- 1School of Life Sciences, The Key Laboratory of Cell Activities and Stress Adaptations, Ministry of Education, Lanzhou University, Lanzhou, China
- 2College of Life Sciences, Zhejiang University, Hangzhou, China
- 3School of Forestry, Beijing Forestry University, Beijing, China
The C-REPEAT BINDING FACTOR signaling pathway is strictly modulated by numerous factors and is essential in the cold response of plants. Here, we show that the DUF793 family gene BYPASS1-LIKE modulates freezing tolerance through the CBFs in Arabidopsis. The expression of B1L was rapidly induced under cold treatment. Comparing to wild type, B1L knockout mutants were more sensitive to freezing treatment, whereas B1L-overexpressing lines were more tolerant. The expression of CBFs and CBF target genes was significantly decreased in b1l mutant. Using yeast two-hybrid screening system, 14-3-3λ was identified as one of proteins interacting with B1L. The interaction was confirmed with bimolecular fluorescence complementation assay and co-immunoprecipitation assay. Biochemical assays revealed that b1l mutation promoted the degradation of CBF3 compared to wild type, whereas 14-3-3κλ mutant and b1l 14-3-3κλ mutant suppressed the degradation of CBF3. Consistently, 14-3-3κλ and b1l 14-3-3κλ mutants showed enhanced freezing tolerance compared to wild type. These results indicate that B1L enhances the freezing tolerance of plants, at least partly through stabilizing CBF. Our findings improve our understanding of the regulation of CBF in response to cold stress.
Introduction
Temperature is one of the most important environmental factors that affect the survival, growth, and reproduction of plants. Plants adapt to freezing stress through multiple physiological and biochemical processes. After exposure to low temperatures above freezing, temperate plants acquire freezing tolerance, a process that is termed cold acclimation (Guy, 1990; Thomashow, 1999). The expression of a class of APETALA2/ETHYLENE-RESPONSIVE FACTOR (AP2/ERF) transcription factor, C-REPEAT BINDING FACTOR/DROUGHT RESPONSE ELEMENT BINDING FACTOR 1B (CBF/DREB1), is rapidly induced under cold stress, playing a central role in the cold acclimation of Arabidopsis (Stockinger et al., 1997; Liu et al., 1998; Thomashow, 1999). CBFs bind to the CRT/DRE element of COLD-REGULATED (COR) genes, inducing their expression and conferring an enhanced freezing tolerance (Yamaguchi-Shinozaki and Shinozaki, 1994; Stockinger et al., 1997; Gilmour et al., 1998; Liu et al., 1998; Thomashow, 1999).
The expression of CBFs is regulated by numerous transcription factors. Using CBF3-LUC transgenic plants, Chinnusamy et al. identified a basic helix-loop-helix (bHLH) transcription factor named INDUCER OF CBF EXPRESSION 1 (ICE1; Chinnusamy et al., 2003). ICE1 promotes the expression of CBF3 through the binding to MYC cis-elements within the promoter region of CBF3 (Chinnusamy et al., 2003). BRASSINAZOLE-RESISTANT 1 (BZR1), LATE ELONGATED HYPOCOTYL (LHY), and CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 3 (CAMTA3) were also found to positively regulate the expression of CBFs (Doherty et al., 2009; Dong et al., 2011; Li et al., 2017a,b). On the other hand, MYB15, ETHYLENE-INSENSITIVE 3 (EIN3), and PHYTOCHROME-INTERACTING FACTOR 3 (PIF3) repress CBF expression (Agarwal et al., 2006; Shi et al., 2012; Jiang et al., 2017).
The posttranslational regulation of CBF is also involved in the plant response to cold stress (Liu et al., 2017; Ding et al., 2018). In this process, 14-3-3 proteins are phosphorylated by COLD-RESPONSIVE PROTEIN KINASE 1 (CRPK1) and translocated from the cytoplasm to the nucleus, where 14-3-3 proteins can interact with CBFs and trigger the degradation of CBFs through the 26S proteasome pathway (Liu et al., 2017). By contrast, BTF3-LIKE (BTF3L) inhibits the degradation of CBFs by interacting with CBF proteins (Ding et al., 2018). However, the proteins that negatively modulate the 14-3-3λ-mediated degradation of CBF remain unknown.
In Arabidopsis, at least 12 proteins contain a conserved DUF793 domain, but only few members have been functionally characterized. BYPASS1 (BPS1) is required to produce a root-sourced signal that moves to the shoot and arrests growth of shoot, through modification of cytokinin signaling (Van Norman et al., 2004, 2011; Lee et al., 2016). ROH1 interacts with the exocyst subunit EXO70A1 and is involved in the localized deposition of seed coat pectin (Kulich et al., 2010). At1g74450 gene affects the plant height, pollen development, and composition of the inner seed coat mucilage layer (Visscher et al., 2015). AT1G18740, which we named as BYPASS1-LIKE (B1L), and AT1G74450 are both responsive to multiple abiotic stresses (Ma and Bohnert, 2007). However, more insight into the molecular function of the DUF793 proteins is in need.
In this study, we found that B1L, which is rapidly induced under cold treatment, modulates freezing tolerance through the CBFs in Arabidopsis. To be specific, B1L reduces the degradation of CBFs through the interaction with 14-3-3λ. Our results indicate that B1L positively modulates plant freezing tolerance, at least partly through stabilizing CBFs.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana Col-0 was used as the wild type. The mutant and transgenic lines that were used in this study were as follows: b1l (SALK_019913), 14-3-3λ (SALK_075219) (Zhou et al., 2014; Liu et al., 2017), 14-3-3κ (SALK_148929) (Van Kleeff et al., 2014), cbfs (Jia et al., 2016), b1l 14-3-3λ, 14-3-3κλ, b1l 14-3-3κλ, b1l cbfs, Super:CBF3-MYC (Liu et al., 2017), Super:CBF3-MYC/b1l, B1L b1l #1, B1L b1l #2, B1L-OE #1, B1L-OE#2, ProB1L:B1L-GFP #1, and ProB1L:GUS.
b1l was obtained from ABRC. 14-3-3κλ was generated by crossing 14-3-3λ and 14-3-3κ. 14-3-3λ, 14-3-3κ, and 14-3-3κλ were kindly provided by the Li Jia laboratory of Lanzhou University. cbfs and Super:CBF3-MYC were kindly provided by the Shu-Hua Yang laboratory from the China Agricultural University. b1l 14-3-3κλ, Super:CBF3-MYC/b1l, and b1l cbfs were generated through genetic crossing.
The ProB1L:B1L-3×FLAG fusion and the b1l restored plants (B1L b1l #1 and #2) were obtained via amplifying the B1L genomic region, including the 2000-bp promoter fragment, and cloning the resulting PCR product into the pMDC302 Gateway binary vector. The ProB1L:B1L-GFP transgenic plants (ProB1L:B1L-GFP #1) were obtained by amplifying the same genomic region and cloning it into the pMDC107 Gateway binary vector. The ProB1L:GUS transgenic plants were obtained by amplifying the ProB1L fragment and cloning it into the pBIB-GUS vector. The 35S:YFP-B1L fusion and the B1L-overexpressing transgenic lines (B1L-OE #1 and #2) were obtained by amplifying B1L cDNA and cloning the resulting PCR product into the pEarlygate104 Gateway binary vector.
Plants were grown at 22°C under long-day conditions (16 h light/8 h dark) in soil or agar plates (1/2 MS, 1% sucrose, and 0.8% agar).
All primer sequences that were used in this study are listed in Supplementary Table S1.
Plant Freezing Assay
The plant freezing assays were performed as previously described (Zhu et al., 2004; Miura et al., 2007) with modifications. Plants were grown in soil at 21°C under long-day (LD) conditions for 3 weeks before the treatments were performed. For each line, the plant freezing assay was performed with four pots of 16 plants. For the treatments without cold acclimation, the pots with different plants were alternately placed in a controlled-temperature chamber (MIR-254; SANYO) for approximately 30 min at 0°C and then for 1 h at 0°C before the temperature was decreased by 1°C/h. The final desired sub-zero temperature was maintained for the indicated period before the temperature was again increased to 4°C. The plants were then kept at 4°C for 12 h before they were returned to 21°C. Survival was scored 5 days later, and those plants able to maintain a green color at the shoot apex were counted as survivors. For the cold acclimation experiments, 3-week-old plants were acclimated to 4°C in the light for 3 days. The freezing treatment was then performed in the same manner as that for the non-acclimated plants, with the final desired freezing temperature maintained for the indicated period. The experiments were conducted with three independent biological replicates.
