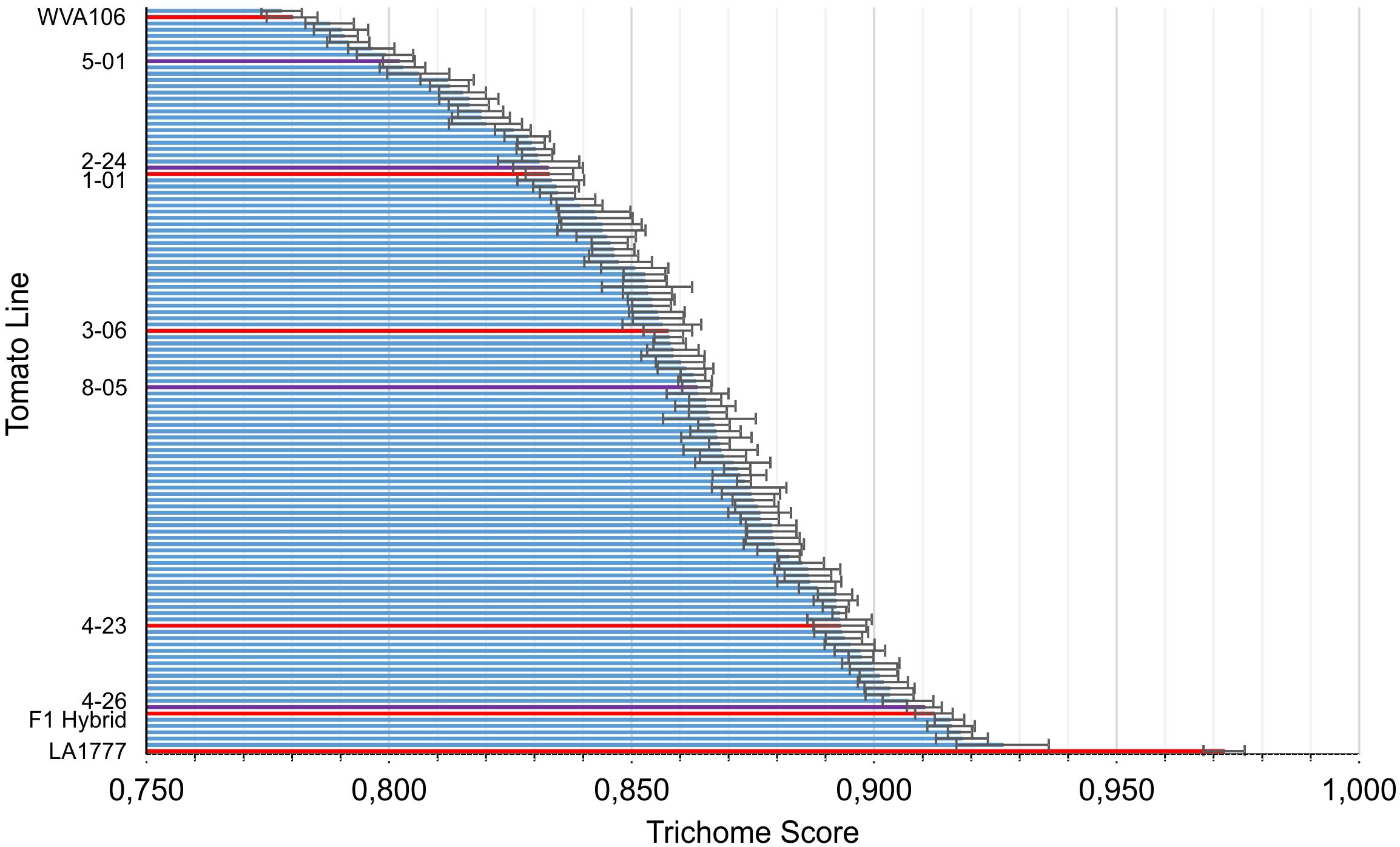
FIGURE 3. The trichome scores across the backcross population. The parental lines (WVa106 and LA1777) are located at the far ends of the scale. For each line, 6–12 trichome heads were scored. Backcross lines that are shown in Figure 1 and the F1 hybrid are highlighted in red. Backcross lines that are shown in Supplementary Figure S3 are highlighted in purple.
Mapping
To perform a QTL analysis to identify genetic regions contributing to the trichome head shape the backcross population was genotyped. This was achieved mainly by using High Resolution Melt analysis (HRM), a technique established to genotype plants (Li et al., 2010) and we supplemented the mapping with CAPS markers and iPLEX sequencingTM. This resulted in 192 markers (see complete list in Supplementary Table S1), which were plotted onto the ITAG2.4 version of the published tomato map (Tomato-Genome-Consortium, 2012). The whole genome is covered with at least 12 markers per chromosome, a maximum marker distance of 14.7 cM and an average of 5.2 cM. The map spans 933 cM in total. The mapping data is provided in Supplementary Table S2 and a graphic representation of the linkage map in Supplementary Figure S4. We note here that there is currently no complete and annotated genome assembly of S. habrochaites, and that the available genomic sequences of S. habrochaites have been aligned onto the reference genome of S. lycopersicum (Aflitos et al., 2014; Lin et al., 2014). Therefore, although there is overall good synteny between S. habrochaites and S. lycopersicum at least based on the observation of synaptonemal complexes (Anderson et al., 2010) and fluorescence in situ hybridization of bacterial artificial chromosomes (BAC-FISH) (Szinay et al., 2012), small chromosome rearrangements are possible. For example at the virus resistance Ty2 locus which comes from S. habrochaites an inversion was detected (Wolters et al., 2015). Thus, we cannot exclude that in the intervals between our markers, the gene order may not be the same.
The trichome scores were used in combination with the mapping data to perform a QTL analysis with the Windows QTL Cartographer software V2.5 using composite interval mapping (CIM) algorithm. This revealed six QTLs on four different chromosomes (Figure 4 and Table 1).
