- 1Strategic Innovation Platform, International Rice Research Institute, Metro Manila, Philippines
- 2Rice Breeding Platform, International Rice Research Institute, Metro Manila, Philippines
- 3National Institute of Crop Science, Rural Development Administration, Jeonju, South Korea
Adaptation of temperate japonica rice varieties to tropical regions is impeded by extremely early flowering probably due to photoperiod change from long to short. However, constant breeding efforts led to development of temperate japonica varieties adapted to tropical/subtropical regions, but the genetic factor underlying this is still elusive. We analyzed the 45 diverse rice accessions and 12 tropical-adapted temperate japonica lines for the allele types of seven major flowering genes Hd1, OsPPR37, DTH8, Ghd7, Ehd1, RFT1, and Hd3a and flowering time under three different field conditions in temperate and tropical locations. The accessions originated from the tropical/subtropical regions preferred the non-functional alleles of Hd1 and not other flowering genes. The genetic effect analysis of each gene showed that only the functional Hd1 caused early flowering in the tropical location. All 12 temperate japonica breeding lines adapted to the tropics possessed the loss-of-function alleles of Hd1 with no change of other flowering genes compared to common Korean temperate japonica varieties. A phylogenetic analysis using 2,918 SNP data points revealed that the genome status of the 12 breeding lines were very similar to Korean temperate japonica varieties. These results indicate that the functional Hd1 alleles of temperate japonica varieties induced extremely early flowering in the tropics and the non-functional hd1 alleles brought about the adaptation of temperate japonica rice to tropical regions.
Introduction
Rice is a major staple food in the world especially in Asian countries. It is also the most rapidly growing food source in African countries (Seck et al., 2012). Various rice genotypes are cultivated in wide ranges of geographical locations in the world (40° S – equator – 53° N). In temperate zones including China (mainly in the northeast regions), Japan, Korea, Central Asia, and Europe in the northern hemisphere and Australia, Chile, and Uruguay in the southern hemisphere, japonica rice types are mostly grown once a year because of the occurrence of frost winter (Jena and Hardy, 2012). In contrast, in tropical zones surrounding the equator (23.2° S – equator – 23.2° N) and in some subtropical zones, indica rice types are widely grown in a double-season rice rotation cropping system because of the mild temperature throughout the year.
Rice is a short-day (SD) plant in which flowering is promoted by a reduced day-length. Flowering time (called as heading date in rice) is an important trait of rice for local domestication and is also one of the priority traits in rice breeding programs because it influences rice grain yield (Wei et al., 2010; Endo-Higashi and Izawa, 2011; Murphy et al., 2011; Yan et al., 2011; Gao et al., 2014; Liu et al., 2015) and other traits such as plant architecture and duration of vegetative phase (Xue et al., 2008; Hori et al., 2015) as well as cropping systems (Toriyama, 1970). The adjustment or fine-tuning of the flowering time of rice varieties can bring about some advantages such as enhanced multiple rice cropping per year, precise rotation cropping systems with other profitable crops, high prices of rice for specific consumers, and the possible improvement of rice field management by avoiding a regular encounter with biotic/abiotic stresses.
To date, more than 40 different genes (>40 gene loci) regulating flowering time in rice have been identified by several approaches including map-based cloning, reverse genetics approach using T-DNA or transposon tagging lines, and control of target gene expression via RNAi and overexpression. Most of the genes were identified and studied in high-latitude field conditions [natural long-day (LD)] or in day-length controlled chambers (Lee and An, 2015a; Hori et al., 2016). Furthermore, intensive research using physiology, genetics, and molecular biology tools quite revealed the molecular mechanisms of rice flowering. The regulatory networks among the flowering-associated genes are monitoring environment factors (mostly day-length in rice) and eventually activating the expression of the florigen genes Heading date 3a (Hd3a) and/or RICE FLOWERING LOCUS T 1 (RFT1) for rice plants to flower. Both Hd3a and RFT1 are mobile proteins that eventually cause the transition from the vegetative shoot apical meristem to the reproductive meristem which is called panicle initiation in rice (Tamaki et al., 2007; Komiya et al., 2008). Hd3a and RFT1 are paralogous to each other with 91% amino acid homology and are very closely located to each other with a physical distance of ∼11 kb on the chromosome 6. The expression of the florigen genes is tightly regulated by several upstream genes. Heading date 1 (Hd1) encoding a homolog of Arabidopsis CONSTANS promotes flowering by activating the transcription of the florigen genes under SD condition, but it suppresses the florigen genes under LD condition (Yano et al., 2000; Hayama et al., 2003). Early heading date 1 (Ehd1) encoding a B-type response regulator protein promotes the expression of Hd3a and RFT1 under both SD and LD conditions (Doi et al., 2004). Grain number, plant height, and heading date7 (Ghd7) encoding a CCT domain protein represses the expression of Ehd1 in LD condition and its functional alleles delay flowering in LD condition (Xue et al., 2008). DTH8 (QTL for days to heading) encoding the NF-Yb subunit of the trimeric NF-Y transcription factor also delays flowering in LD condition by suppressing the expression of Ehd1 (Wei et al., 2010) while its allelic mutant ds9 exhibited early flowering in LD and delayed flowering in SD, respectively (Xing et al., 2015). Another upstream gene of Ehd1, OsMADS51, is a flowering activator in SD with major effect and in LD with minor effect (Kim et al., 2007). Recent study by Chen et al. (2018) identified that OsMADS51 is also involved in high temperature-induced flowering of rice. Oryza sativa Pseudo-Response Regulator 37 (OsPRR37)/Hd2 also known as Ghd7.1 (Liu et al., 2013) functions as a flowering suppressor in both SD and LD conditions independently of Hd1 and Ehd1 pathways (Koo et al., 2013). The alleles of the identified flowering genes, especially natural variations, need to be more actively utilized in rice breeding programs to optimize the flowering time of local rice cultivars.
The indica rice cultivars were extensively utilized for the breeding of high-yielding lines in the temperate zone through shuttle breeding between the International Rice Research Institute (IRRI), Philippines (14.2° N) and the Rural Development Administration (RDA) of Korea. As a consequence, Tongil-type cultivars derived from the indica x japonica cross were successfully developed and brought the Green Revolution to Korea in the 1970s (Kim et al., 2012). The reverse adaptation of rice cultivars (from temperate zones to the tropics) is less known. In the 1930s, japonica rice breeding was conducted in Taiwan (22°N∼25°N) located at the boundary between the subtropics and tropics (called Tropics of Cancer) using Japanese temperate varieties which led to the development of the variety Taichung 65 that had long basic vegetative growth periods; since then, this variety has been widely utilized in japonica rice breeding programs in Taiwan (Wasano, 1982; Hsing, 2014). Another breeding program, the Germplasm Utilization Value Added (GUVA) project has initiated to breed high quality and high-yielding temperate japonica rice varieties in the tropics at IRRI since 1992. For several years at the beginning of the project, most of the Korean temperate japonica varieties were tested at IRRI fields to observe their performance. However, no varieties were found to be adequate for practical cultivation in the tropics mainly because of their extremely early flowering due to reduced day-lengths (Kang, 2010). Nonetheless, a handful of breeding lines showing good performance at IRRI were developed through the conventional breeding methods without recognition of the genetic factors associated with extreme early flowering. Among them, four lines were registered as temperate japonica rice varieties in the Philippines. However, the genetic factors influencing the adaptation of temperate japonica to tropical and subtropical environments are unclear.
In this study, the phenotypes of temperate japonica varieties in the tropical environment of the Philippines were examined and the key gene for a geographical adaptation was elucidated through allele typing of the major heading date genes and observation of flowering time in three different natural field conditions. Furthermore, the use of allele types of the major flowering genes for the fine-tuning of heading date in local rice varieties was discussed.
Materials and Methods
Plant Materials
A total of 45 diverse rice accessions were chosen for this study, including 20 IRRI diverse core collections (McNally et al., 2009), temperate japonica varieties from Japan, Korea, and Taiwan, indica-genome based Tongil-type cultivars of Korea, and popular indica varities developed by IRRI, Philippines. These 45 accessions covered the diverse geographical origins and four different rice genotypes including aus, indica, tropical japonica, and temperate japonica (Supplementary Table S1). In addition, 12 temperate japonica breeding lines/varieties developed by the GUVA project of IRRI-RDA collaborations in the Philippines were included (Supplementary Table S2). Briefly, in every winter season of Korea, many genetic materials including Korean varieties, germplasm and breeding lines were sent to the Philippines and grown at the IRRI field for seeds multiplication and advancing breeding cycles. The temperate japonica lines showing good performance in the Philippines were continuously selected and the lines were further improved by conventional breeding processes including crossing with other japonica or indica accessions (Kang, 2010; Ha et al., 2011). Since the initiation of the GUVA project in 1992, four breeding lines (IRRI 142, IRRI 152, IRRI 157, and IRRI 202) that have high grain quality and improved grain yield (>4.5t/ha) were approved as japonica rice varieties by the National Seed Industry Council of the Philippines and released in the Philippines.
Testing Locations
All the 57 accessions were tested at Los Baños, the Philippines and Suwon, Sotuh Korea. Los Baños lies in the tropical zone with latitude 14.2° N, 121.2° E and an altitude of 23 m where rice is cultivated generally in two cycles per year, dry season (DS) and wet season (WS), because of yearly mild temperature. In contrast, Suwon, with latitude 37.2° N and 126.6° E and an altitude of 37 m, lies in the temperate region where rice is grown in only one cultivation cycle or a short-duration rice variety with other crops per year because of frost winter.