Electrolyte Leakage Assay
Electrolyte leakage of detached leaves from 3-week-old plants was measured as previously described (Zhao et al., 2017) with modifications. Plants were grown in soil at 21°C in LD conditions, and the fifth oldest leaf was used. The leaves were placed in tubes containing 100 μl of deionized water. Ice chips were added to the tubes, and the tubes were kept at 0°C for 30 min, followed by a temperature decrease of 1°C/h. The samples were removed at the indicated temperature points and immediately placed on ice. Ten milliliters of deionized water were added to each tube, and the samples were incubated for 1 h at 21°C under gentle shaking, after which the conductivity of the solution was determined via a conductivity meter (DDSJ-308A; INESA). The tubes were then autoclaved at 120°C for 30 min, and the conductivity of the solution was measured again after the samples were cooled to 21°C. Electrolyte leakage was quantified as a percentage of the conductivity after the treatment relative to total conductivity. The experiments were conducted with three independent biological replicates.
qRT-PCR and RT-PCR Assays
Total RNA was extracted with a RNAprep pure plant kit (TIANGEN) and treated with DNaseI to digest the DNA. First-strand cDNA was synthesized from 1 μg of RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) according to the manufacturer’s instructions.
For the qRT-PCR assay, 12-day-old seedlings were grown on agar plates and treated at 4°C in the light, and the plant material was collected in a time-course manner. qRT-PCRs were performed with the SsoFast EvaGreen Supermix (Bio-Rad) using the CFX96 Real-Time System (Bio-Rad). Actin2/8 was used for the normalization of the results (Shi et al., 2012; Liu et al., 2017; Li et al., 2017a). qRT-PCRs were typically performed with at least three independent biological samples, and each was measured with at least three technical repeats. The statistical significance of the differences between two samples was assessed using a Student’s t test.
For the RT-PCR analysis, the roots and whole seedlings were collected from 12-day-old seedlings; the leaves and stems were collected from 5-week-old plants; and the flowers and siliques were collected from 8-week-old plants. β-TUBULIN was used as an internal control. Ethidium bromide staining was used to detect the PCR products.
Confocal Microscopic Analysis
B1L cDNA was amplified and cloned into the pEarleygate104 Gateway binary vector. The plasmid was introduced into Agrobacterium tumefaciens GV3101 and transiently expressed in N. benthamiana leaves. Two days after infiltration, the YFP fluorescence signal was detected with a confocal microscope (Leica SP8). The 5-day-old ProB1L:B1L-GFP and B1L-OE with YFP tag seedlings were also used for a subcellular localization assay.
Histochemical GUS Reporter Gene Expression Analysis
The GUS staining assay was performed as previously described (Chen et al., 2016) with T3 ProB1L:GUS transgenic plants of 1-day-old seedlings, 2-day-old seedlings, 10-day-old seedlings, and 8-week-old mature plants. After staining, the samples were rinsed with acetic acid/methanol [1:3 (v/v)]. The images were collected on a stereomicroscope (Nikon SMZ800).
Y2H Screening and Assay
AH109 was used as a host strain. B1L cDNA was subcloned into the pGBKT7 Gateway binary vector. The transcriptional activation of B1L-pGBKT7 was detected, and B1L-pGBKT7 was used as bait to screen an Arabidopsis thaliana cDNA library. The transformation was performed according to the Clontech Yeast Protocols Handbook (PT3024), with selection on media lacking leucine (Leu), tryptophan (Trp), histidine (His), and adenine (Ade). The positive clones were isolated and sequenced. To determine the interaction between B1L and 14-3-3λ in yeast, B1L-N terminal, B1L-C terminal, B1LS213A, and B1LS213D were amplified and cloned into the pGBKT7 Gateway binary vector. The coding sequence of 14-3-3λ was amplified and cloned into the pGADT7 Gateway binary vector. The coding sequence of 14-3-3ψ was also amplified and cloned into the pGADT7 Gateway binary vector to analyze the interaction between B1L and 14-3-3ψ. The yeast transformation and growth assays were performed as described above.
BiFC Assay
To determine the interaction between B1L and 14-3-3λ, B1L cDNA, B1LS213A, and B1LS213D were amplified and cloned into PNYFP-X, and 14-3-3λ cDNA was amplified and cloned into the PCCFP-X Gateway binary vector. Plasmids containing YFPN-B1L and YFPC-14-3-3λ, YFPN-B1L and YFPC, YFPN and YFPC-14-3-3λ, YFPN-B1LS213D and YFPC-14-3-3λ, or YFPN-B1LS213A and YFPC-14-3-3λ were introduced into A. tumefaciens GV3101 and transformed to N. benthamiana leaves. Two days after infiltration, the YFP fluorescence signal was detected using a Leica SP8 confocal microscope.
CoIP Assay
35S:B1L-FLAG/35S:14-3-3λ-MYC or 35S:FLAG/35S:14-3-3λ-MYC were co-expressed into N. benthamiana leaves. Total protein was subsequently extracted in IP buffer containing 50 mM Tris–HCl, pH 7.6; 150 mM NaCl; 10% glycerol; and 1× Cocktail. The cell debris were removed via two 12-min centrifugations at 16,000 g at 4°C. The supernatant was collected and incubated with anti-FLAG agarose (Abmart) overnight at 4°C. After washing with IP buffer five times, the co-immunoprecipitated products were separated by SDS-PAGE and detected with anti-MYC (1:5,000, Abcam) and anti-FLAG (1:10,000, Abmart) antibodies.
Protein Degradation Assay
Protein degradation assays were performed as previously described (Wang et al., 2009; Liu et al., 2010) with modifications. Arabidopsis seedlings were harvested and ground to a fine powder in liquid nitrogen. Total protein was subsequently extracted in degradation buffer containing 25 mM Tris-HCl, pH 7.5; 10 mM NaCl; 10 mM MgCl2; 5 mM DTT; and 1× Cocktail, and the protein concentration was determined.
For the cell-free degradation assay, 100 μg purified MBP-CBF3 recombinant proteins were incubated with the total proteins that were extracted from wild type, b1l, B1L-OE #1, 14-3-3κλ, or b1l 14-3-3κλ plants in the presence of 1 mM ATP (Sigma) at 25°C for different time courses, and the CBF3-MBP proteins were detected via immunoblotting with an anti-MBP antibody (1:10,000; Abcam). MG132 (Sigma) was added to the various degradation assays, as indicated.
Software Availability
Band intensity quantifications of CBF3 were performed using the ImageJ1. The motifs within B1L protein that are likely to be phosphorylated and bound by 14-3-3 proteins were predicted with Scansite 42. The diagram for the B1L protein was performed using the IBS3.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL/Swiss-Prot databases under the following accession numbers: B1L (AT1G18740), CBF1 (AT4G25490), CBF2 (AT4G25470), CBF3 (AT4G25480), COR15a (AT2G42540), COR15b (AT2G42530), COR47 (AT1G20440), RD29A (AT5G52310), ERF4(AT3G15210), ERF11(AT1G28370), 14-3-3λ (AT5G10450), 14-3-3κ (AT5G65430), and 14-3-3ψ (AT5G38480).
Results
Cold Induces the Expression of B1L, Which Positively Modulates Freezing Tolerance
To investigate the function of B1L in plant freezing tolerance, a T-DNA insertion mutant (b1l) was obtained from the Arabidopsis Biological Resource Center (ABRC), and the complete disruption of B1L expression in the b1l mutant was verified (Supplementary Figure S1A). Three-week-old plants showing similar growth and development in wild type and b1l were used to examine the freezing tolerance (Supplementary Figure S1B). The b1l mutant was more sensitive to freezing treatment than wild type under cold-acclimated (CA) conditions (Figures 1A,B). ProB1L:B1L-3×FLAG was then transformed to b1l mutant, and the complementation lines (B1L b1l) fully restored the freezing sensitivity of b1l (Supplementary Figure S2). Transgenic plants overexpressing B1L-YFP (B1L-OE) were also used to examine the role of B1L (Supplementary Figure S3). B1L-OE #1 and #2 plants were more freezing tolerant compared to wild type, particularly under non-acclimated (NA) conditions (Figures 1C–F). Consistently, ion leakage in the b1l mutant was higher than that in wild type after freezing treatment (Figure 1G), whereas the ion leakage of the B1L-OE #1 and #2 plants was lower than that of wild type after freezing treatment (Figure 1H). A quantitative real-time PCR (qRT-PCR) assay was used to investigate the expression of B1L under cold treatment (4°C). The expression of B1L quickly increased after 1 h and reached a peak after 6 h (Figure 1I). These data indicate that B1L acts as a positive regulator of freezing tolerance in Arabidopsis.