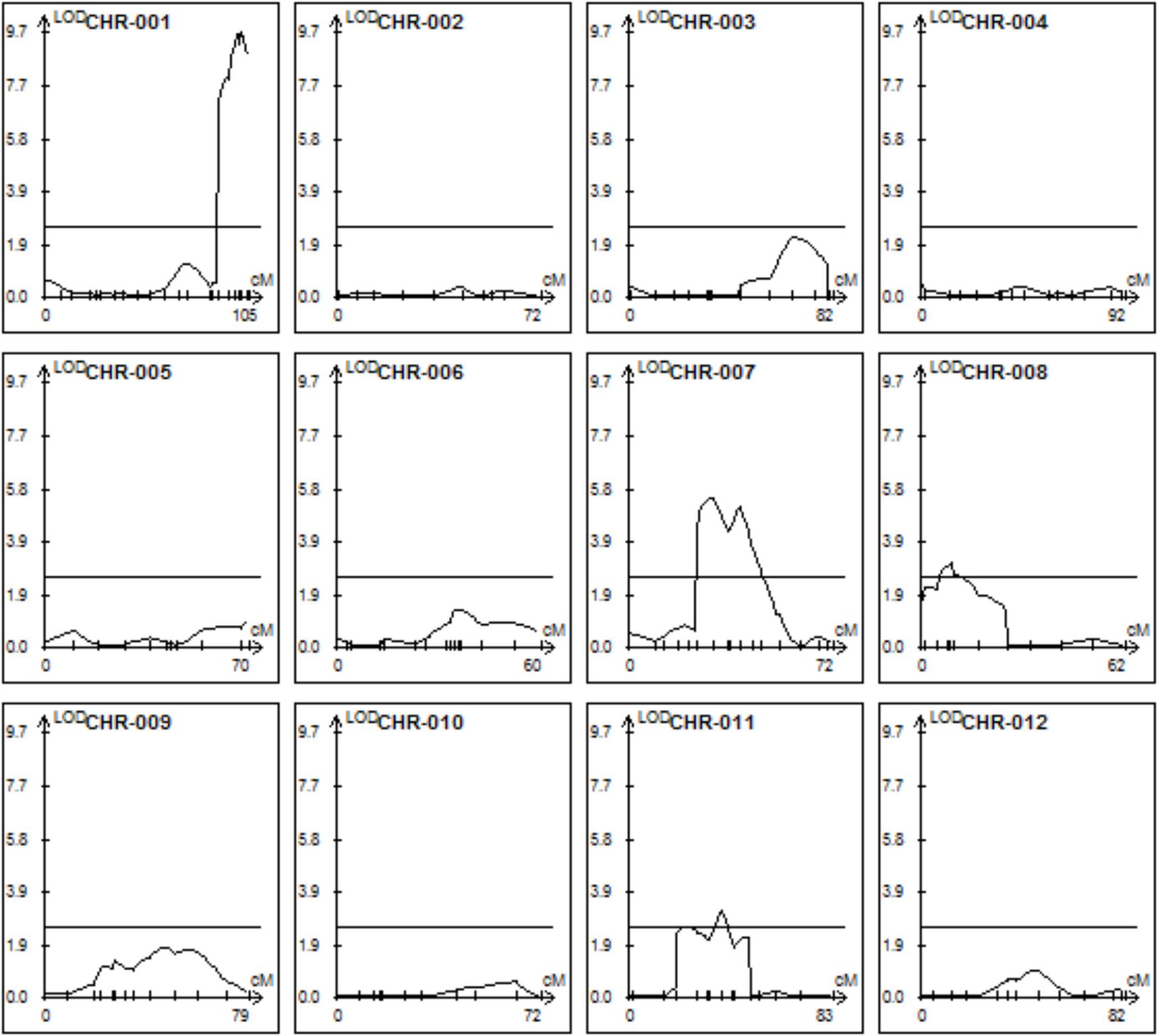
FIGURE 4. Quantitative trait locus (QTLs) determining trichome shape on the 12 tomato chromosomes. CIM mapping with a LOD threshold of 2.5.
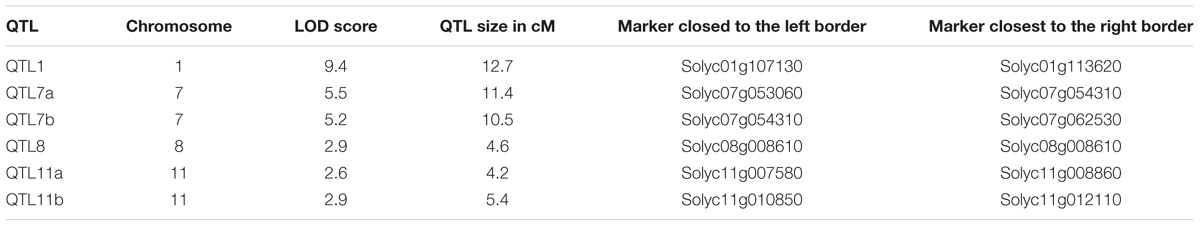
TABLE 1. Quantitative trait locus (QTLs) supporting trichome head shape identified using CIM mapping with a LOD threshold of 2.5.
All QTLs provide an additive effect with the strong QTL on chromosome I being by far the strongest one and thus the major contributor. In order to see if epistatic interactions underlie the trichome shape we used the QTLNetwork software version 2.1. No significant epistasis could be found.
Phenotyping and Mapping the Progeny of Selected BC Plants
To support the claim that a QTL on Chromosome I is the main contributor to the trichome shape we looked at a number of self-pollinated backcross lines. Line 4-06 showed one of the highest trichome scores with a value of 0.896 ± 0.011 and was heterozygous for markers covering the associated QTLs on chromosomes I and VII. The 98 individual self-pollinated progeny of 4-06 shows again a segregating trichome score distribution comparable to the backcross population itself with some lines scoring at the cultivated level and some lines around 0.9 but no line reaching wild type levels. When genotyping the QTL on chromosome I we found that all of the 40 selfed plants with the highest trichome scores carry at least one copy of the wild DNA at the QTL on chromosome I while none of the lowest scoring lines is homozygous for LA1777 and 8 of the 10 the lowest scoring lines are homozygous cultivated. The nine top scoring lines are all homozygous wild for at least one of the two strongest QTLs on chromosome I or VII. The average trichome score for the lines homozygous for the S. habrochaites markers within the QTL on Chromosome I is 0.873 ± 0.024, significantly higher (t-test: p-value < 0.05) than the heterozygous (0.856 ± 0.025) or homozygous cultivated lines (0.829 ± 0.023). Unfortunately, no novel recombination event could be detected in the QTL regions on either chromosome I or VII.