Plant Growth
For testing in the low-latitude tropical climate, all the 57 accessions were grown at the IRRI fields (for two regular cropping seasons in the DS and WS) following the standard IRRI rice cultivation protocol. Dates of seeding/transplanting were on November 22, 2016/December 15, 2016 for the 2017 DS and May 29, 2017/June 20, 2017 for the 2017 WS. For each accession, the plants were transplanted at one plant per hill with 20 cm × 20 cm spaces and a total of five rows with 25 hills per row. Fertilizers (t/ha) were applied at a rate of 120-17.5-33-5 (N-P-K-Zn) in the 2017 DS and 90-13-25-5 (N-P-K-Zn) in the 2017 WS according to the seasonal and local standard fertilizer methods. For testing in the high-latitude temperate climate, all materials were grown at the RDA field of Suwon station in Korea according to the local normal rice cultivation method. The accessions were seeded in a seedling bed on April 30, 2016 and the seedling plants were transplanted in the field on May 30, 2016. Transplanting space was 30 cm × 15 cm among hills (one plant per hill) and five rows with 26 hills per accession. The rate of fertilizer applied was 90-45-57 (N-P-K).
Phenotype Measurement
Days to heading (DTH) is defined by the days from seeding to heading in 50% of total plants. The temperate japonica varieties under tropical conditions showed very irregular flowering among plants as well as among tillers in a single plant. They also frequently showed extremely early flowering in the main tiller (∼45 days from seeding). In this study, the DTH for the secondary tillers emerged after the main tiller was measured. The culm length and the grain yield were measured from the 10 Korean japonica varieties at the IRRI field in the 2017 DS. The average culm length was calculated from those of 10 plants and grain yield in paddy (t/ha) was calculated from those of 50 plants when the seed moisture was 14%. The phenotype data of the 10 Korean japonica varieties in Korea were obtained from the rice variety information released by the RDA. Grain yield in paddy (t/ha) in Korea was deduced by multiplying the yield of milled rice by 1.281.
Allele Determination of the Heading Date Genes
Leaf tissues were collected at the IRRI field and genomic DNAs were extracted using the modified CTAB method (Kim et al., 2011). Several sets of PCR primers for each gene were designed for the amplification of the protein-coding regions of the genes Hd1, OsPRR37, DTH8, and Ghd7 and the promoter region of the Hd3a. For identification of the known functional nucleotide polymorphism (FNP) of Ehd1 and RFT1 genes, the FNP containing region was amplified by PCR. PCRs were conducted by using the designed primers for each gene from the 57 rice accessions. The PCR products were then directly sequenced through the Sanger method-based sequencing using the AB3730xl DNA analyzer (Applied Biosystems2) by Macrogen3. Obtained sequences were analyzed using a web-based BLAST tool4 and the BioEdit software5. The polymorphic nucleotide positions among accessions were confirmed by manual checking of the sequencing chromatogram files. A large insertion/deletion on the heading date genes (Hd1, Hd3a, and OsMADS51) was determined by PCR amplification. The primer positions mapped on the gene structure and its sequences are presented in Supplementary Figure S1 and Supplementary Table S3, respectively.
Phylogenetic Analysis
Genome-wide genotyping was conducted using the Illumina Infinium 6K SNP chip (Thomson et al., 2017) at the IRRI Genotyping Service Laboratory6 according to the manufacturer’s instruction. The produced SNP genotype data were further processed in Microsoft Excel software to remove the meaningless SNP data points including monomorphic SNPs, rare allele SNPs, high frequency of heterozygous SNPs, or no data calling. Finally, the phylogenetic tree was constructed using PowerMarker software (version 3.25) (Liu and Muse, 2005) with 2,918 SNP data points. The similarity distance matrix of the alleles was measured using the C.S. Chord 1967 method (Cavalli-Sforza and Edwards, 1967) while the gene distribution tree was constructed using the neighbor-joining method.
Results
Performance of the Temperate Japonica Rice Varieties in the Tropical Location
We grew 10 Korean temperate japonica varieties at the IRRI field in the Philippines in DS of 2017. The remarkable phenotypic difference of the varieties between in Korea and in the Philippines was the heading date. Except for Jinmi and Unkwang, eight varieties showed extremely early flowering (<63 DTH) (Figure 1A). These eight varieties exhibited a tendency of irregular flowering time among plants as well as among tillers in a single plant: the embryo-derived main tiller flowered very quickly (∼45 days) and the following tillers flowered in order (Figure 1B). Eventually, these plants became stunted at the reproductive stage (Figures 1C,D) and showed very low grain yield (Figure 1E). As observed, quite early flowering of the elite temperate japonica varieties in the tropical zone is a major constraint in breeding temperate japonica rice varieties for the tropics. This might have been caused by the reduced day-length according to the geographical transition from a high-latitude location, Korea, to a low-latitude location, the Philippines (Figure 1F). In the Philippines, most of the days in a year belong to the SD condition with a range of day-length from 11.2 to 13 h. In contrast, the basic vegetative phase of rice plants in Korea is under the LD condition (May and June: 13.6 ∼ 14.6 h).
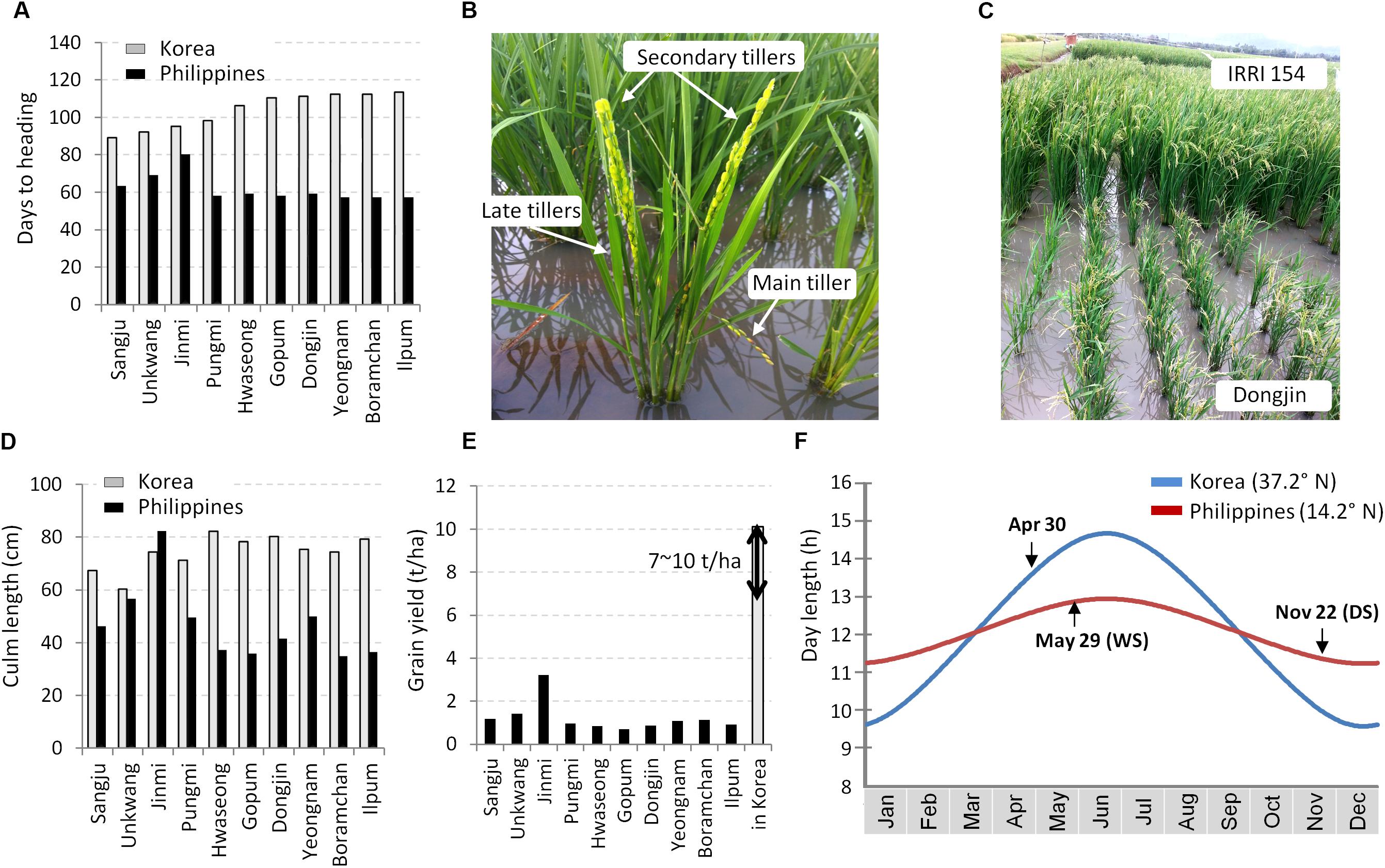
Figure 1. Phenotypes of the temperate japonica varieties in the tropical region. Ten Korean temperate japonica varieties were tested in the dry season in Los Baños (14.2° N), Philippines and in Suwon (37.2° N), Korea. (A) Comparison of days to heading (DTH). (B) Irregular flowering among tillers (Dongjin variety). (C) Photo of the maturation stage. (D) Comparison of culm length. (E) Comparison of grain yield. Yield data in Korea were obtained from the rice variety information released by the RDA. (F) Seasonal day-length (h) of the RDA Suwon station and the IRRI headquarter in Los Baños. The dates of seeding are depicted in the graph. DS, dry season; WS, wet season.