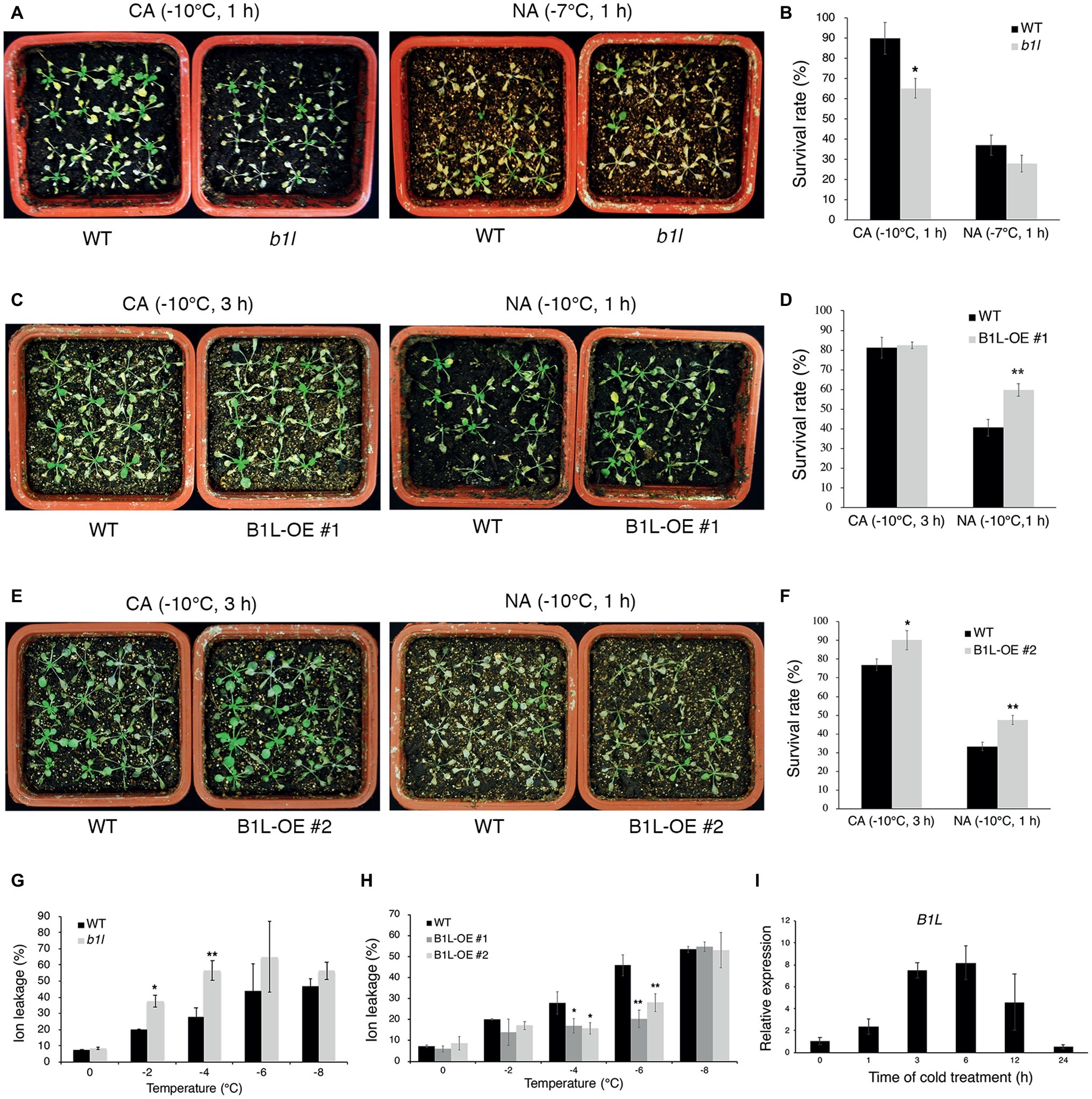
Figure 1. B1L positively modulates freezing tolerance in Arabidopsis. (A–F) Freezing tolerance (A,C,E) and survival rates (B,D,F) of wild type (WT), b1l mutant, and B1L-overexpressing plants under non-acclimated (NA) or cold-acclimated (CA) conditions. The 3-week-old plants were treated at −7 or −10°C for 1 h (NA) or were pretreated at 4°C for 3 days and then treated at −10°C for 1 or 3 h (CA). For each line, the survival rate assay was performed with four pots of 16 plants and scored 5 days later. The photos presented one pot of each line. The data are shown as means of three independent biological replicates ± SD. Asterisks indicate significant differences (*p < 0.05, and **p < 0.01) from wild type. (G,H) Ion leakage of wild type, b1l mutant, and B1L-overexpressing plants in (A,C,E) after exposure to the temperature indicated. Data are means ±SD. n = 4 leaves, each from a different plant. Asterisks indicate significant differences (*p < 0.05 and **p < 0.01) from wild type. (I) Expression of B1L in wild type under cold treatment. Total RNA was extracted from 12-day-old seedlings treated at 4°C for 0, 1, 3, 6, 12, 24 h and then subjected to qRT-PCR. Actin2/8 was used as a control. The expression of B1L in untreated wild type was set to 1. The data are shown as means of three independent biological replicates ± SD.
B1L Was Shown to Modulate the Expression of Genes in the CBF Pathway
To examine whether B1L participates in plant cold acclimation, the expression of CBF pathway genes that perform important roles in cold acclimation was examined through qRT-PCR. The qRT-PCR results showed that both the CBFs and CBF target genes were dramatically reduced in the b1l mutants compared to wild type (Figure 2). These results indicate that B1L positively regulates the expression of CBF pathway genes.
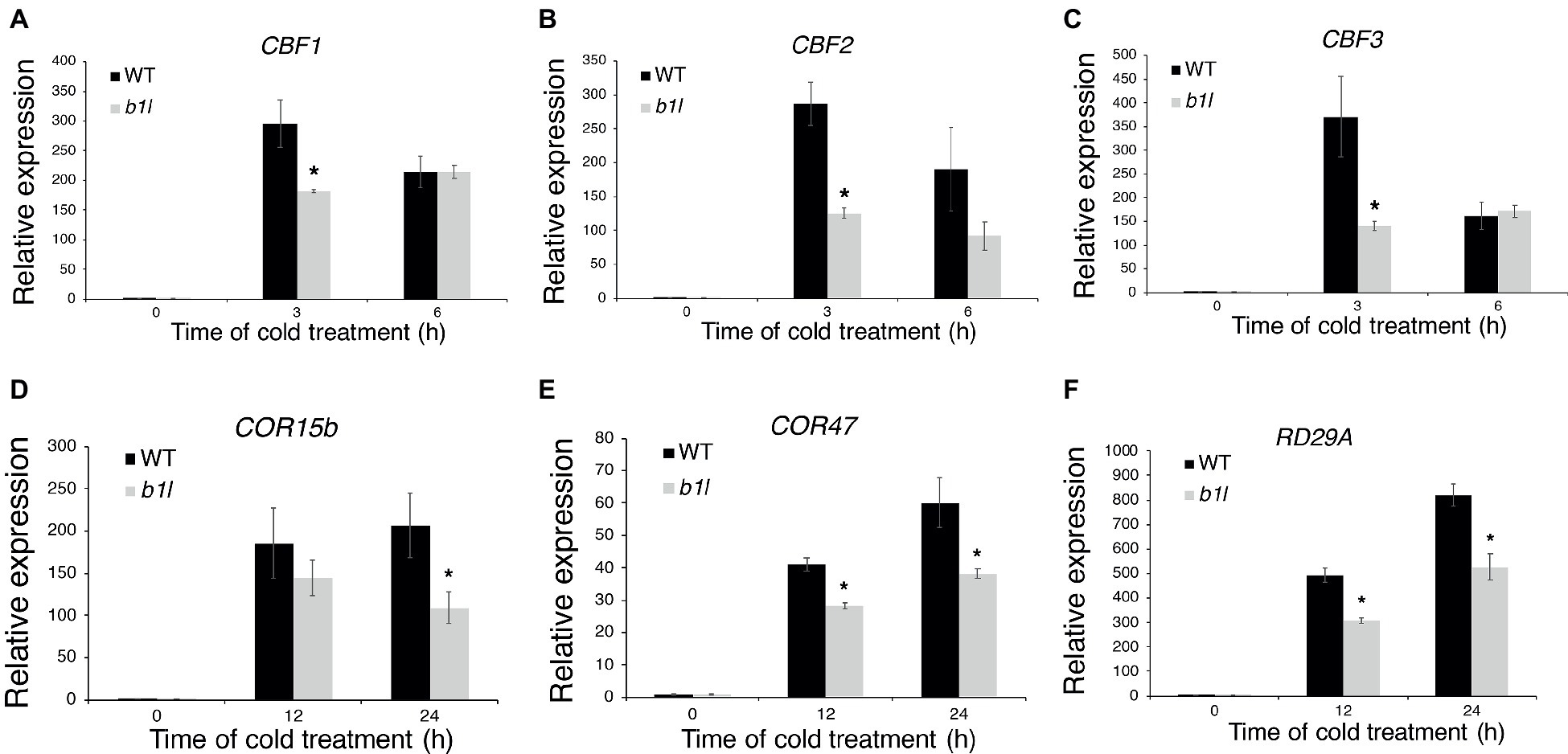
Figure 2. B1L is a positive regulator of CBF pathway genes. The expression of CBF1, CBF2, and CBF3 (A–C) and CBF target genes COR15b, COR47, and RD29A (D–F) in wild type and b1l seedlings was determined via qRT-PCR analyses. Total RNA was extracted from 12-day-old seedlings treated at 4°C for 0, 3, 6 h for CBFs and 0, 12, 24 h for CBF target genes and then subjected to qRT-PCR. The data are shown as means of three independent biological replicates ± SD. Asterisks indicate significant differences (*p < 0.05) from wild type.