In contrast, the backcross lines 2–11 had a low trichome score of 0.831 ± 0.020 and was homozygous for S. lycopersicum markers over the QTL on chromosome I. Individuals of the self-pollinated progeny (n = 67) do not have higher trichome scores, with an average TS of 0.828 ± 0.028.
Prospective Candidate Genes in the Identified QTLs
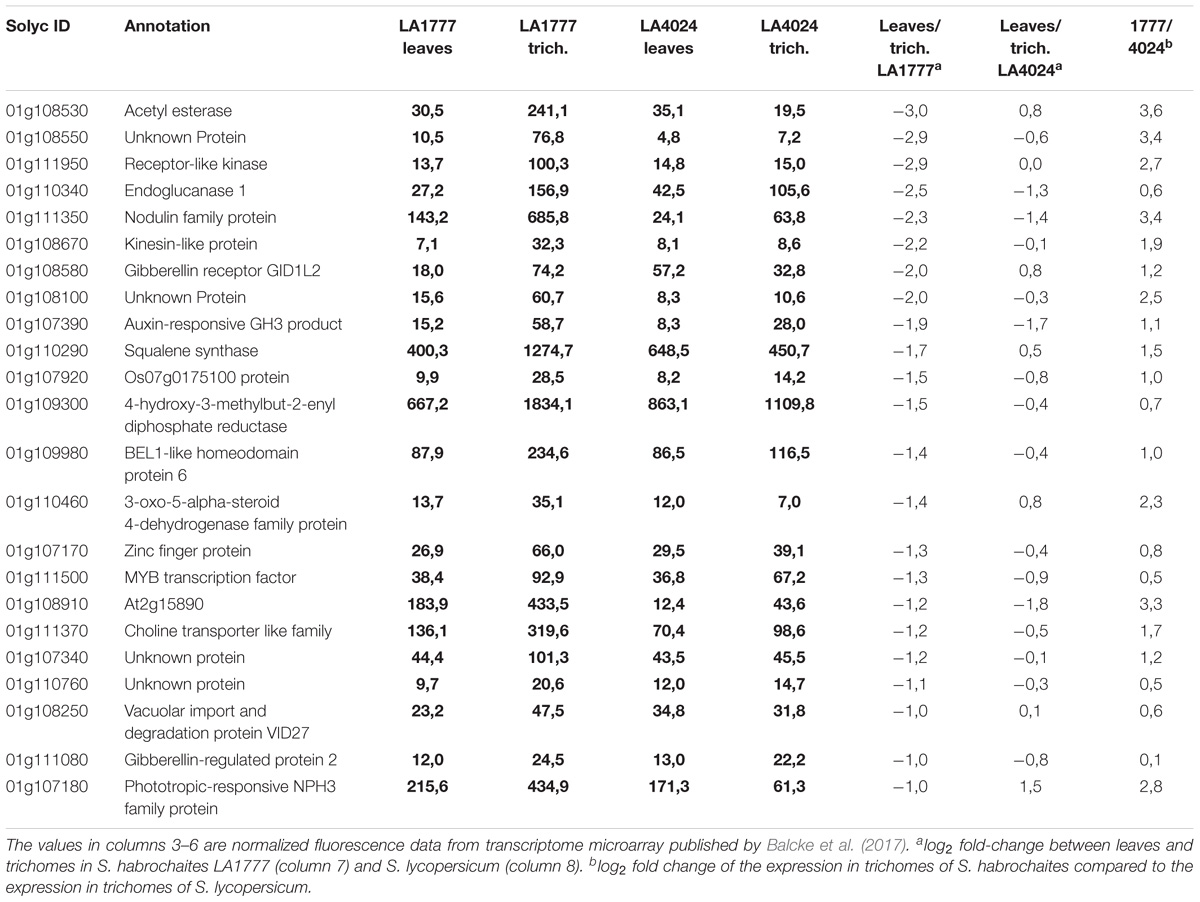
The major QTL on chromosome I spans a genetic interval of 12.7 cM in a relatively gene rich region containing 370 genes (see Supplementary Table S3). Assuming that genes responsible for the different shape of type VI trichomes in S. habrochaites would be likely to be on the one hand more strongly expressed in trichomes and on the other hand more strongly expressed in LA1777 compared to LA4024, a reduced list of potential candidate genes can be established (see Table 2). The 22 genes thus selected encode proteins with quite diverse functional classes, including four unknown proteins, three transcription factors, two cell wall metabolism enzymes, and four metabolic enzymes.