Allele Typing of the Major Flowering Genes From the 45 Diverse Rice Accessions
To elucidate the genes causing extremely early flowering in the tropics, the allele types of the major heading date genes were analyzed for the 45 diverse rice accessions including 17 temperate japonica, 4 tropical japonica, 4 aus, and 20 indica varieties from different geographical origins (Supplementary Table S1). We sequenced the full coding regions of Hd1, OsPRR37, DTH8, and Ghd7, and the 2-kb promoter region of Hd3a (Supplementary Figure S1). In addition, the PCR products containing the known FNP of the Ehd1 (G/A SNP causing G221R; Saito et al., 2009) and RFT1 (G/A SNP causing E105K; Ogiso-Tanaka et al., 2013) were sequenced. Sequences of the genes showed that most allelic variations occurred at the nucleotide positions previously reported. Therefore, the classification of allele types and their functionalities followed those described by Takahashi et al. (2009) for Hd1 and Hd3a promoter, Gao et al. (2014) for OsPRR37, Wei et al. (2010) for DTH8, and Lu et al. (2012) for Ghd7. However, we found new alleles in Hd1, OsPRR37, and Hd3a and named them as ‘NT (new type).’ In total, 11 allele types of Hd1, 10 of OsPRR37, 8 of DTH8, and 7 of Gdh7 were found from the 45 accessions (Figures 2A–D). Each allele type was divided into two groups in terms of protein functionality: functional and non-functional alleles. At first, we examined the presence of the prevalent allele types or allele combinations of heading date genes in the tropical location-adapted varieties. Regardless of rice types, most accessions had functional alleles for OsPRR37 (40 accession), DTH8 (37 accession), Ghd7 (43 accession), Ehd1 (43 accession) and RFT1 (39 accession), suggesting that these genes might not be the major determinants of flowering time which discriminates between plants grown in temperate zones and in the tropics. In the case of the Hd3a promoter, Type 1 allele among the six types was the most frequent in indica (13 out of 20), tropical japonica (3 out of 4), and temperate japonica (16 out of 17) rice types while the other allele types were very rare (Figure 2E). The new allele Hd3a-NT2 found in IR8, Saducho, and Zhenshan 97B possessed an insertion of a mobile DNA sequence (∼4.9 kb) near the translation start codon (-292 bp from ATG) (Supplementary Figures S1, S2). However, a certain level of allele prevalence was found for only Hd1. The non-functional hd1 alleles were more frequently found in the tropical/subtropical-adapted accessions (indica, aus, and tropical japonica types) (23 out of 28). In contrast, the temperate japonica varieties mostly possessed the functional alleles of Hd1 (13 out of 17). Furthermore, we examined the regulatory region of Hd1 using PCR method because the non-expressed Hd1 allele caused by insertion of a mobile DNA element at the regulatory region was found (Goretti et al., 2017). However, none of the lines tested harbored the mobile DNA element at Hd1 promoter region (Supplementary Figure S3).

Figure 2. Allele types of the five major flowering genes in the 45 diverse accessions. The allele frequencies in each rice type are presented at the right panel (blue letter). The nucleotide position referred to the protein coding sequence (CDS) or promoter sequence. (A) Hd1 alleles. Reference: Ginbouzu (GenBank Accession No. AB041840.1). (B) OsPRR37 alleles. Reference: Nipponbare (MSU ID: Os07g49460). (C) DTH8 alleles. Reference: Nipponbare (MSU ID: Os08g07740). (D) Ghd7 alleles. Reference: Minghui 63 (GenBank Accession No. EU286801.1). (E) Hd3a promoter alleles. Reference: Nipponbare (MSU ID: Os06g06320). FS, frame shift; ind, indica; j-tr, tropical japonica; j-te, temperate japonica.
Genetic Effects of Individual Flowering Genes in Natural SD and LD Conditions
We examined flowering time of the 45 varieties in the natural SD condition at the IRRI field in the Philippines for two cropping seasons, DS and WS, as well as in the natural LD condition at the RDA field in Korea. Because rice is a SD plant, all 45 varieties were found to flower earlier in natural SD than in natural LD without exceptions (Figure 3). This result supports the fact that rice is a SD plant regardless of types and geographical origins. However, the degree of early flowering was most significant for the Korean temperate japonica varieties and three Japanese temperate varieties Nipponbare, Koshikari, and Kitaake at the IRRI field. To examine the genetic effects of each flowering gene, all the accessions were divided into functional and non-functional allele groups for each gene, and the mean values of DTH between these two groups in three different environments were compared (Supplementary Figure S4). The functional alleles of Hd1 and RFT1 significantly reduced DTH only in the natural SD conditions (both DS and WS). Except for this, no significant genetic effect of the flowering genes was observed in all three environments.
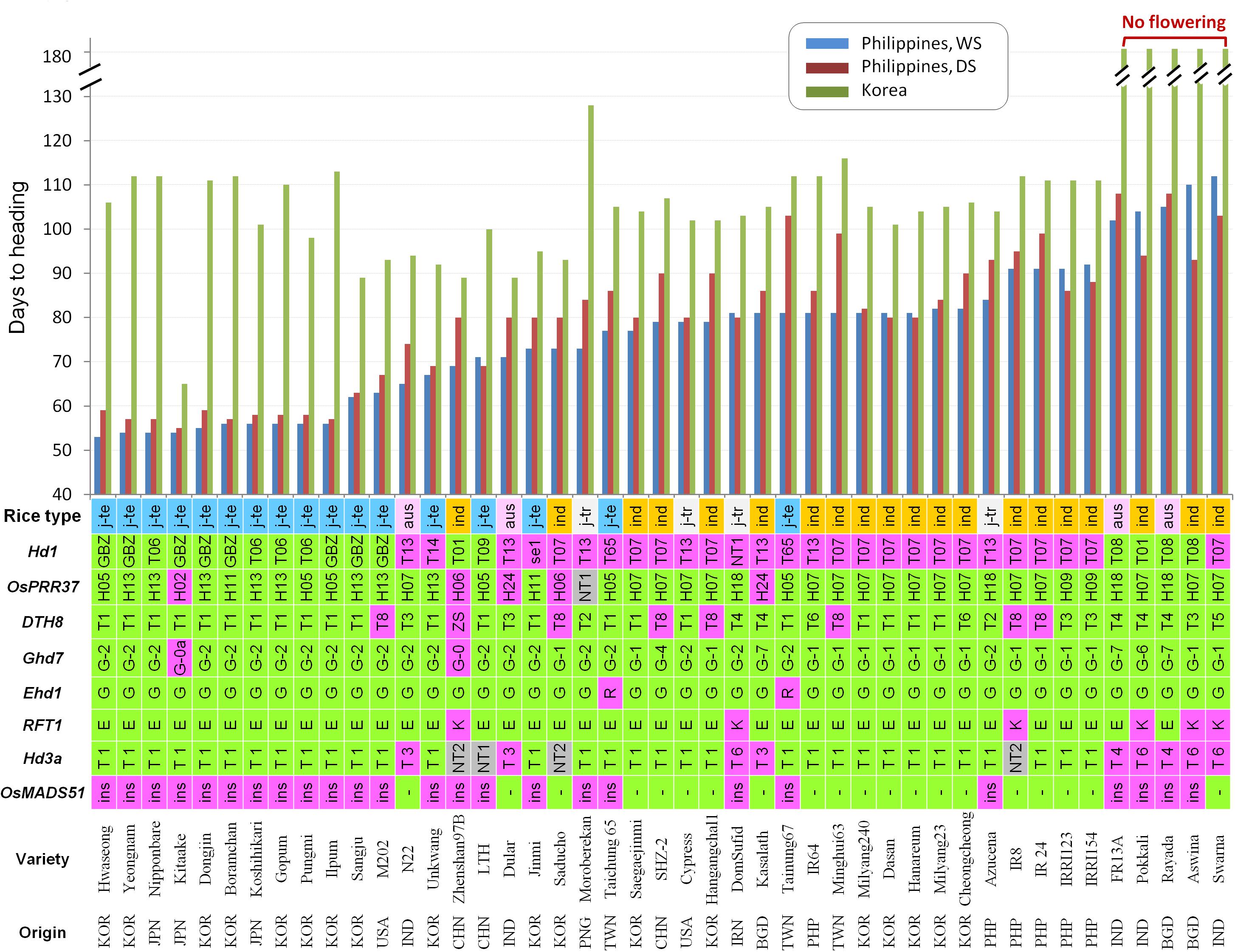
Figure 3. Days to heading (DTH) and the allele types of the major seven heading date genes from 45 diverse accessions. DTH was observed in the three different environments and the samples were sorted by DTH from early to late heading at the IRRI field in the wet season. In the cases of the Ehd1 and RFT1, the FNPs are presented with the corresponding amino acid (G221R of Ehd1 and E105K of RFT1). The tolerance allele to high temperature-induced flowering caused by 9.5-kb insertion at 1st intron of OsMADS51 is presented by ‘ins.’ The allele types with their functionality to each gene are depicted (green: functional allele and pink: non-functional allele). In the case of the Hd3a promoter, Group A and Group B are colored with green and pink, respectively, according to the previous research (Takahashi et al., 2009). New allele types (NT) with unknown functionality are shown with gray color. j-te, temperate japonica; j-tr, tropical japonica; ind, indica.