B1L Is Expressed in Most Tissues and B1L Protein Localizes to the Cytoplasm and the Nucleus
To analyze the expression pattern of B1L, the total RNA from different tissues of wild type was collected and tested. Semi-quantitative RT-PCR results showed that B1L was expressed in most tissues; the mRNA from b1l seedlings was used as a negative control (Figure 3A). GUS staining assay using ProB1L:GUS transgenic plants revealed that B1L was predominately expressed in the roots, leaves, and flowers (Figures 3B–F).
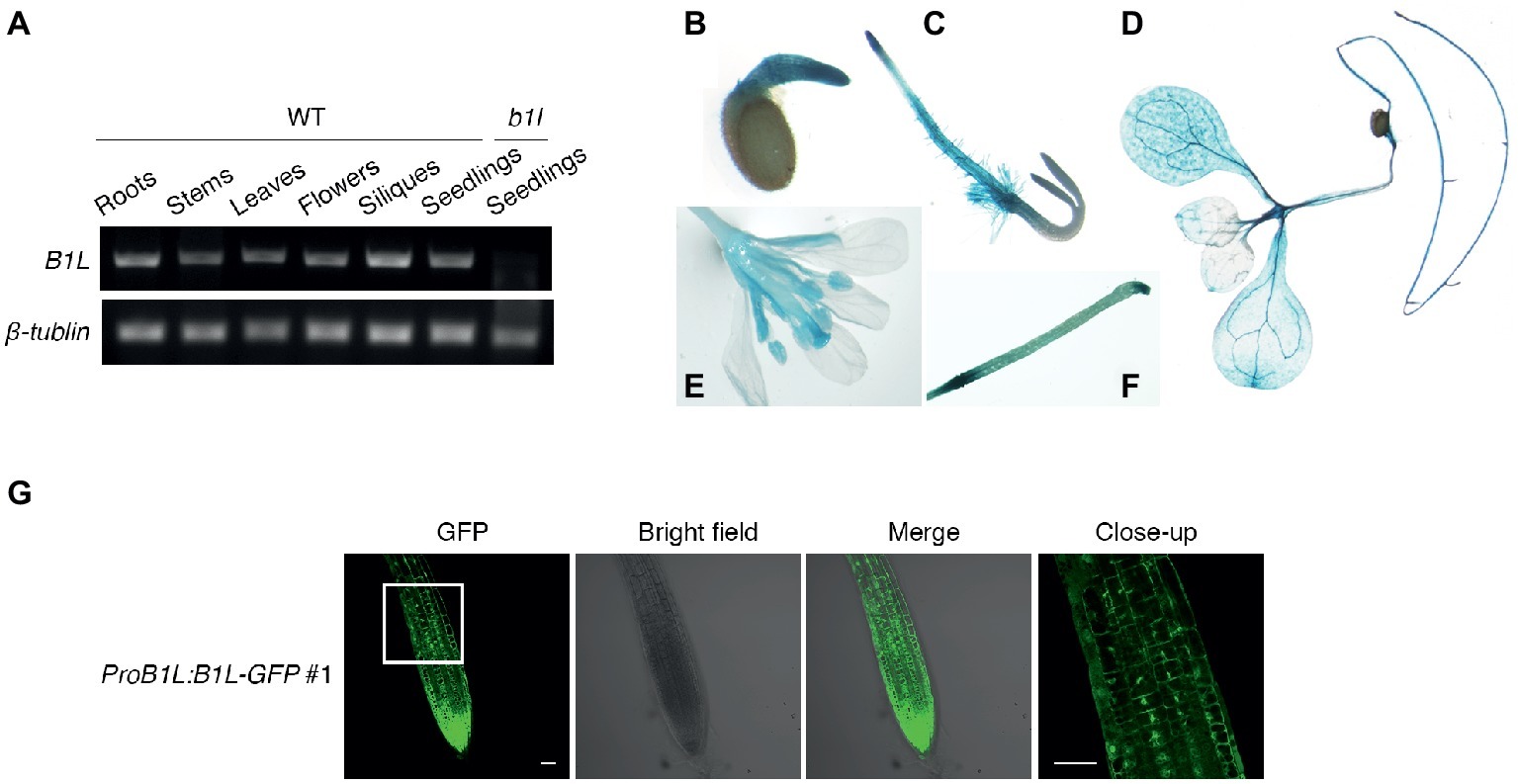
Figure 3. The expression of B1L in different tissues and the subcellular localization of B1L. (A) RT-PCR analysis of B1L transcripts in the different tissues of wild type. The B1L transcript in the b1l mutant seedlings was used as a negative control. β-TUBULIN was used as a loading control. (B–F) Histochemical analysis of the GUS reporter gene expression driven by the B1L promoter. The GUS signal was detected in 1-day-old seedlings (B), 2-day-old-seedlings (C), 10-day-old seedlings (D), and 8-week-old mature plants (E,F). (G) Localization of B1L in transgenic plants expressing B1L-GFP driven by its native promoter (ProB1L:B1L-GFP #1). The signals in the root tips of 5-day-old plants were visualized. Bar = 50 μm.
To examine the subcellular localization of B1L, A ProB1L:B1L-GFP construct was generated and transformed to b1l mutant plants (ProB1L:B1L-GFP #1). Under microscope, GFP fluorescence signal was detected in both the cytoplasm and nucleus (Figure 3G). The transient expression of 35S:YFP-B1L in Nicotiana benthamiana and the stable expression of 35S:YFP-B1L in Arabidopsis were also used to show that B1L localizes to the cytoplasm and nucleus (Supplementary Figure S4).
B1L Directly Interacts With 14-3-3λ Both in vitro and in vivo
To further investigate B1L function in the freezing tolerance of plants, a yeast two-hybrid (Y2H) screening system was used. A 14-3-3 family protein, 14-3-3λ, was identified. A bimolecular fluorescence complementation (BiFC) assay was used to confirm the interaction, and the results showed that B1L interacts with 14-3-3λ under both cold treatment (4°C, 6 h) and normal conditions (Figure 4A). A co-immunoprecipitation (coIP) assay using co-expressed B1L-FLAG and 14-3-3λ-MYC was also performed and the result verified that B1L directly interacts with 14-3-3λ in vivo (Figure 4B).