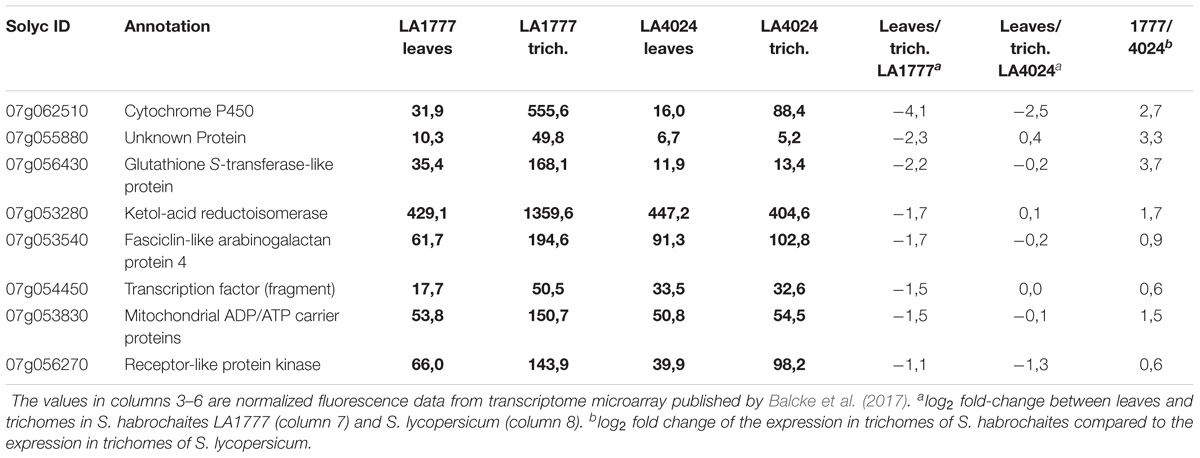
For the QTL on chromosome VII, this selection leads to only eight genes, with diverse functional annotations, including a transcription factor, a cell wall protein, two metabolic enzymes (P450 and a ketol-acid reductoisomerase), an ATP/ADP carrier protein and a receptor kinase (see Table 3).
Discussion
The shape of the glandular type VI trichomes is significantly different between wild and cultivated tomato species. In particular, the S. habrochaites accessions have a spherical trichome head while cultivated tomatoes like S. lycopersicum var. cerasiforme WVa106 have the shape of a four-leaf clover. To quantify this trait we designed a trichome score that reflects the presence of constrictions between the glandular cells in the cultivated tomato. We could show that the shape is a quantifiable trait. All other tested wild tomato species such as S. pennellii, S. chilense or S. arcanum show an intermediate trichome phenotype closer to S. lycopersicum than to S. habrochaites but clearly distinguishable from both. S. lycopersicum and S. pimpinellifolium accessions, including S. lycopersicum var. cerasiforme WVa 106 which is considered to be an admixture of S. lycopersicum and S. pimpinellifolium (Ranc et al., 2008). Notably, S. habrochaites is the only tomato species we could identify with this typical spherical type VI trichome shape. This raises the question of the evolution of the type VI trichomes within the tomato genus: was the spherical type VI trichome an ancestral form, which was conserved in S. habrochaites and lost in the other species, or on the contrary is it a specific evolution of S. habrochaites? S. habrochaites, together with species of the S. arcanum group is more closely related to S. lycopersicum than S. peruvianum or S. chilense for example (Zuriaga et al., 2009). This would support the hypothesis of a recent evolution of this trait in S. habrochaites rather than a loss in the other species.
We created a BC1 population of the S. habrochaites LA1777 and S. lycopersicum WVa 106 parents and could quantify the shape of the trichome heads using the trichome score. This showed that the shape of the different lines based on this score follows a normal distribution, with the parents on either ends, indicating that there are multiple genes contributing to type VI trichome shape determination. None of the backcross lines reaches a score comparable to that of S. habrochaites, neither do the F1 plants. As, by design, none of the backcross lines are homozygous for the S. habrochaites genotype in any genetic loci, a gene dosage effect might play a role in preventing the backcross lines from reaching wild type like scores. However, in the progeny of one of the BC lines, plants homozygous for the QTLs on chromosomes I and VII showed high trichome scores but still not as high as that of S. habrochaites. The absence of S. habrochaites homozygosis also prevents the detection of any recessive trait originating from the wild parent.
The extensive sequence information available for both parents allowed a mapping which is essentially only limited by the population size and the recombination rates. Although the genomes of S. habrochaites and S. lycopersicum are largely syntenic (Anderson et al., 2010; Szinay et al., 2012), there are multiple lines of evidence for the presence of chromosome rearrangements, such as inversions, deletions, or mispositioning of kinetochores as shown by the observation of synaptonemal complexes of hybrids (Anderson et al., 2010). The sequencing and detailed annotation of some specific loci of S. habrochaites confirmed this. For example the Cf-4 locus that confers resistance to Cladosporium fulvum contains an expanded set of genes encoding Leucine Rich Repeat proteins on chromosome 1 (Parniske et al., 1997) and the locus on chromosome 8 that contains the santalene/bergamotene synthase gene shows significant differences with the homologous locus from S. lycopersicum (Matsuba et al., 2013). Perhaps most problematic for mapping is the presence of inversions as shown in the case of the Ty2 locus, which confers resistance to the tomato yellow leaf curl virus (Wolters et al., 2015). Such inversions lead to significantly reduced recombination and therefore prevent high-resolution mapping. Because recombination in our QTL, particularly the one on chromosome 1 seems to be suppressed, we cannot exclude that the order or number of the genes in these intervals in S. habrochaites is different to those of S. lycopersicum. The QTL analysis reveals that there are several QTLs contributing to the spherical trichome head. Three of the six QTLs have an LOD score only slightly above the defined significance level of 2.5, indicating that the actual contribution of these loci to type VI trichome shape has to be taken with caution. Two closely located QTLs have been predicted on both chromosomes VII and XI. However, a higher population size and finer mapping would be needed to fully exclude the possibility of there only being one and not two QTLs.