For the more critical evaluations of the flowering genes, the genetic effects were re-analyzed under the same genotype backgrounds of the major flowering genes tested. The group of the functional Hd1 alleles (n = 10) showed quite early flowering in natural SD conditions (24.5 days in WS and 24.7 days in DS) than the group of the non-functional hd1 alleles (n = 14) under the same genotype background: the functional alleles for OsPRR37, DTH8, Ghd7, Ehd1, RFT1 genes and Hd3a-Type 1 allele (Figure 4A). However, between the two groups, there was no difference in flowering time in natural LD. These data indicate that Hd1 is a strong flowering activator in natural SD but its effect is insignificant in natural LD. Similar analysis showed that the group of non-functional dth8 alleles (n = 4) flowered later than the group of functional DTH8 alleles (n = 14) in all three environments (2.2, 11.5, and 3.3 days in WS, DS, and natural LD, respectively) (Figure 4B). The Ghd7-2 allele is known as a mild allele compared to the Ghd7-1 allele (Xue et al., 2008), thus the effect of these two alleles on flowering time within the accessions having the non-functional hd1 alleles was compared. The plants with Ghd7-1 allele (n = 9) flowered significantly later than those with the Ghd7-2 allele (n = 5) in SD of WS but there was no difference in SD of DS and LD conditions (Figure 4C), suggesting that the two alleles are not much different to each other in terms of the regulation of flowering time. The effects of other gene/allele combinations were not analyzed because of the limited number of samples (less than 3).
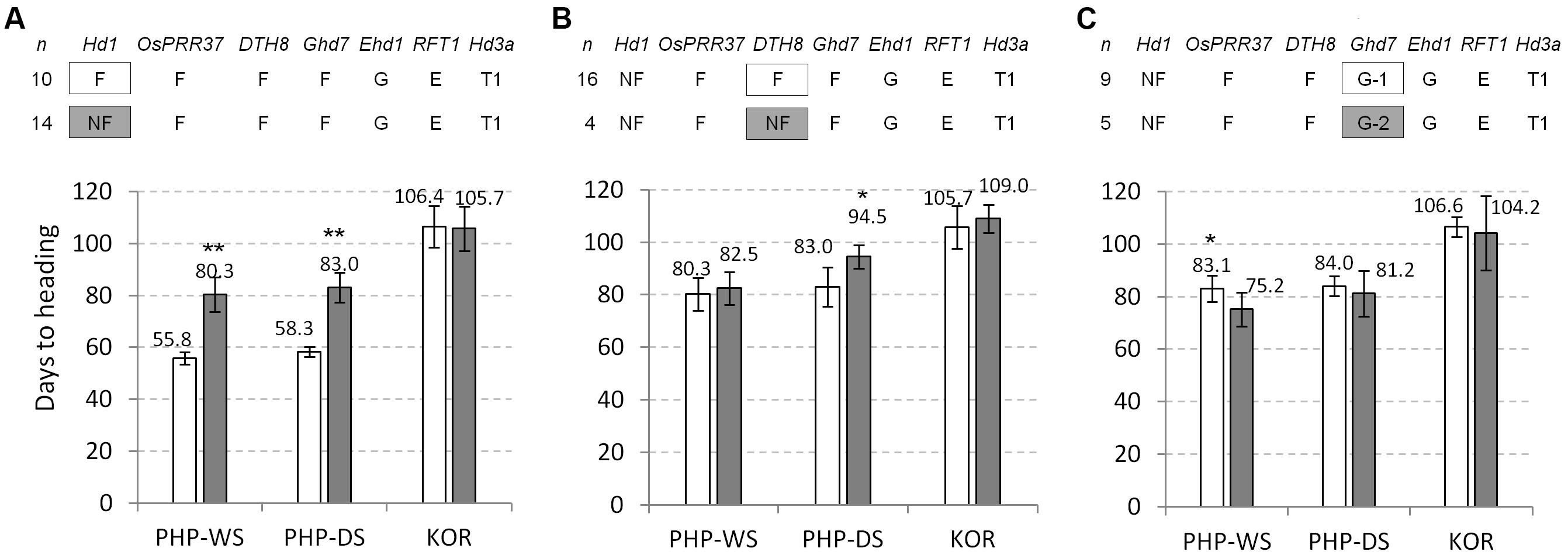
Figure 4. Genetic effects of the major flowering genes on flowering time in three different field conditions. Allele types of each flowering gene and the number of samples are presented on the top panel. (A) Genetic effect of Hd1. (B) Genetic effect of DTH8. (C) Comparison of the genetic effects between Ghd7-1 (G-1) and Ghd7-2 (G-2) alleles. A significant difference between two groups was calculated based on Student’s t-test (∗α = 0.05 and ∗∗α = 0.01). PHP, Philippines; KOR, Korea. DS, dry season; WS, wet season.
Examination of Temperature-Response Flowering Gene of Rice
Temperature is also considered as a major environmental factor in plant flowering although the clear genetic factors have not been demonstrated yet in rice. Recently, the day-length independent flowering QTL, qHd1 encoding OsMADS51 showed reduced heading date in response to increased temperature (Chen et al., 2018). A large insertion (∼9.5 kb) at the first intron of OsMADS51 significantly reduced gene expression and caused tolerance to high temperature-induced flowering. Overall a temperature during cropping season was higher in the tropics than in the temperate region (Supplementary Figure S5). We performed PCRs to identify the presence/absence of the large insertion from the 45 diverse accessions and the 12 japonica-type breeding lines adapted to the tropics (Supplementary Figure S6). Interestingly, all 17 temperate japonica accessions already possessed the thermo tolerance allele having a large insertion (Figure 3). Similarly, 11 out of the 12 japonica breeding lines still kept the large insertion allele (Figure 5A). These results support that the temperature tolerant allele of OsMADS51 is not associated with early flowering of temperate japonica varieties in the tropics and did not contribute to adaptation of temperate japonica rice to the tropics.
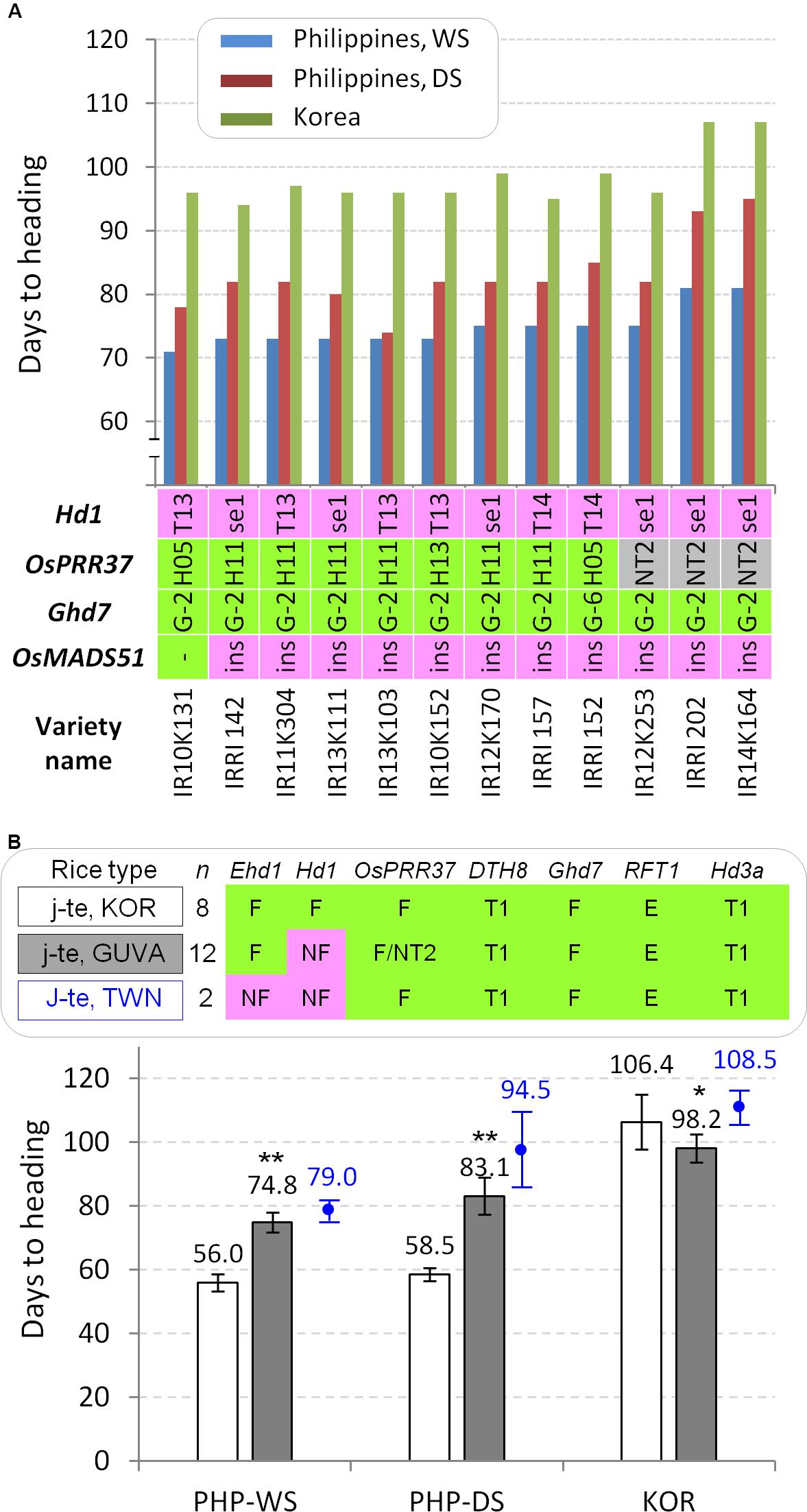
Figure 5. The 12 GUVA temperate japonica lines adapted to the tropics. (A) DTH and allele types of each flowering gene. Varieties/lines are sorted by DTH at the IRRI in the wet season. All 12 lines have DTH8-Type 1, functional Ehd1, functional RFT1, and Hd3a-Type 1 alleles. (B) Comparison of DTH between Korean temperate japonica varieties (j-te, KOR) and the 12 tropical zone-adapted temperate japonica varieties (j-te, GUVA) in three environmental conditions. Student’s t-test (∗ α = 0.05 and ∗∗ α = 0.01). As references, two Taiwanese temperate japonica varieties Taichung 65 and Tainung 67 (j-te, TWN) were presented (labeled by blue color).