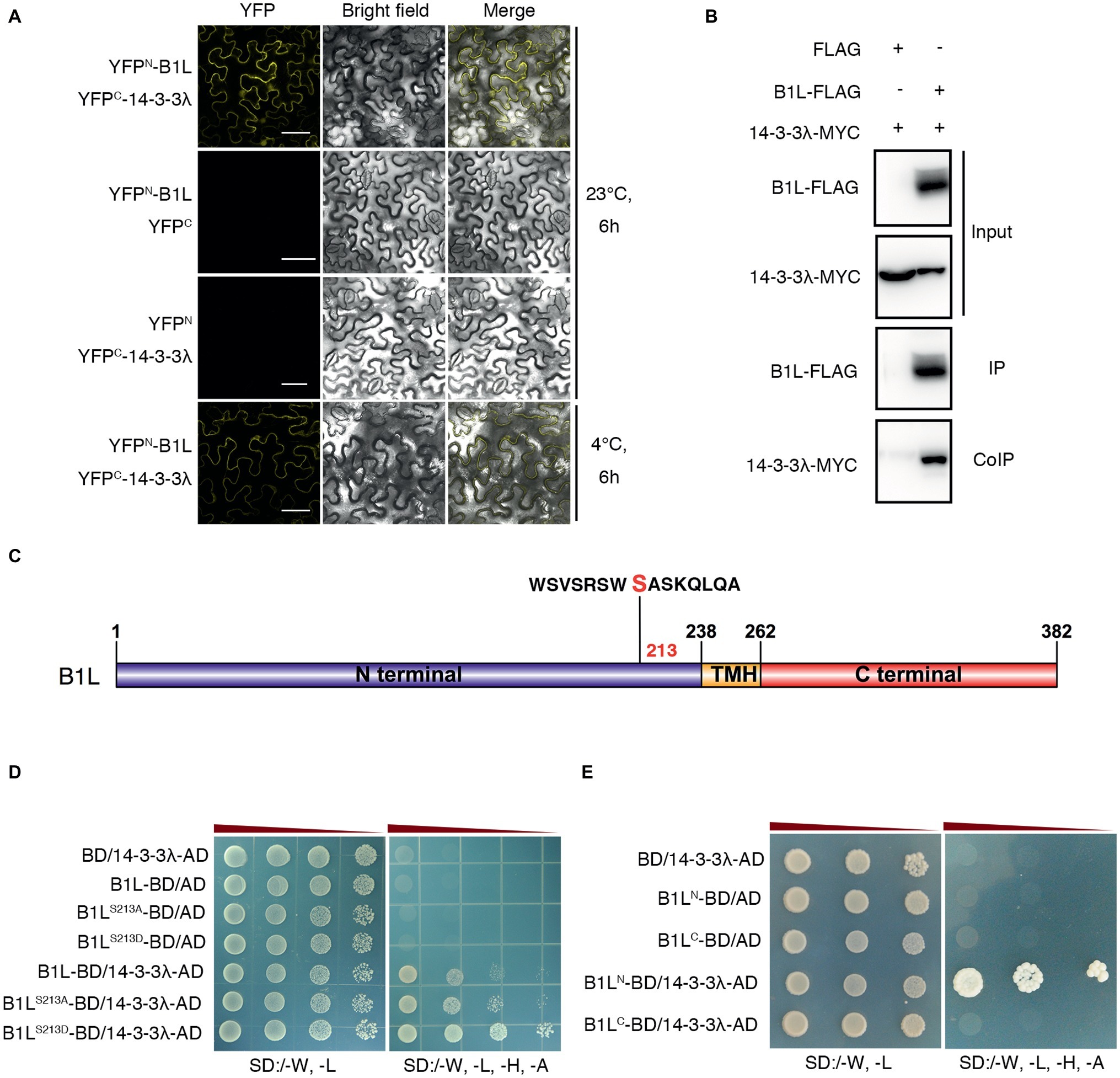
Figure 4. B1L interacts with 14-3-3λ, and serine 214 in the N-terminal of B1L modulates the interaction between B1L and 14-3-3λ. (A) BiFC analysis in N. benthamiana showing the interaction between B1L and 14-3-3λ. The plants were pretreated with or without 4°C for 6 h, and then, the YFP signals were detected. Bar = 50 μm. (B) CoIP assay showing the interaction between B1L and 14-3-3λ in plants. Total proteins were extracted from transformedN. benthamiana leaves, immunoprecipitated with an anti-FLAG antibody, and detected with an anti-MYC antibody and an anti-FLAG antibody. (C) Diagram of B1L-truncated proteins and B1L-mutated proteins used for the Y2H assays. Serine 213 in the N-terminal of B1L was a candidate phosphorylated site that can be recognized by 14-3-3 proteins. This site was mutated to alanine (B1LS213A) and aspartic acid (B1LS213D) to mimic nonphosphorylation and autophosphorylation of B1L, respectively. The N-terminal (amino acids 1-238) and the C-terminal (amino acids 262-382) of B1L were used to identify the binding domain of B1L that interacts with 14-3-3λ. (D,E) Y2H analysis of the interaction between 14-3-3λ and B1L native protein, B1L-mutated proteins or B1L-truncated proteins. Panels show yeast serial decimal dilutions.
The 14-3-3 proteins are well-known to bind to many proteins that are phosphorylated by recognizing phosphoserine or phosphothreonine within their conserved binding motifs (Jaspert et al., 2011; Wang et al., 2011; Yoon and Kieber, 2013; Zhou et al., 2014; Huang et al., 2018). To search for 14-3-3 motifs in B1L sequence, the Scansite 44 was used to predict the candidate site, and Serine 213 in the N-terminal of B1L was identified (Figure 4C). This site was then mutated to alanine (B1LS213A) and aspartic acid (B1LS213D) to mimic nonphosphorylation and autophosphorylation, respectively. The Y2H assays showed that both B1LS213A and B1LS213D could interact with 14-3-3λ (Figure 4D). The BiFC assay was also performed and showed the same results (Supplementary Figure S5). These results suggest that B1LS213A and B1LS213D may not be sufficient to affect the interaction between B1L and 14-3-3λ. Then, the N-terminal (amino acids 1-238) and the C-terminal (amino acids 262-382) of B1L were used to identify the binding domain of B1L that interacts with 14-3-3λ. The Y2H assays showed that the N-terminal of B1L, but not the C-terminal of B1L, is sufficient for the interaction (Figure 4E).
B1L Regulates Freezing Tolerance, Possibly Through a 14-3-3λ-Dependent Pathway
To understand the genetic interaction between B1L and 14-3-3λ, the b1l 14-3-3λ double mutant was generated. The 14-3-3λ mutant displayed freezing tolerance similar to wild type (Supplementary Figures S6A,B), consistent with a previous report (Liu et al., 2017). The b1l 14-3-3λ mutant displayed a similar freezing sensitivity to b1l (Supplementary Figures S6C,D). 14-3-3κ is a close homologue to 14-3-3λ, and the 14-3-3κλ double mutant showed enhanced freezing tolerance in the previous report (Liu et al., 2017), as well as in our study (Figures 5A,B). A b1l 14-3-3κλ triple mutant was then generated, and this mutant showed enhanced freezing tolerance compared to that in wild type (Figures 5C,D). Consistently, the expression of COR genes was higher in 14-3-3κλ and b1l 14-3-3κλ plants than in wild type (Figures 6A–C). Intriguingly, the expression of CBF3 was significantly higher in 14-3-3κλ plants than in wild type (Figure 6F), which is consistent with the results from a previous study (Liu et al., 2017). qPCR results also showed that the reduced expression of all three CBF genes in the b1l mutants was rescued in b1l 14-3-3κλ plants (Figures 6D–F), indicating that B1L and 14-3-3 proteins may also participate in modulating the expression of CBFs. These results reveal that B1L regulates freezing tolerance possibly through 14-3-3λ.