The strongest QTL is located on chromosome I and with the current mapping encompasses about 500 annotated genes (see Supplementary Tables S3, S4). Based on RNA sequencing data of leaves and trichomes previously published (Balcke et al., 2017), we could identify 370 genes expressed in trichomes and leaves of LA1777. Of these, only 23 genes have a more than twofold higher abundance in isolated trichomes compared to whole leaves in LA1777 and with a higher expression in LA1777 trichomes than in LA4024 trichomes (Table 2). One of the strongest regulated genes is Solyc01g110340.2.1, coding for an endoglucanase. This cell wall modifying enzyme is a good candidate as the formation of the intercellular storage cavity, which appears to be correlated to the round shape of the type VI trichomes, is likely to involve cell wall remodeling. In addition, there are two highly regulated transcription factors of the homeodomain (Solyc01g109980.2.1) and MYC (Solyc01g111500.2.1) families as well a receptor-like kinase (Solyc01g111950.2.1). The closest Arabidopsis homolog of Solyc01g109980 is BLH6 (BEL-LIKE HOMEODOMAIN6), which was shown to be involved in a negative regulatory complex of secondary cell wall (Liu et al., 2014). Since the outer envelope of the trichomes is likely to play a determining role in the shape, this is a potentially interesting candidate. Solyc01g111500 encodes a MYB factor whose closest Arabidopsis homolog is AtMYB4 (At4g38620), a transcriptional repressor of phenolics biosynthesis. A closely related MYB homolog from maize (Zm 38) is involved in repression of anthocyanin biosynthesis (Franken et al., 1994), indicating this group of MYB factors share conserved functions in the repression of phenolic compounds. Such functions could also have an impact on the formation of the outer envelope of the trichomes by regulating its phenolic content. Another potentially interesting candidate is the gene encoding 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR), the last enzyme of the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway, which supplies isoprenyl-diphosphate precursors for terpenoids in the plastids. It is significantly more expressed in trichomes of LA1777, which is in line with the larger amounts of terpenes produced in trichomes this species. Overexpression of genes involved in the biosynthesis of methylketones in S. habrochaites sp. glabratum, which also has round type VI trichomes, has been observed by Ben-Israel et al. (2009). Thus, one possibility is that the flux of metabolites exported into the cavity contributes to the shape of the trichome. However, unlike deoxy-xylulose phosphate synthase (DXS), HDR is not known to be a regulatory bottleneck in the flux through the MEP pathway. Also, Ben-Israel et al. (2009) proposed another explanation, namely that the biosynthesis of MKs, which are fatty acid derivatives, diverts a pool of fatty acid acids away from the very long chain fatty acids that are incorporated in the cuticle, thereby influencing the shape of the trichome. In the case of S. habrochaites LA1777, this is unlikely to be the case because sesquiterpenoids are the major products of type VI trichomes and therefore should not alter the composition of the cuticle. Another potentially interesting candidate is Solyc01g108670.2.1, which encodes a protein of the phragmoplast-associated kinesin family. In Arabidopsis there are two homologs of this type of kinesin, PHRAGMOPLAST ORIENTING KINESIN 1 and 2, whose combined mutation leads to defects in the orientation of the cell division plane (Lipka et al., 2014).
With the same criteria, only seven genes from the QTL on chromosome VII could be retained (Table 3). There is one transcription factor (Solyc07g054450.2.1) with similarity to ULTRAPETALA1, a repressor of inflorescence and flower meristem size, a receptor like kinase (Solyc07g056270.2.1), a fasciclin-like arabinogalactan protein (Solyc07g053540.1.1) and a protein of unknown function (Solyc07g055880.1.1). The gene with the strongest differential expression (Solyc07g062510.2.1) encodes a cytochrome P450 oxidase of the CYP72 family, whose function is, however, unclear. Also of note in this list is the presence of a ketol-acid reductoisomerase, an enzyme involved in branched chain amino acid and in pantothenate and coenzyme A biosynthesis. How these genes could contribute in any way, if any, to type VI trichome shape is unclear and will need further functional analysis.
Furthermore, because the trichome preparation used for our transcriptome data is made up of mature trichomes and the shape of the trichomes is a developmental trait, it is possible that we are missing genes that are only expressed in a specific stage of trichome development. We recently described a method to isolate such young trichomes (Bergau et al., 2016). Transcriptome data from these developing trichomes is being produced and will be used to mine for differentially expressed genes that are located in the QTLs identified in this study.
Identification of the causal gene(s) will require a combination of refined mapping, use of developmental stage-specific transcriptomics, and functional testing of the genes, for example by virus-induced gene silencing as shown by (Besser et al., 2009) or by gene inactivation with CRISPR/Cas9 (Ito et al., 2015).
All QTLs detected here contribute in the same direction and the additive effects toward a rounder trichome head are driven by the presence of the S. habrochaites alleles within the QTLs. This is mirrored by the fact that S. habrochaites lines show the highest and S. lycopersicum the lowest trichome scores. We could not detect strong epistasis effects regulating the QTLs showing the contributors are independent from each other. These results suggest that the type VI trichome phenotype is mostly contributed by the dominant expression of genes from the S. habrochaites genome and that these different genes are not part of a unique pathway but individually contribute to this phenotype.
The effect of the QTL on chromosome I could be confirmed by genotyping the self-pollinated progeny of two backcross lines. A BC1 line with a low trichome score and without S. habrochaites genetic information within the QTL produced offspring with a low score, while a BC1 line that was heterozygous for S. habrochaites markers spanning the chromosome I QTL produced offspring with a further segregating trichome phenotype with the top-scoring plants retaining the S. habrochaites genotype on Chromosome I and the bottom-scoring plants losing it. Even in plants that were homozygous for S. habrochaites markers within the QTL, the trichome score did not reach S. habrochaites levels, confirming that there are additional contributing factors.
One of the factors might be the accumulation of sesquiterpenoids derived from α-santalene and various forms of bergamotene. While S. lycopersicum does not make these compounds but instead essentially monoterpenes, they are accumulating to high amounts (over 10 mg/g leaf fresh weight) in S. habrochaites LA1777 (Coates et al., 1988). The enzymes needed to synthesize santalene and bergamotene in LA1777 for example are located in a cluster on chromosome VIII (Sallaud et al., 2009), close to the weak QTL identified on that chromosome. Furthermore, lines in the backcross population accumulating santalene have a slightly higher trichome score compared to lines that do not accumulate the sesquiterpene. The accumulation of these secondary metabolites within the trichome heads could lead to an increased pressure thereby influencing the trichome shape and thereby the trichome score we measure. A similar hypothesis was proposed in a previous study investigating the correlation between the type VI trichome shape and genes involved in the biosynthesis of methylketones in S. habrochaites ssp. glabratum (Ben-Israel et al., 2009). It should be noted, however, that this does not appear to be a major contributing factor and that the major QTLs we identified on chromosome I and VII do not contain genes that are involved in the biosynthesis of these sesquiterpenoids.