Introgression of the Loss-of-Function Alleles of Hd1 Might Enable Breeding of Temperate Japonica Varieties Adapted to the Tropical Region
Through the japonica rice breeding project of GUVA, the japonica-type breeding lines adapted to the tropics were continuously selected from the several cross combinations (Supplementary Table S2). We grew 12 breeding lines both in Korea and the Philippines. In the Philippines, all the tropical-adapted lines flowered later than common Korean japonica varieties during both the cropping seasons (71–81 days in WS and 74–95 days in DS) (Figures 5A,B) and these lines did not show an irregular flowering time among tillers in a single plant. Allele typing of the major heading date genes showed that all the tropical-adapted breeding lines had the functional alleles for the OsPRR37, DTH8, Ghd7, Ehd1, and RFT1 genes and Type 1 promoter for Hd3a gene, as observed from common Korean temperate japonica varieties, except for Hd1 alleles. Interestingly, all 12 tropical-adapted lines had non-functional hd1 alleles [Type 13, Type 14, and photoperiod sensitivity 1 (se1)] (Figure 5A). Furthermore, the eight Korean temperate japonica varieties and 12 GUVA tropical-adapted lines were compared for their average DTH. The GUVA-derived japonica lines exhibited quite delayed flowering (18.8 days in WS and 24.6 days in DS) compared to the Korean temperate japonica varieties (Figure 5B), but they flowered earlier (8.2 days) than the Korean temperate japonica varieties in natural LD condition in Korea. These results indicate that the functional Hd1 alleles of temperate japonica varieties cause very early flowering with an irregular flowering pattern among tillers in natural SD condition. Thus, the loss-of-function alleles of Hd1 might have been strongly selected as key genes during the breeding process and its introgression might have enabled adaptation of temperate japonica varieties to the tropics.
Phylogenetic Analysis of the Rice Accessions
In terms of phenotypes, all the GUVA tropical-adapted lines showed characteristics of japonica varieties such as short grains and sticky texture of cooked rice (data not shown). However, during the japonica rice breeding process, the indica-genome-added lines were also used as parents. To examine the genome status of the breeding lines, a phylogenetic analysis was conducted using a chromosome-wide 2,918 SNP data points. Based on the classification by McNally et al. (2009), the materials were grouped into four groups: aus, indica, tropical japonica, and temperate japonica types. All 12 GUVA breeding lines were clustered with the temperate japonica varieties including Dongjin and Nipponbare (Figure 6), indicating that they are highly close to the temperate japonica rice type at the genome level. This result suggested that an introgression of the non-functional alleles of Hd1 into the temperate japonica background, together with a conventional plant selection breeding method (this might cause a small change in the genome of the temperate japonica background), enabled the breeding of japonica varieties for the tropics.
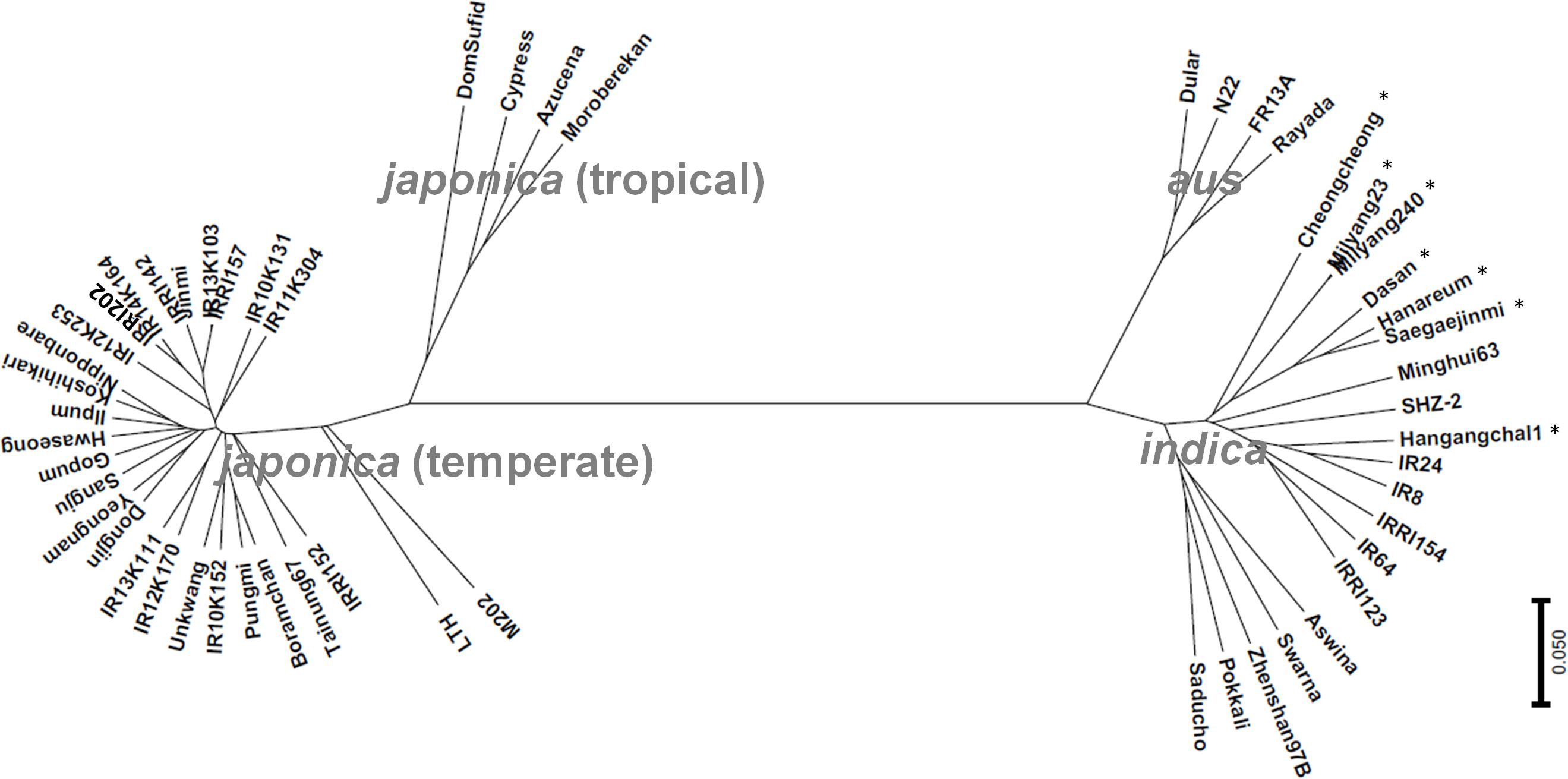
Figure 6. Phylogenetic analysis. All materials, except for three accessions (Kitaake, Taichung 65, and Kasalath), were analyzed by 6K SNP chip and the phylogenetic tree was constructed by using 2,918 SNP data points. The samples were grouped into aus, indica, tropical japonica, and temperate japonica types consistent with the previous report (McNally et al., 2009). Asterisk (∗) refers to Tongil-type rice cultivars.
Discussion
Currently, more than 40 flowering-related genes have been cloned in the rice genome and the process of gene identification and its functional studies were mostly done in controlled conditions like a growth chamber or room and/or at a paddy field in high-latitude locations (natural LD) (Lee and An, 2015a,b; Hori et al., 2016). In addition, through allele typing of major rice flowering genes from diverse collections, the major alleles/genes for the geographical adaptation/expansion, geographical favorable alleles, and diversification of alleles were studied in mostly temperate zone (Naranjo et al., 2014; Gómez-Ariza et al., 2015; Itoh et al., 2018; Ye et al., 2018) where japonica rice is cultivated. In terms of breeding purposes, evaluations of the genetic effects of the identified flowering genes at the natural field conditions and the further use of the proper allele types of the genes, especially those derived from the natural variations not transgenic-based identified genes, are important to optimize flowering time of local rice varieties. In this study, we examined the allele types of major seven flowering genes and DTH in three different natural environments from the 45 diverse accessions. Through this, we analyzed allele preference of each flowering gene among the rice types and the genetic effect of each gene on flowering time in the temperate and tropical locations, respectively. Furthermore, we compared DTH and allele types of flowering genes between common temperate japonica varieties and the 12 GUVA breeding lines and we identified the key genetic factor which contributed to adaptation of japonica rice to the tropics.