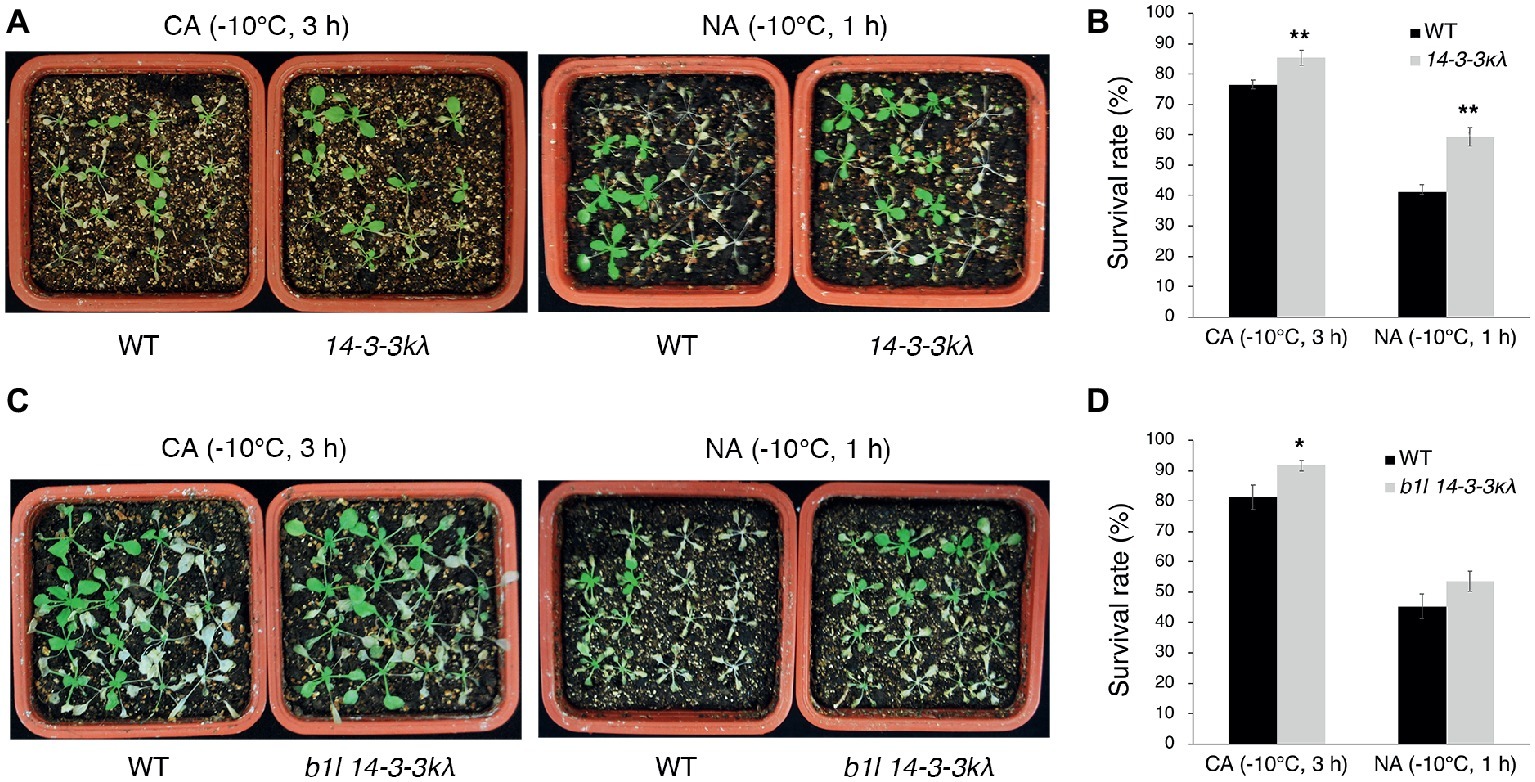
Figure 5. B1L regulates freezing tolerance via a 14-3-3λ-dependent way. Freezing tolerance (A,C) and survival rates (B,D) of 14-3-3kλ mutants and b1l 14-3-3kλ mutants under NA or CA conditions. The assays were performed as in Figure 1. The data are shown as means of three independent biological replicates ± SD. Asterisks indicate significant differences (*p < 0.05, and **p < 0.01) from wild type.
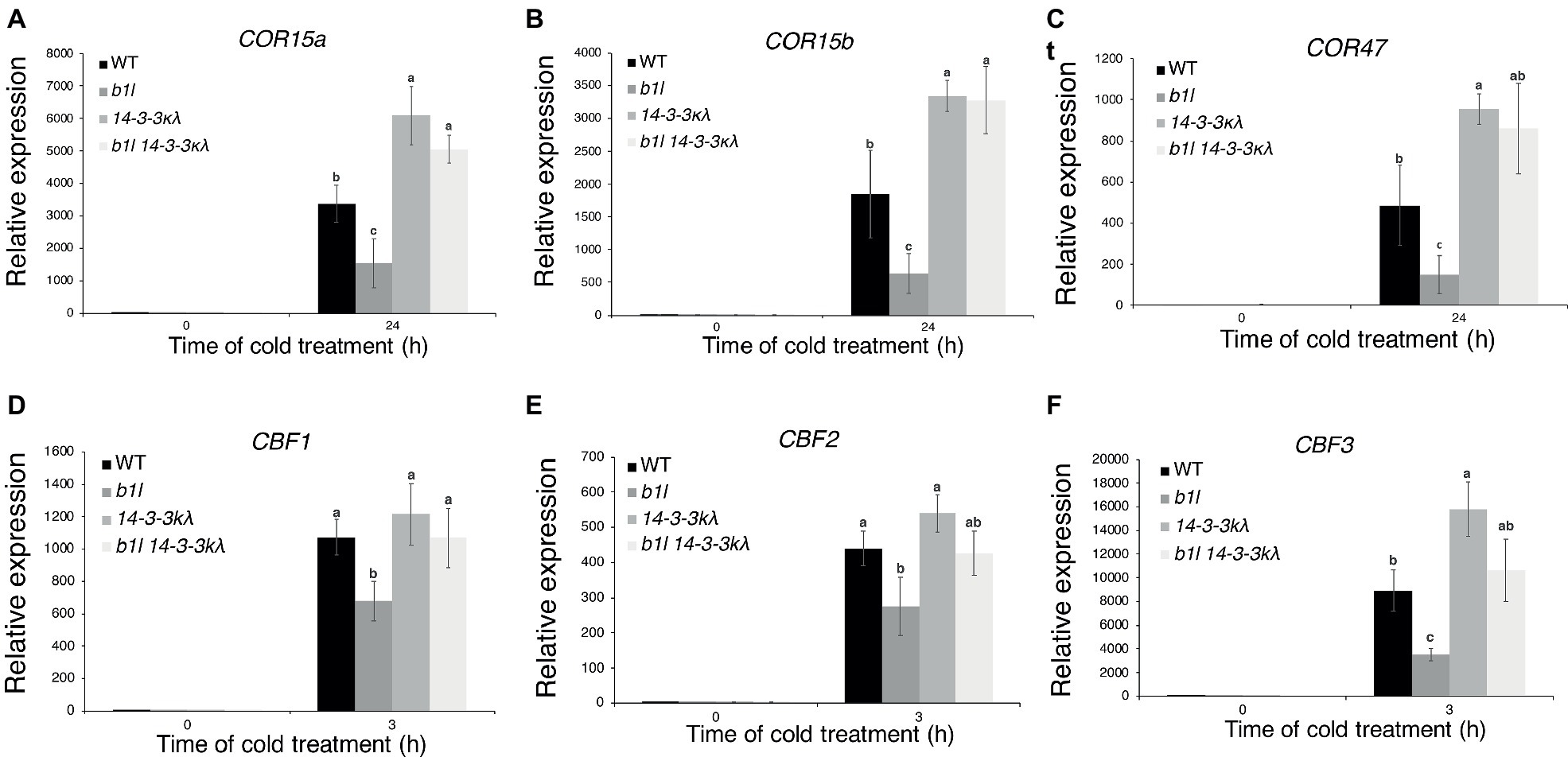
Figure 6. The expression of CBFs and COR genes in 14-3-3kλ and b1l 14-3-3kλ under cold treatment. The expression of the CBF target genes COR15a, COR15b, and COR47 (A–C) and CBF genes (D–F) in wild type, b1l, 14-3-3kλ, and b1l 14-3-3kλ seedlings was determined by qRT-PCR analyses. The data are shown as means of three independent biological replicates ± SD. Significant differences (p < 0.05) are indicated by different lowercase letters.
B1L Suppresses the Ubiquitin-Mediated Degradation of CBF3 via 14-3-3λ
The 14-3-3 proteins were shown to interact with and destabilize CBFs in the ubiquitin/26S proteasome pathway under cold stress (Liu et al., 2017). To verify whether B1L affects the degradation of CBFs, several CBF3 degradation assays were performed.
First, an in vitro cell-free degradation assay was performed using purified CBF3-MBP proteins that were expressed in Escherichia coli. Western blotting results showed that the degradation of the CBF3-MBP increased when it was treated with the b1l mutant total proteins; however, that the degradation of the CBF3-MBP was suppressed when it was treated with B1L-OE #1 total proteins (Figures 7A,B). The degradation could be inhibited by MG132 (an inhibitor of 26S proteasome degradation) (Figures 7C,D). These results indicate that the stability of CBF3 is mediated by B1L.