Our findings show that the shape of glandular trichomes in tomato is a quantifiable trait. The genes behind these morphological differences remain elusive, but additional transcriptomics data from developing trichomes combined with medium-throughput VIGS or targeted deletions of sub-intervals of the QTL with genome-editing tools should help us identify the causal genes. This should provide crucial data to breed trichome-based insect resistance traits in cultivated tomato.
Author Contributions
SB performed all the experiments except the fluorescence microscopy. NB performed the fluorescence microscopy. AT conceived the project and contributed to the experimental design. SB and AT analyzed the results and wrote the manuscript. All authors read and approved the manuscript before submission.
Funding
This project was funded in part by grant number TI 800/1 from the Deutsche Forschungsgemeinschaft to AT.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tina Clauss for help in making the crosses between LA1777 and WVa 106 and the greenhouse staff of the IPB, Petra Janssen, Philip Plato, and Sabine Voigt for assistance in maintaining the back-cross population. We also thank Prof. Albrecht Melchinger for advice on QTL mapping.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01421/full#supplementary-material
FIGURE S1 | Fluorescence microscopy of isolated type VI trichomes from Solanum habrochaites LA1777 and S. lycopersicum WVa106.
FIGURE S2 | Examples of type VI trichome shape from several wild tomato species, other than S. habrochaites.
FIGURE S3 | Correlation between trichome score and size of the intercellular storage cavity. Type VI trichomes images were taken with a laser-scanning confocal microscope (see section “Materials and Methods”) at the z-position where the area of the cavity is the largest. The scale bar represents 20 μm. These images were used to calculate the trichome score (see Figure 1) and the area of the cavity. Ten images were taken per line, except for Wva106 where three images were taken. The area of the cavity was then normalized to the total area of the trichome. The data were then plotted and curve-fitted in Sigma Plot version 10. The best fit was obtained with a second degree polynomial curve (see equation on the graph) with a r2 of 0.93. The dashed line represents the fitted curve. The solid lines represent the curves for a 99% confidence interval.
FIGURE S4 | Linkage map with the relative (on the left; in cM) and absolute (on the right; in MB) positioning of the markers.
TABLE S1 | Molecular markers used in this study.
TABLE S2 | Mapping data.
TABLE S3 | Genes of the QTL on chromosome I.
TABLE S4 | Examples of polymorphisms found in 5 genes randomly selected in the QTL of chromosome 1.
Footnotes
References
Aflitos, S., Schijlen, E., de Jong, H., de Ridder, D., Smit, S., Finkers, R., et al. (2014). Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 80, 136–148. doi: 10.1111/tpj.12616
Anderson, L. K., Covey, P. A., Larsen, L. R., Bedinger, P., and Stack, S. M. (2010). Structural differences in chromosomes distinguish species in the tomato clade. Cytogenet. Genome Res. 129, 24–34. doi: 10.1159/000313850
Balcke, G. U., Bennewitz, S., Bergau, N., Athmer, B., Henning, A., Majovsky, P., et al. (2017). Multi-omics of tomato glandular trichomes reveals distinct features of central carbon metabolism supporting high productivity of specialized metabolites. Plant Cell 29, 960–983. doi: 10.1105/tpc.17.00060
Ben-Israel, I., Yu, G., Austin, M. B., Bhuiyan, N., Auldridge, M., Nguyen, T., et al. (2009). Multiple biochemical and morphological factors underlie the production of methylketones in tomato trichomes. Plant Physiol. 151,1952–1964. doi: 10.1104/pp.109.146415
Bergau, N., Bennewitz, S., Syrowatka, F., Hause, G., and Tissier, A. (2015). The development of type VI glandular trichomes in the cultivated tomato Solanum lycopersicum and a related wild species S. habrochaites. BMC Plant Biol. 15:289. doi: 10.1186/s12870-015-0678-z
Bergau, N., Navarette Santos, A., Henning, A., Balcke, G. U., and Tissier, A. (2016). Autofluorescence as a signal to sort developing glandular trichomes by flow cytometry. Front. Plant Sci. 7:949. doi: 10.3389/fpls.2016.00949
Besser, K., Harper, A., Welsby, N., Schauvinhold, I., Slocombe, S., Li, Y., et al. (2009). Divergent regulation of terpenoid metabolism in the trichomes of wild and cultivated tomato species. Plant Physiol. 149, 499–514. doi: 10.1104/pp.108.126276
Brown, G. D. (2010). The biosynthesis of artemisinin (Qinghaosu) and the phytochemistry of Artemisia annua L. (Qinghao). Molecules 15, 7603–7698. doi: 10.3390/molecules15117603
Brückner, K., Bozic, D., Manzano, D., Papaefthimiou, D., Pateraki, I., Scheler, U., et al. (2014). Characterization of two genes for the biosynthesis of abietane-type diterpenes in rosemary (Rosmarinus officinalis) glandular trichomes. Phytochemistry 101, 52–64. doi: 10.1016/j.phytochem.2014.01.021
Channarayappa, C., Shivashankar, G., Muniyappa, V., and Frist, R. H. (1992). Resistance of Lycopersicum species to Bemisia tabaci, a tomato leaf curl virus vector. Can. J. Bot. 70, 2184–2192. doi: 10.1139/b92-270
Coates, R. M., Denissen, J. F., Juvik, J. A., and Babka, B. A. (1988). Identification of alpha-santalenoic and endo-beta-bergamotenoic acids as moth oviposition stimulants from wild tomato leaves. J. Org. Chem. 53, 2186–2192. doi: 10.1021/jo00245a012
Croteau, R. B., Davis, E. M., Ringer, K. L., and Wildung, M. R. (2005). (−)-Menthol biosynthesis and molecular genetics. Naturwissenschaften 92, 562–577. doi: 10.1007/s00114-005-0055-0
Dal Cin, V., Kevany, B., Fei, Z., and Klee, H. J. (2009). Identification of Solanum habrochaites loci that quantitatively influence tomato fruit ripening-associated ethylene emissions. Theor. Appl. Genet. 119, 1183–1192. doi: 10.1007/s00122-009-1119-x
Fahn, A. (2000). “Structure and function of secretory cells,” in Plant Trichomes, eds D. L. Hallahan and J. C. Gray (New York, NY: Academic Press), 37.