The Hd1 gene was identified by fine mapping of the Hd1 QTL derived from the Nipponbare x Kasalath cross and it was further confirmed that Hd1 is allelic to Se1 by the sequence analysis of the photoperiod sensitivity 1 (se1) mutant having a 43-bp deletion in the first exon of Hd1 and its background variety Ginbouzu (Se1) (Yano et al., 2000). The near-isogenic line (NIL) having non-functional hd1-Kasalath allele showed late and early flowering under controlled SD and LD conditions, respectively, compared to the background cultivar Nipponbare, indicating that Hd1 promotes flowering under SD but inhibits it under LD (Lin et al., 2000). Takahashi et al. (2009) showed high associations between Hd1 functionality and flowering under the controlled SD by studying the six major SD-pathway flowering genes in 64 rice germplasm accessions. In our study, a strong effect of Hd1 on flowering promotion under the natural SD condition was clearly observed. In the 45 diverse accessions, the functional Hd1 alleles showed quite early flowering under the natural SD condition dependent on Figure 4A, or independent of Supplementary Figure S4A, the allele types of the other flowering genes genotyped. This result means that the functional Hd1 allele is less influenced by the other genetic factors, and is able to enforce early flowering under the SD condition.
Under an LD condition, Hd1 represses flowering through the suppression of Hd3a and RFT1 transcription. Although Hd1 is regarded as an evolutionally conserved transcriptional activator, its interaction with DTH8 directly represses Hd3a expression (Du et al., 2017). Alternatively, suppression of Ehd1 by Hd1-Ghd7 complex affects transcription of florigen genes in LD (Nemoto et al., 2016). In other words, when the Hd1 exists with the non-functional dth8 or ghd7, Hd1 functions as transcriptional activator for Hd3a or Ehd1 respectively, resulted in flowering promotion. In consistent with above studies, the germplasm possessing the Hd1/dth8 alleles (M202 and Zhenshan 97B) and the Hd1/ghd7 alleles (Kitaake and Zhenshan 97B) showed early flowering tendency under the natural LD condition compared to the other accessions (Supplementary Figure S7). The NIL having the hd1 allele of Kasalath and the T-DNA-inserted hd1 mutant on the background cultivar Dongjin (DTH8/Ghd7 alleles) showed significantly early flowering by 17 and 16 days, respectively, under natural LD field conditions (Lin et al., 2000; Lee et al., 2016). In our study, the single genetic effect analysis of the Hd1 gene under the LD condition did not show a significant difference in flowering time between the Hd1 and the hd1 groups in both complex genotype background of flowering genes and the same genetic backgrounds (OsPRR37/DTH8/Ghd7/Ehd1/RFT1/Hd3a) (Figure 4A). However, in the closer genetic backgrounds between Korean temperate japonica varieties and the 12 GUVA lines, the hd1 plants (GUVA lines) significantly flowered early (∼8.2 days) under the natural LD condition (Figure 5B). These results suggest that the function of Hd1 under LD is influenced by other genetic factors in addition to known DTH8 and Ghd7. In conclusion, the functionality of Hd1 might be one of the major domestication traits in the tropical area whereas Hd1 with some other genetic factors might be co-selected in the temperate zone during domestication or breeding process.
All 12 GUVA tropical-adapted japonica breeding lines possessed the hd1 alleles without changes of functionality for the other flowering genes compared to Korean common japonica varieties. This data clearly support that Hd1 causes too early flowering of temperate japonica varieties in the tropics and the introgression of hd1 contributed for breeding of temperate japonica rice to the tropics. Six out of the 12 GUVA breeding lines had the se1 allele (Figure 5A). The Korean japonica cultivar Jinmi harboring the se1 allele was used as the immediate parent of the IRRI 142 and IR14K164 lines (Supplementary Table S2). The intermediate breeding lines having the Jinmi genome segments were also used for development of 10 GUVA breeding lines, resulted in a high-frequency introgression of the se1 allele in the GUVA breeding lines. However, other non-functional hd1 alleles, Type 13 and Type 14, were also selected during conventional breeding process although Jinmi or Jinmi-derived lines were highly used. This result supports that any types of hd1 alleles will be effective for the breeding of temperate japonica rice varieties for the tropics.
Testing and adaptation of temperate japonica varieties in low latitudes are less studied. A successful adaptation of a japonica variety in Taiwan was achieved through the breeding of Taichung 65 and since then, this variety was highly utilized for the development of other Taiwanese japonica varieties (Hsing, 2014). A previous phylogenetic analysis constructed by using 119 simple sequence repeat markers showed that most Taiwan japonica varieties including Taichung 65 and Tainung 67, which were also used in this study, were clustered into Japanese temperate japonica varieties (Lin et al., 2012). Consistently, the genome-wide 2,918 SNP markers analysis in this study indicated that the genome status of Tainung 67 is highly close to those of Japanese and Korean temperate japonica varieties (Figure 6), confirming that the varieties having temperate japonica genomes have successfully adapted to tropical and subtropical regions. A genetic analysis by Doi et al. (2004) revealed that the non-functional allele of Ehd1 (also called Early flowering 1 (Ef1)) of Taichung 65 in combination with the hd1 delayed flowering in an SD condition. Similarly, the ehd1 (ef1) allele markedly delayed flowering when it coexisted with the hd1 allele in a controlled SD condition (Nishida et al., 2002). The ehd1 allele was frequently found in Taiwanese japonica rice varieties including Tainung 67 but not in other japonica and indica varieties (Saito et al., 2009; Takahashi et al., 2009; Wei et al., 2016), suggesting that the ehd1 allele caused long basic vegetative growth periods in low-latitude regions. Similar to the previous reports, only two Taiwanese varieties (Taichung 65 and Tainung 67) possessed the ehd1 allele in our diverse panel. Thus, we compared the flowering time among three temperate japonica types (Korean, GUVA-developed, and Taiwanese). Taiwanese cultivars showed slightly delayed flowering compared to the GUVA lines in the Philippines. However, the GUVA lines (hd1/Ehd1) showed quite delayed flowering compared to the Korean japonica cultivars (Hd1/Ehd1) (Figure 5B), suggesting that the functional Ehd1 of the GUVA lines did not cause extremely early flowering in the natural SD condition. As another supporting examples in our study, the varieties having the hd1/Ehd1 alleles like the GUVA lines showed late flowering at the IRRI field (Figure 3). To conclude, the photoperiod insensitivity of Taichung 65 might have been achieved firstly by the introgression of the hd1 allele and then the introduction of the ehd1 allele for further delaying of flowering. The ehd1 allele needs to be tested in the GUVA line background for further yield enhancement through extending duration of vegetative growth.
In contrast to the adaptation of temperate japonica varieties to the tropics, the indica germplasm was well-utilized to breed high-yielding rice varieties (called as Tongil type rice) under the temperate climate in Korea. Since the 1970s, many Tongil-type varieties were used as rice varieties or breeding materials in several Asian countries (Kim et al., 2012; Dingkuhn et al., 2015) because of their wide geographical adaptability. In this study, most of the tropical-origin varieties showed normal flowering (89–116 days) in Suwon, Korea (Figure 3), suggesting that the flowering time of the indica germplasm may not be a major constraint for breeding of indica type varieites for the temperate zones. The genome status of all seven Tongil-type varieties was highly close to the indica genome (Figure 6) although a japonica variety was used as one of the parents. Interestingly, all seven Tongil varieties and all four IRRI indica varieties (IR8, IR24, IRRI 123, and IRRI 154) had the non-functional Hd1-Type 7 allele (Figure 3). This result suggests that the Hd1-Type 7 of Tongil varieties might have originated from IR8 or IR24 (called the Green Revolution rice varieties) because these two IRRI varieties were frequently used as donors for the breeding of Tongil-type varieties (Kim et al., 2012). This non-functional hd1 allele might also have contributed to a wide geographical adaptation of the Tongil varieties to tropical and temperate regions.
DTH8 is known as a flowering repressor under LD conditions and its function is unclear under SD conditions (Wei et al., 2010; Yan et al., 2011; Du et al., 2017). The functionality of DTH8 did not significantly influence flowering time in the natural LD condition in the single gene effect analysis (Supplementary Figure S4C), and when the DTH8 and the dth8 alleles were compared in the same genotype background (hd1/OsPRR37/Ehd1/RFT1/Ghd7) (Figure 4B). In contrast, in the natural SD condition, the plants with the dth8 allele showed delayed flowering (2–11.5 days) compared to the ones with DTH8 in hd1 backgrounds (Figure 4B), suggesting that DTH8 may promote flowering in the absence of Hd1 in the SD condition. Consistent with our results, the genetic effect of DTH8 might be influenced by other genetic factors including Hd1 (Lin et al., 2003; Yan et al., 2011).
OsPRR37 and Ghd7 are flowering repressors in LD conditions (Hori et al., 2016; Nemoto et al., 2016). The non-functional alleles for these genes were rare alleles found only in five and two accessions, respectively, in the diversity panel (Figure 2). These non-functional alleles also co-existed with the non-functional alleles of another flowering genes (Figure 3). However, the single genetic factor analysis showed that both non-functional alleles of OsPRR37 and Ghd7 were involved in the early flowering tendency in all three conditions (Supplementary Figure S4), especially in the natural LD condition. In the case of OsPRR37, the varieties having the non-functional allele types (Kitaake, Zhenshan 97B, Dular, and Saducho), except Kasalath, flowered early in the natural LD condition (Supplementary Figure S7). Kitaake and Zhenshan 97B having the ghd7 and osprr37 exhibited very early flowering in the natural LD condition (Supplementary Figure S7). Consistently, the japonica varieties having non-functional alleles of both genes flowered extremely early under a natural LD condition because of the additive genetic effect of the genes (Kim et al., 2013) and these allele types might have contributed to adaptation of rice to the northernmost regions of rice cultivation (∼53° N) (Koo et al., 2013). In the single gene effect analysis using the T-DNA-inserted mutants in the Dongjin background, the ghd7 and osprr37 mutants flowered earlier by 34 and 7 days, respectively, than Dongjin in a natural LD field condition (Koo et al., 2013; Lee et al., 2016), suggesting that the Ghd7-Ehd1-Hd3a/RFT1 pathway is a major flowering pathway in natural LD conditions.