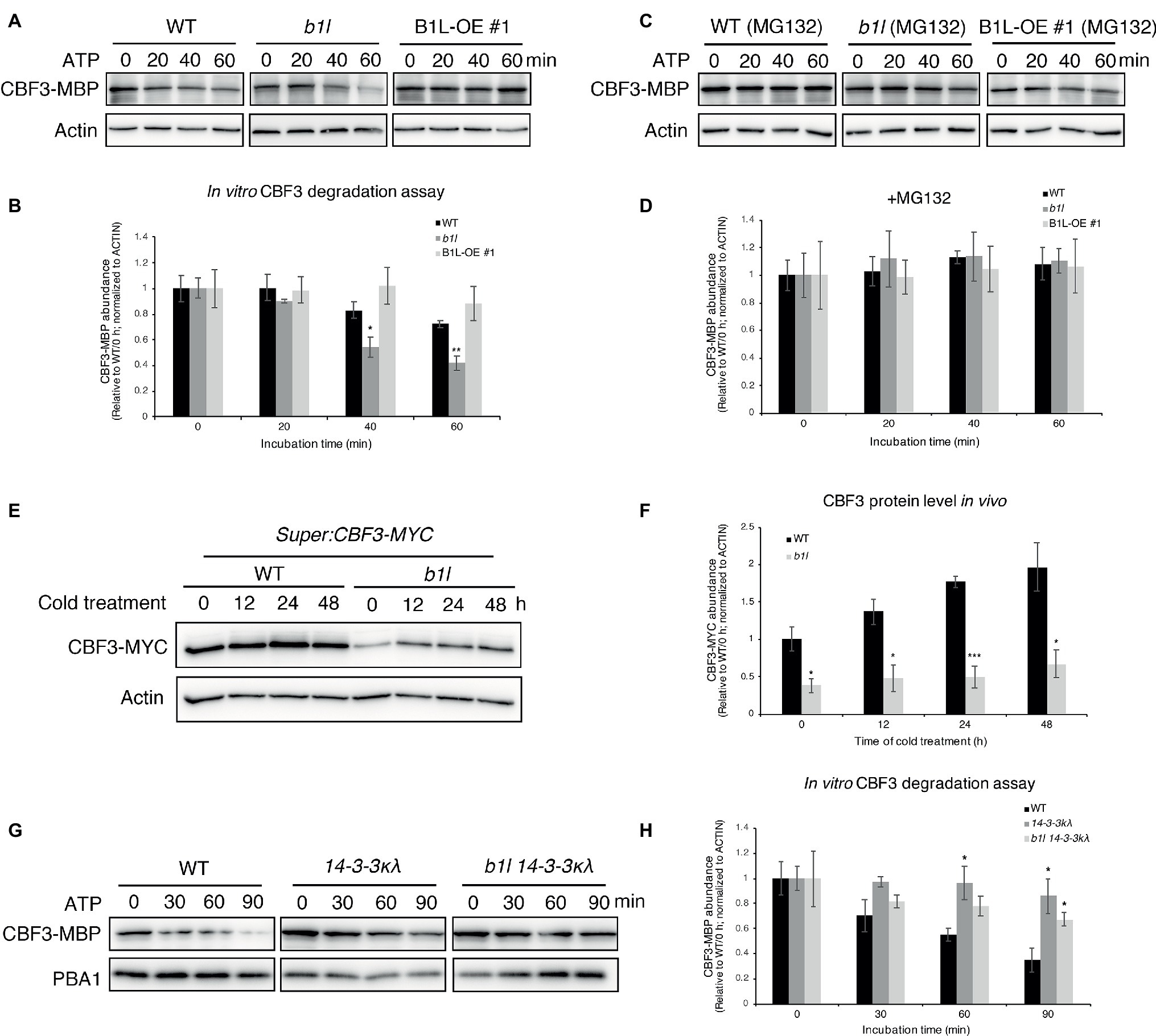
Figure 7. B1L inhibits the ubiquitin-mediated degradation of CBF3 via a 14-3-3λ-dependent way. (A–D) In vitro cell-free degradation assay showing that B1L inhibits the degradation of CBF3. The recombinant purified CBF3-MBP proteins were incubated with the total proteins extracted from wild type, b1l or B1L-OE #1 seedlings in the presence of ATP. These total proteins were pretreated with or without 50 μM MG132 (an inhibitor of 26S proteasome degradation) for 3 h. Typical immunoblotting results were shown (A,C), along with quantification (B,D). Actin was used as a control. The ratio of the band intensity of CBF3-MBP to actin without the ATP treatment was set to 1. The data are shown as means of three independent biological replicates ± SE. Asterisks indicate significant differences (*p < 0.05, and **p < 0.01) from wild type. (E,F) In vivo degradation assay showing that the CBF3 is stabilized by B1L under cold stress. Super:CBF3-MYC transgenic plants were crossed with b1l to generate Super:CBF3-MYC/b1l transgenic plants, and then the protein level of CBF3-MYC in both wild type background and b1l background was tested after different times of cold treatment. Actin served as a control. The ratio of the band intensity of CBF3 to actin without the cold treatment in Super:CBF3-MYC plants was set to 1. The data are shown as means of four independent biological replicates ± SE. Asterisks indicate significant differences (*p < 0.05, and ***p < 0.001) from Super:CBF3-MYC plants. (G,H) In vitro cell-free degradation assays showing that B1L inhibits the degradation of CBF3 via 14-3-3λ. The recombinant purified CBF3-MBP proteins were incubated with the total proteins extracted from wild type, 14-3-3λ or b1l 14-3-3λ seedlings in the presence of ATP. PBA1 was used as a control. The ratio of the band intensity of CBF3-MBP to actin without the ATP treatment was set to 1. The data are shown as means of three independent biological replicates ± SE. Asterisks indicate significant differences (*p < 0.05) from wild type.
Then, the protein level of CBF3 in vivo was examined. Super:CBF3-MYC transgenic plants (Liu et al., 2017) were crossed with b1l, and the protein levels of CBF3-MYC in both wild type background and b1l background were analyzed after different time of cold treatment. The protein level of CBF3-MYC was much lower in b1l mutants than wild type before and after cold treatment (4°C) (Figures 7E,F). These results further reveal that the stability of CBF3 is mediated by B1L.
To explore the role of 14-3-3 proteins in the B1L-mediated degradation of CBFs, the purified CBF3-MBP proteins were used to test the stability of CBF3 in 14-3-3κλ and b1l 14-3-3κλ mutants. Western blotting results showed that the 14-3-3κλ mutant suppressed the degradation of CBF3-MBP compared to wild type (Figures 7G,H), as previously reported (Liu et al., 2017), and that the b1l 14-3-3κλ mutant also inhibited the degradation of the CBF3 (Figures 7G,H). These results show that B1L regulates CBF3 degradation via 14-3-3λ.
B1L Acts Upstream of CBFs, Positively Regulating Plant Freezing Tolerance
To further examine the relationship between B1L and CBFs, b1l mutant was crossed with cbfs triple mutant (Jia et al., 2016). The cbfs mutant was more sensitive to freezing treatment than wild type (Figures 8A,B), consistent with previous studies (Jia et al., 2016; Liu et al., 2017). The b1l cbfs quadruple mutant did not aggravate the freezing sensitivity of cbfs (Figures 8C,D). These results further indicate that B1L positively regulating plant freezing tolerance via CBF proteins.
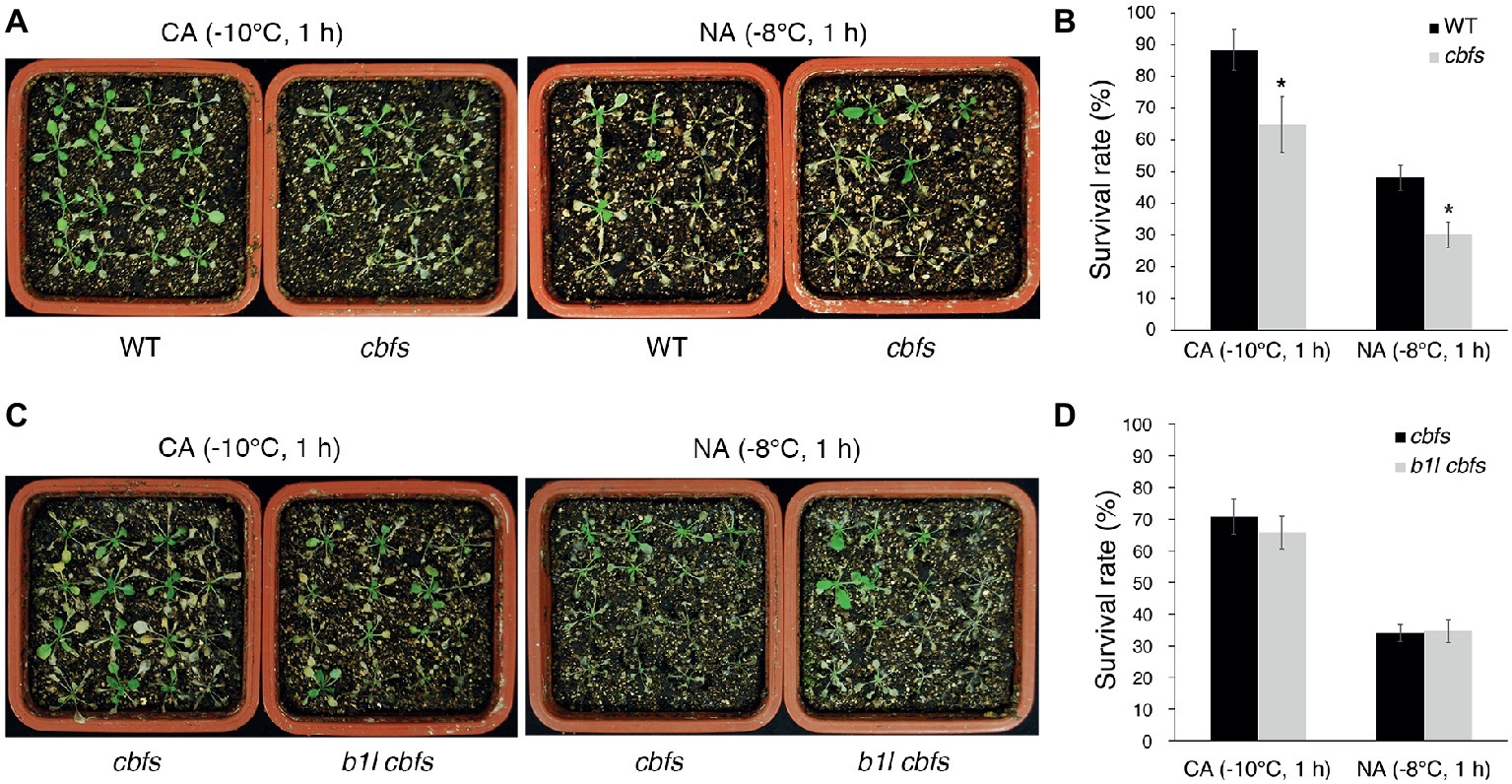
Figure 8. B1L acts upstream of CBFs to regulate cold signaling. Freezing tolerance (A,C) and survival rates (B,D) of cbfs mutants and b1l cbfs mutants under NA or CA conditions. The assays were performed as in Figure 1. The data are shown as means of three independent biological replicates ± SD. Asterisks indicate significant differences (*p < 0.05) from wild type.