Fan, P., Miller, A. M., Schilmiller, A. L., Liu, X., Ofner, I., Jones, A. D., et al. (2016). In vitro reconstruction and analysis of evolutionary variation of the tomato acylsucrose metabolic network. Proc. Natl. Acad. Sci. U.S.A. 113, E239–E248. doi: 10.1073/pnas.1517930113
FAOSTAT (2017). The Food and Agricultural Organization of the United Nations. Avaiable at: http://www.fao.org/faostat/en/#data/QC [accessed April 04, 2017].
Firdaus, S., van Heusden, A. W., Hidayati, N., Supena, E. D., Mumm, R., de Vos, R. C., et al. (2013). Identification and QTL mapping of whitefly resistance components in Solanum galapagense. Theor. Appl. Genet. 126, 1487–1501. doi: 10.1007/s00122-013-2067-z
Franken, P., Schrell, S., Peterson, P. A., Saedler, H., and Wienand, U. (1994). Molecular analysis of protein domain function encoded by the myb-homologous maize genes C1, Zm 1 and Zm 38. Plant J. 6, 21–30. doi: 10.1046/j.1365-313X.1994.6010021.x
Glas, J. J., Schimmel, B. C., Alba, J. M., Escobar-Bravo, R., Schuurink, R. C., and Kant, M. R. (2012). Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. Int. J. Mol. Sci. 13, 17077–17103. doi: 10.3390/ijms131217077
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M., and Toki, S. (2015). CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467, 76-82. doi: 10.1016/j.bbrc.2015.09.117
Kang, J. H., Campos, M. L., Zemelis-Durfee, S., Al-Haddad, J. M., Jones, A. D., Telewski, F. W., et al. (2016). Molecular cloning of the tomato hairless gene implicates actin dynamics in trichome-mediated defense and mechanical properties of stem tissue. J. Exp. Bot. 67, 5313–5324. doi: 10.1093/jxb/erw292
Kang, J. H., Liu, G. H., Shi, F., Jones, A. D., Beaudry, R. M., and Howe, G. A. (2010). The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol. 154, 262–272. doi: 10.1104/pp.110.160192
Kang, J. H., McRoberts, J., Shi, F., Moreno, J. E., Jones, A. D., and Howe, G. A. (2014). The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 164, 1161–1174. doi: 10.1104/pp.113.233395
Lander, E., and Kruglyak, L. (1995). Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11, 241–247. doi: 10.1038/ng1195-241
Leckie, B. M., De Jong, D. M., and Mutschler, M. A. (2012). Quantitative trait loci increasing acylsugars in tomato breeding lines and their impacts on silverleaf whiteflies. Mol. Breed. 30, 1621–1634. doi: 10.1007/s11032-012-9746-3
Li, S. F., Milliken, O. N., Pham, H., Seyit, R., Napoli, R., Preston, J., et al. (2009). The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21, 72–89. doi: 10.1105/tpc.108.063503
Li, Y. D., Chu, Z. Z., Liu, X. G., Jing, H. C., Liu, Y. G., and Hao, D. Y. (2010). A cost-effective high-resolution melting approach using the EvaGreen dye for DNA polymorphism detection and genotyping in plants. J. Integr. Plant Biol. 52, 1036–1042. doi: 10.1111/j.1744-7909.2010.01001.x
Lin, T., Zhu, G. T., Zhang, J. H., Xu, X. Y., Yu, Q. H., Zheng, Z., et al. (2014). Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46, 1220–1226. doi: 10.1038/ng.3117
Lipka, E., Gadeyne, A., Stockle, D., Zimmermann, S., De Jaeger, G., Ehrhardt, D. W., et al. (2014). The phragmoplast-orienting kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell 26, 2617–2632. doi: 10.1105/tpc.114.124933
Liu, Y., You, S., Taylor-Teeples, M., Li, W. L., Schuetz, M., Brady, S. M., and Douglas, C. J. (2014). Bel1-like homeodomain6 and knotted Arabidopsis thaliana 7 interact and regulate secondary cell wall formation via repression of revoluta. Plant Cell 26, 4843–4861. doi: 10.1105/tpc.114.128322
Lloyd, C. (2011). Dynamic microtubules and the texture of plant cell walls. Int. Rev. Cell Mol. Biol. 287, 287–329. doi: 10.1016/B978-0-12-386043-9.00007-4
Luckwill, L. C. (1943). The genus Lycopersicon: a historical, biological and taxonomic survey of the wild and cultivated tomato. Aberd. Univ. Stud. 120, 1–44.
Maes, L., Van Nieuwerburgh, F. C. W., Zhang, Y. S., Reed, D. W., Pollier, J., Casteele, S., et al. (2011). Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol. 189, 176–189. doi: 10.1111/j.1469-8137.2010.03466.x
Majdi, M., Malekzadeh-Mashhady, A., Maroufi, A., and Crocoll, C. (2017). Tissue-specific gene-expression patterns of genes associated with thymol/carvacrol biosynthesis in thyme (Thymus vulgaris L.) and their differential changes upon treatment with abiotic elicitors. Plant Physiol. Biochem. 115, 152–162. doi: 10.1016/j.plaphy.2017.03.016
Matsuba, Y., Nguyen, T. T., Wiegert, K., Falara, V., Gonzales-Vigil, E., Leong, B., et al. (2013). Evolution of a complex locus for terpene biosynthesis in Solanum. Plant Cell 25, 2022–2036. doi: 10.1105/tpc.113.111013
McDowell, E. T., Kapteyn, J., Schmidt, A., Li, C., Kang, J. H., Descour, A., et al. (2011). Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol. 155, 524–539. doi: 10.1104/pp.110.167114
Micheli, F. (2001). Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6, 414–419. doi: 10.1016/S1360-1385(01)02045-3
Momotaz, A., Scott, J. W., and Schuster, D. J. (2010). Identification of quantitative trait loci conferring resistance to Bemisia tabaci in an F-2 population of Solanum lycopersicum x Solanum habrochaites accession LA1777. J. Am. Soc. Horticult. Sci. 135, 134–142.