In the regulatory region of Hd3a, a large mobile DNA sequence (∼4.9 kb) was inserted at the -292 bp position from the translation start codon (ATG) in Hd3a-NT2 allele (Supplementary Figures S1, S2). Probably, the large insertion did not cause the non-expressed Hd3a allele because another florigen gene RFT1 is already non-functional in Zhenshan 97B. However, the influences of the large insertion to Hd3a transcription need to be tested in the future. Recent studies of the regulatory elements for Hd3a expression characterized several cis-elements at different positions on Hd3a promoter. Pasriga et al. (2018) identified the regulatory region for vascular tissue-specific expression of Hd3a (-397 to -197 region from ATG) using GUS reporter system and Du et al. (2017) identified two cis-elements of Hd3a (-1,475 to -1,274 and -526 to -313 regions from ATG) for Hd1 binding using yeast one-hybrid assay. Another report by Goretti et al. (2017) identified CO Response Element 2 (CORE2) (-176 to -168 from ATG) using electrophoretic mobility shift assay. This new Hd3a-NT2 allele will be useful material to study a regulatory element of Hd3a promoter.
Under the natural LD condition in Suwon, three indica varieties (Swarna, Aswina, and Pokkali) and two aus varieties (Rayada and FR13A) did not flower before frost winter (> 180 days) (Figure 3). No flowering was observed in a natural LD condition when rice plants had the ehd1/rft1 (Zhao et al., 2015) or the Hd1/rft1 alleles combination (Ogiso-Tanaka et al., 2013). Two accessions (Pokkali and Aswina) possessing Hd1/rft1 are expected for no flowering based on the above research but the others had Hd1/RFT1 (FR13A and Rayada) or hd1/rft1 (Swarna) alleles (Figure 3). In the cases of FR13A and Rayada, Hd1-mediated flowering suppression (Hd1-DTH8 complex and Hd1-Ghd7 complex via Ehd1 suppression) will be functional, suggesting that transcriptional defect of RFT1 or unknown strong flowering repressor are involved in no flowering in LD. In contrast, Swarna had both non-functional hd1/rft1 alleles, assuming presence of unknown strong repressor of Hd3a. Interestingly, all these five no flowering accessions showed the latest flowering even in the SD conditions, especially in WS (Figure 3). These five accessions and Dom Sufid had unique Type 4 or Type 6 promoters of Hd3a. The relationship between Hd3a types and late flowering under SD needs to be studied in the future.
All the 45 tester accessions and 12 GUVA japonica breeding lines showed early flowering in the natural SD conditions (Figures 3, 5A). This result supports the previous finding that a conventional SD treatment like placing pots containing rice plants into a dark room may be effective to promote flowering and, in turn, to shorten the duration of a breeding cycle. In the natural SD conditions at IRRI, most of the accessions flowered earlier in WS than in DS (Supplementary Figure S8). Around 30–40 days from seeding (basic vegetative growth phase), the day-length is slightly decreasing at IRRI during WS whereas increasing during DS, although the actual day-length on the seeding date is longer in WS than in DS (Figure 1F). This observation suggests that the rice plants are continuously monitoring a minor change of day-length (becoming shorter) not the absolute day-length (h) although most of days during both cropping seasons are close to the SD condition. Similarly, in the natural LD condition in Suwon, rice plants might perceive shorter day-lengths in early July (the absolute day-length is the longest), and trigger the flowering pathway genes. In contrast, the few accessions having the Hd3a-Type 6 allele flowered slightly late in DS than in WS (Figure 3). However, the influences of some other environmental factors such as daily radiation and temperature as well as those of other genetic factors such as the Hd3a-Type 6 allele need to be examined to elucidate the biological/genetic mechanisms causing a minor flowering difference between the two cropping seasons at the same location.
Many genetic factors regulating flowering have been identified at the gene level. Through marker-assisted selection, the putative favorable alleles of the identified genes can be tested and utilized to adjust flowering in elite rice varieties, which can improve yields, cropping systems, and farmer’s income. Here, the association of Hd1 with the adaptation of japonica varieties in the tropical regions was examined. For further improvement of the GUVA lines by extension of vegetative growth phase, non-functional alleles for Ehd1 and DTH8 loci are needed to be tested in the future.
Author Contributions
S-RK, U-SY, O-YJ, and J-SL conceived the research. GT and M-RY, JK, and C-KL designed and conducted field experiments in the Philippines and Korea, respectively. S-RK, I-RC, and J-SL conducted allele typing of rice flowering genes. GP performed phylogenetic analysis. S-RK, I-RC, KJ, and J-SL analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
Funding
This research was funded by the GUVA Phase II project (Grant No. PJ012101) of Rural Development Administration (RDA), Korea and International Rice Research Institute (IRRI).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to RDA and IRRI scientists who have been involved in the GUVA project since 1992, Mr. Mario Lapiz and Mr. Renan Belen for general field works and Ms. Graciana Clave for DNA works at IRRI. We thank Prof. Gynheung An (Kyung Hee University, South Korea) for revising the manuscript, Dr. Fiona Hay and Dr. Jae-Sung Lee (IRRI) for providing seeds of 20 diverse rice collections, and Ms. Anne Lasam for editing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01827/full#supplementary-material
Footnotes
- ^http://www.knowledgebank.irri.org
- ^http://www.appliedbiosystems.com/
- ^http://dna.macrogen.com/
- ^https://blast.ncbi.nlm.nih.gov/
- ^http://www.mbio.ncsu.edu/BioEdit/
- ^http://gsl.irri.org/
References
Cavalli-Sforza, L. L., and Edwards, A. W. F. (1967). Phylogenetic analysis: models and estimation procedures. Evolution 21, 550–570. doi: 10.1111/j.1558-5646.1967.tb03411.x
Chen, J. Y., Zhang, H. W., Zhang, H. L., Ying, J. Z., Ma, L. Y., and Zhuang, J. Y. (2018). Natural variation at qHd1 affects heading date acceleration at high temperatures with pleiotropism for yield traits in rice. BMC Plant Biol. 18:112. doi: 10.1186/s12870-018-1330-5
Dingkuhn, M., Laza, M. R. C., Kumar, U., Mendez, K. S., Collard, B., Jagadish, K. S. V., et al. (2015). Improving yield potential of tropical rice: achieved levels and perspectives through improved ideotypes. Field Crop Res. 182, 43–59. doi: 10.1016/j.fcr.2015.05.025
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. doi: 10.1101/gad.1189604
Du, A., Tian, W., Wei, M., Yan, W., He, H., Zhou, D., et al. (2017). The DTH8-Hd1 Module mediates day-length-dependent regulation of rice flowering. Mol. Plant 10, 948–961. doi: 10.1016/j.molp
Endo-Higashi, N., and Izawa, T. (2011). Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice. Plant Cell Physiol. 52, 1083–1094. doi: 10.1093/pcp/pcr059
Gao, H., Jin, M., Zheng, X. M., Chen, J., Yuan, D., Xin, Y., et al. (2014). Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 16337–16342. doi: 10.1073/pnas.1418204111
Gómez-Ariza, J., Galbiati, F., Goretti, D., Brambilla, V., Shrestha, R., Pappolla, A., et al. (2015). Loss of floral repressor function adapts rice to higher latitudes in Europe. J. Exp. Bot. 66, 2027–2039. doi: 10.1093/jxb/erv004
Goretti, D., Martignago, D., Landini, M., Brambilla, V., Gómez-Ariza, J., Gnesutta, N., et al. (2017). Transcriptional and post-transcriptional mechanisms limit heading date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 13:e1006530. doi: 10.1371/journal.pgen.1006530
Ha, W.-G., Torollo, G. V., Kang, K.-H., Lapiz, M., and Padolina, T. (2011). Breeding temperate japonica rice MS11 for the tropics. Philipp. J. Crop Sci. 36:111.
Hayama, R., Yokoi, S., Tamaki, S., Yano, S., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Hori, K., Matsubara, K., and Yano, M. (2016). Genetic control of flowering time in rice: integration of Mendelian genetics and genomics. Theor. Appl. Genet. 129, 2241–2252. doi: 10.1007/s00122-016-2773-4
Hori, K., Nonoue, Y., Ono, N., Shibaya, T., Ebana, K., Matsubara, K., et al. (2015). Genetic architecture of variation in heading date among Asian rice accessions. BMC Plant Biol. 15, 1–16. doi: 10.1186/s12870-015-0501-x
Hsing, Y. I. C. (2014). “Rice in taiwan,” in Encyclopaedia of the History of Science, Technology, and Medicine in Non-Western Cultures, ed. H. Selin (Berlin: Springer), 3769–3772.
Itoh, H., Wada, K. C., Sakai, H., Shibasaki, K., Fukuoka, S., Wu, J., et al. (2018). Genomic adaptation of flowering-time genes during the expansion of rice cultivation area. Plant J. 94, 895–909. doi: 10.1111/tpj
Jena, K. K., and Hardy, B. (2012). Advances in Temperate Rice Research. Manila: International Rice Research Institute.