Parniske, M., Hammond-Kosack, K. E., Golstein, C., Thomas, C. M., Jones, D. A., Harrison, K., et al. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91, 821–832. doi: 10.1016/S0092-8674(00)80470-5
Pattanaik, S., Patra, B., Singh, S. K., and Yuan, L. (2014). An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front. Plant Sci. 5:259. doi: 10.3389/fpls.2014.00259
Ranc, N., Munos, S., Santoni, S., and Causse, M. (2008). A clarified position for Solanum lycopersicum var. cerasiforme in the evolutionary history of tomatoes (solanaceae). BMC Plant Biol. 8:130. doi: 10.1186/1471-2229-8-130
Rozen, S., and Skaletsky, H. J. (2000). “Primer3 on the WWW for general users and for biologist programmers,” in Bioinformatics Methods and Protocols: Methods in Molecular Biology, eds S. Misener and S. A. Krawetz (New York, NY: Humana Press Inc.), 365–386.
Sallaud, C., Giacalone, C., Topfer, R., Goepfert, S., Bakaher, N., Rosti, S., et al. (2012). Characterization of two genes for the biosynthesis of the labdane diterpene Z-abienol in tobacco (Nicotiana tabacum) glandular trichomes. Plant J. 72, 1–17. doi: 10.1111/j.1365-313X.2012.05068.x
Sallaud, C., Rontein, D., Onillon, S., Jabes, F., Duffe, P., Giacalone, C., et al. (2009). A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21, 301–317. doi: 10.1105/tpc.107.057885
Scheler, U., Brandt, W., Porzel, A., Rothe, K., Manzano, D., Božiæ, D., et al. (2016). Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast. Nat. Commun. 7:12942. doi: 10.1038/ncomms12942
Schilmiller, A. L., Gilgallon, K., Ghosh, B., Jones, A. D., and Last, R. L. (2016). Acylsugar acylhydrolases: carboxylesterase-catalyzed hydrolysis of acylsugars in tomato trichomes. Plant Physiol. 170,1331–1344.
Schilmiller, A. L., Last, R. L., and Pichersky, E. (2008). Harnessing plant trichome biochemistry for the production of useful compounds. Plant J. 54, 702–711. doi: 10.1111/j.1365-313X.2008.03432.x
Schilmiller, A. L., Schauvinhold, I., Larson, M., Xu, R., Charbonneau, A. L., Schmidt, A., et al. (2009). Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl. Acad. Sci. U.S.A. 106, 10865–10870. doi: 10.1073/pnas.0904113106
Serna, L., and Martin, C. (2006). Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 11, 274–280. doi: 10.1016/j.tplants.2006.04.008
Szinay, D., Wijnker, E., van den Berg, R., Visser, R. G., de Jong, H., and Bai, Y. (2012). Chromosome evolution in Solanum traced by cross-species BAC-FISH. New Phytol. 195, 688–698. doi: 10.1111/j.1469-8137.2012.04195.x
Tissier, A. (2012). Glandular trichomes: what comes after expressed sequence tags? Plant J 70, 51–68. doi: 10.1111/j.1365-313X.2012.04913.x
Tomato-Genome-Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Van Sandt, V. S., Guisez, Y., Verbelen, J. P., and Vissenberg, K. (2007). Xyloglucan endotransglycosylase/hydrolase (XTH) is encoded by a multi-gene family in the primitive vascular land plant Selaginella kraussiana. Plant Biol. 9, 142–146. doi: 10.1055/s-2006-924661
Wang, S., Basten, C. J., and Zeng, Z.-B. (2012). Windows QTL Cartographer 2.5. Raleigh, NC: Department of statistics, North Carolina State University.
Wittwer, C. T., Reed, G. H., Gundry, C. N., Vandersteen, J. G., and Pryor, R. J. (2003). High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 49, 853–860. doi: 10.1373/49.6.853
Wolters, A.-M. A., Caro, M., Dong, S., Finkers, R., Gao, J., Visser, R. G. F., et al. (2015). Detection of an inversion in the Ty-2 region between S. lycopersicum and S. habrochaites by a combination of de novo genome assembly and BAC cloning. Theor. Appl. Genet. 128, 1987–1997. doi: 10.1007/s00122-015-2561-6
Yang, J., Hu, C., Hu, H., Yu, R., Xia, Z., Ye, X., et al. (2008). QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24, 721–723. doi: 10.1093/bioinformatics/btm494
Keywords: tomato, type VI glandular trichomes, trichome morphology, Solanum habrochaites, Solanum lycopersicum, QTL mapping
Citation: Bennewitz S, Bergau N and Tissier A (2018) QTL Mapping of the Shape of Type VI Glandular Trichomes in Tomato. Front. Plant Sci. 9:1421. doi: 10.3389/fpls.2018.01421
Received: 30 April 2018; Accepted: 06 September 2018;
Published: 26 September 2018.
Edited by:
Delphine Laurinda Fleury, University of Adelaide, AustraliaReviewed by:
Jose M. Jimenez-Gomez, INRA UMR1318 Institut Jean-Pierre Bourgin, FranceJohn Larkin, Louisiana State University, United States
Copyright © 2018 Bennewitz, Bergau and Tissier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alain Tissier, alain.tissier@ipb-halle.de orcid.org/0000-0002-9406-4245
†Present address: Stefan Bennewitz, VEROVACCiNES GmbH, Halle, Germany
 Stefan Bennewitz†
Stefan Bennewitz† Alain Tissier
Alain Tissier