Kim, S. D., Moon, H. P., Kim, J. K., and Park, D. G. (2012). The Green Revolution in Korea: Development and Dissemination of Tongil-type Rice Varieties. Seoul: Ministry of Strategy and Finance.
Kim, S. L., Choi, M., Jung, K. H., and An, G. (2013). Analysis of the early-flowering mechanisms and generation of T-DNA tagging lines in Kitaake, a model rice cultivar. J. Exp. Bot. 64, 4169–4182. doi: 10.1093/jxb/ert226
Kim, S. L., Lee, S., Kim, H. J., Nam, H. G., and An, G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145, 1484–1494. doi: 10.1104/pp.107.103291
Kim, S. R., Jeon, J. S., and An, G. (2011). Development of an efficient inverse PCR method for isolating gene tags from T-DNA insertional mutants in rice. Methods Mol. Biol. 678, 139–146. doi: 10.1007/978-1-60761-682-5_11
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S., and Shimamoto, K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774. doi: 10.1242/dev.008631
Koo, B. H., Yoo, S. C., Park, J. W., Kwon, C. T., Lee, B. D., An, G., et al. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6, 1877–1888. doi: 10.1093/mp/sst088
Lee, Y. S., and An, G. (2015a). Complex regulatory networks of flowering time in rice. Rice Res. 3, 1–9.
Lee, Y. S., and An, G. (2015b). Regulation of flowering time in rice. J. Plant Biol. 58, 353–360. doi: 10.1007/s12374-015-0425-x
Lee, Y. S., Yi, J., Jung, K. H., and An, G. (2016). Comparison of rice flowering-time genes under paddy conditions. J. Plant Biol. 59, 238–246. doi: 10.1007/s12374-016-0029-0
Lin, H., Liang, Z.-W., Sasaki, T., and Yano, M. (2003). Fine mapping and characterization of quantitative trait loci Hd4 and Hd5 controlling heading date in rice. Breed. Sci. 53, 51–59. doi: 10.1270/jsbbs.53.51
Lin, H. X., Yamamoto, T., Sasaki, T., and Yano, M. (2000). Characterization and detection of epistaic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines. Theor. Appl. Genet. 101, 1021–1028. doi: 10.1007/s001220051576
Lin, H. Y., Wu, Y. P., and Hour, A. L. (2012). Genetic diversity of rice germplasm used in Taiwan breeding programs. Bot. Stud. 53, 363–376.
Liu, C., Song, G., Zhou, Y., Qu, X., Guo, Z., Liu, Z., et al. (2015). OsPRR37 and Ghd7 are the major genes for general combining ability of DTH, PH and SPP in rice. Sci. Rep. 5:12803. doi: 10.1038/srep12803
Liu, K., and Muse, S. V. (2005). Power marker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129. doi: 10.1093/bioinformatics/bti282
Liu, T., Liu, H., Zhang, H., and Xing, Y. (2013). Validation and characterization of Ghd7. 1, a major quantitative trait locus with pleiotropic effects on spikelets per panicle, plant height, and heading date in rice (Oryza sativa L.). J. Integr. Plant Biol. 55, 917–927. doi: 10.1111/jipb.12070
Lu, L., Yan, W. H., Xue, W., Shao, D., and Xing, Y. (2012). Evolution and association analysis of Ghd7 in rice. PLoS One 7:e34021. doi: 10.1371/journal.pone.0034021
McNally, K. L., Childs, K. L., Bohnert, R., Davidson, R. M., Zhao, K., Ulat, V. J., et al. (2009). Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl. Acad. Sci. U.S.A. 106, 12273–12278. doi: 10.1073/pnas.0900992106
Murphy, R. L., Klein, R. R., Morishge, D. T., Brady, J. A., Rooney, W. L., Miller, F. R., et al. (2011). Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. U.S.A. 108, 16469–16474. doi: 10.1073/pnas.1106212108
Naranjo, L., Talón, M., and Domingo, C. (2014). Diversity of floral regulatory genes of japonica rice cultivated at northern latitudes. BMC Genomics 15:101. doi: 10.1186/1471-2164-15-101
Nemoto, Y., Nonoue, Y., Yano, M., and Izawa, T. (2016). Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 86, 221–233. doi: 10.1111/tpj.13168
Nishida, H., Inoue, H., Okumoto, Y., and Tanisaka, T. (2002). A novel gene ef1-h conferring an extremely long basic vegetative growth period in rice. Crop Sci. 42, 348–354. doi: 10.2135/cropsci2002.3480
Ogiso-Tanaka, E., Matsubara, K., Yamamoto, S., Nonoue, Y., Wu, J., Fujisawa, H., et al. (2013). Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS One 8:e75959. doi: 10.1371/journal.pone.0075959
Pasriga, R., Cho, L. H., Yoon, J., and An, G. (2018). Identification of the regulatory region responsible for vascular tissue-specific expression in the rice Hd3a promoter. Mol. Cells 41, 342–350. doi: 10.14348/molcells
Saito, H., Yuan, Q., Okumoto, Y., Doi, K., Yoshimura, A., Inoue, H., et al. (2009). Multiple alleles at Early flowering 1 locus making variation in the basic vegetative growth period in rice (Oryza sativa L.). Theor. Appl. Genet. 119, 315–323. doi: 10.1007/s00122-009-1040-3
Seck, P. A., Diagne, A., Mohanty, S., and Wopereis, M. C. S. (2012). Crops that feed the world 7: rice. Food Secur. 4, 7–24. doi: 10.1007/s12571-012-0168-1
Takahashi, Y., Teshima, K. M., Yokoi, S., Innan, H., and Shimamoto, K. (2009). Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. U.S.A. 106, 4555–4560. doi: 10.1073/pnas.0812092106
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Thomson, M. J., Singh, N., Dwiyanti, M. S., Wang, D. R., Wright, M. H., Perez, F. A., et al. (2017). Large-scale deployment of a rice 6 K SNP array for genetics and breeding applications. Rice 10:40. doi: 10.1186/s12284-017-0181-2
Toriyama, K. (1970). Breeding non-seasonal short duration rice varieties in southern Japan. JARQ 5, 1–4.
Wasano, K. (1982). Japan’s Role in Tropical Rice Research: Usefulness of Japonica Varieties as Breeding Material. Manila: International Rice Research Institute.
Wei, F. J., Tsai, Y. C., Wu, H. P., Huang, L. T., Chen, Y. C., Chen, Y. F., et al. (2016). Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice. Plant Sci. 242, 187–194. doi: 10.1016/j.plantsci.2015.09.005
Wei, X. J., Xu, J. F., Guo, H. N., Jiang, L., Chen, S. H., Yu, C. Y., et al. (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153, 1747–1758. doi: 10.1104/pp.110.156943
Xing, Q., Zheng, Z., Zhou, X., Chen, X., and Guo, Z. (2015). Ds9 was isolated encoding as OsHAP3H and its C-terminus was required for interaction with HAP2 and HAP5. J. Plant Biol. 58, 26–37. doi: 10.1007/s12374-014-0191-1
Xue, W. Y., Xing, Y. Z., Weng, X. Y., Zhao, Y., Tang, W. J., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet 40, 761–767. doi: 10.1038/ng.143
Yan, W. H., Wang, P., Chen, H. X., Zhou, H. J., Li, Q. P., Wang, C. R., et al. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330. doi: 10.1093/mp/ssq070
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene Constans. Plant Cell 12, 2473–2483. doi: 10.1105/tpc.12.12.2473
Ye, J., Niu, X., Yang, Y., Wang, S., Xu, Q., Yuan, X., et al. (2018). Divergent Hd1, Ghd7, and DTH7 alleles control heading date and yield potential of japonica rice in Northeast China. Front. Plant Sci. 9:35. doi: 10.3389/fpls.2018.00035
Zhao, J., Chen, H., Ren, D., Tang, H., Qiu, R., Feng, J., et al. (2015). Genetic interactions between diverged alleles of early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ Rice Flowering locus T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol. 208, 936–948. doi: 10.1111/nph.13503
Keywords: Hd1, heading date, flowering time, temperate japonica, tropics, genetic diversity, rice
Citation: Kim S-R, Torollo G, Yoon M-R, Kwak J, Lee C-K, Prahalada GD, Choi I-R, Yeo U-S, Jeong O-Y, Jena KK and Lee J-S (2018) Loss-of-Function Alleles of Heading date 1 (Hd1) Are Associated With Adaptation of Temperate Japonica Rice Plants to the Tropical Region. Front. Plant Sci. 9:1827. doi: 10.3389/fpls.2018.01827
Received: 09 July 2018; Accepted: 26 November 2018;
Published: 10 December 2018.
Edited by:
Giampiero Valè, Cereal Research Centre, CRA-CER, ItalyReviewed by:
Fabio Fornara, University of Milan, ItalyFan Chen, Institute of Genetics and Developmental Biology (CAS), China
Yongzhong Xing, Huazhong Agricultural University, China
Copyright © 2018 Kim, Torollo, Yoon, Kwak, Lee, Prahalada, Choi, Yeo, Jeong, Jena and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeom-Sig Lee, leejsyr@korea.kr
 Sung-Ryul Kim
Sung-Ryul Kim Gideon Torollo
Gideon Torollo Mi-Ra Yoon3
Mi-Ra Yoon3 Jieun Kwak
Jieun Kwak G. D. Prahalada
G. D. Prahalada Il-Ryong Choi
Il-Ryong Choi Un-Sang Yeo
Un-Sang Yeo Kshirod K. Jena
Kshirod K. Jena Jeom-Sig Lee
Jeom-Sig Lee