Discussion
CBF signaling pathway has important roles in cold acclimation. The expression of CBFs is regulated by numerous transcription factors. However, studies about the posttranslational regulation of CBFs are limited. Here, we show that B1L participates in regulating freezing tolerance partly through repressing the degradation of CBFs (Figure 9). Several lines of evidence were provided: (1) B1L promotes the expression of CBF pathway genes and therefore freezing tolerance in Arabidopsis. (2) B1L directly interacts with 14-3-3λ. (3) B1L inhibits the 14-3-3 protein-mediated degradation of CBFs. (4) The b1l cbfs quadruple mutant displayed a freezing sensitivity similar to cbfs.
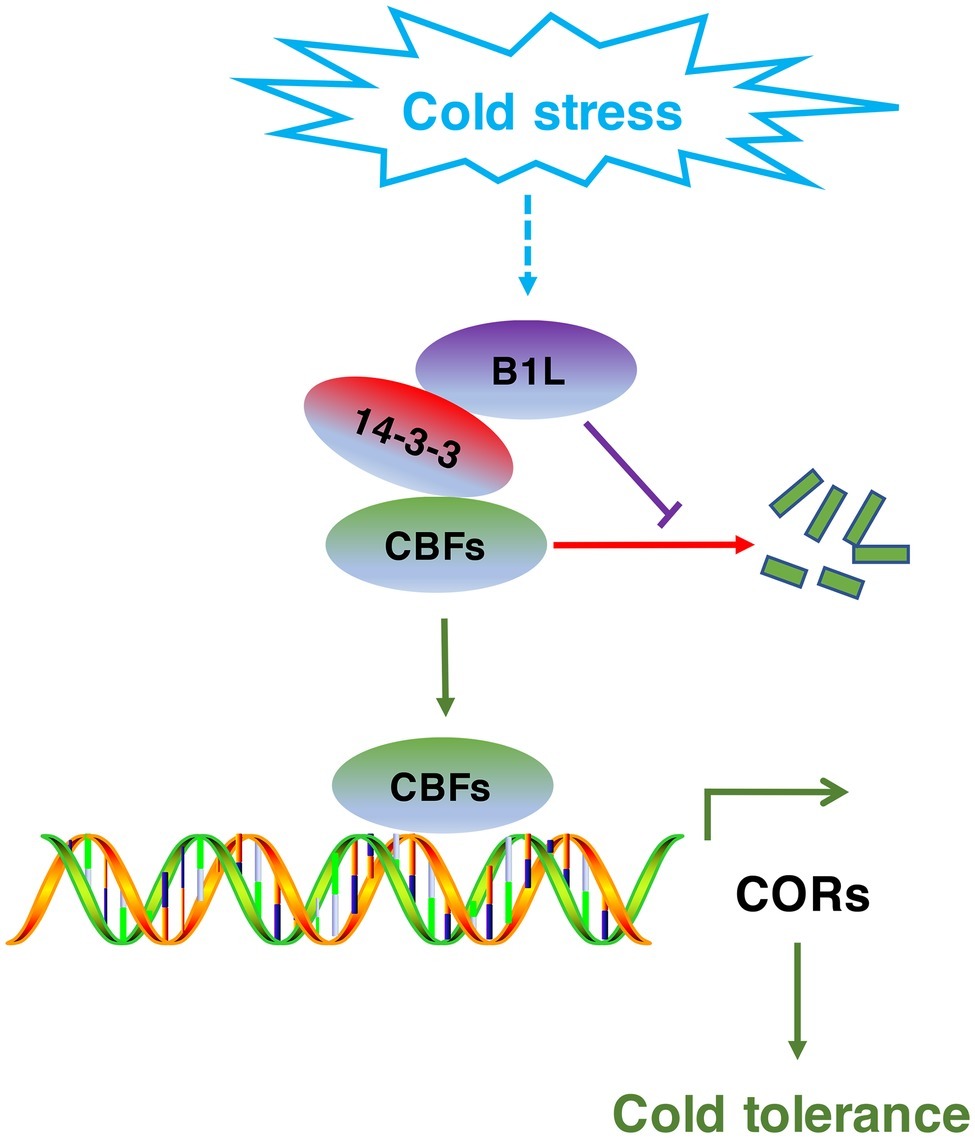
Figure 9. Model for the regulation of the CBF signaling pathway by B1L under cold stress. The expression of B1L is induced by cold stress. Then, B1L interacts with 14-3-3λ to reduce the 14-3-3λ-mediated degradation of CBFs. As a result, CBFs induce the expression of their target genes such as COR15a, COR47, and RD29A to enhance plant freezing tolerance.
The 14-3-3 proteins serve important roles in many processes, such as stomata movements, phytohormone regulation, biotic stress, and abiotic stress (Cotelle et al., 2000; Gampala et al., 2007; Wang et al., 2011; de Boer et al., 2013; Catala et al., 2014; Kaundal et al., 2017; Keicher et al., 2017; Liu et al., 2017). These proteins mostly interact with phosphorylated proteins and affect the subcellular localization, protein stability, enzymatic activity of target proteins or the interaction between the target proteins and other proteins (Jaspert et al., 2011; Wang et al., 2011; Yoon and Kieber, 2013; Zhou et al., 2014; Huang et al., 2018). Our results showed that B1L interacted with 14-3-3λ (Figure 4), indicating that B1L may also be a phosphorylated protein. Serine 213 in the N-terminal of B1L is a potential phosphorylated site that can be recognized by 14-3-3 proteins (Figure 4C), and the N-terminal of B1L is sufficient for the interaction between B1L and 14-3-3λ. However, mutating Serine 213 to alanine (B1LS213A) did not abolish the interaction between B1L and 14-3-3λ, indicating the possible involvement of new sites to affect the interaction between B1L and 14-3-3λ (Figure 4D; Supplementary Figure S5). The phosphorylated site of B1L that affects the interaction between B1L and 14-3-3λ and the biological roles of these sites need to be further investigated.
The 14-3-3 protein 14-3-3ψ (also known as RARE COLD INDUCIBLE 1A or GRF3) has also been shown to participate in the regulation of freezing tolerance via an ethylene biosynthesis pathway (Catala et al., 2014). It interacts with ACS6 and inhibits the expression of ethylene-related genes and CBFs, leading to decreased freezing tolerance (Catala et al., 2014). Although the expression of CBFs was reduced in b1l mutants under cold treatment (Figure 2), the expression of ethylene-related genes (ERF4 and ERF11) was not significantly changed in b1l mutants compare to wild type (Supplementary Figures S7A,B). The interaction between B1L and 14-3-3ψ was also not observed in the Y2H system (Supplementary Figure S7C). These results indicate that B1L may specifically interact with 14-3-3λ, modulating freezing tolerance via an ethylene-independent pathway.
The stability of CBFs has been shown to be modulated by 14-3-3 proteins and BTF3L via a ubiquitin/26S proteasome pathway (Liu et al., 2017; Ding et al., 2018). HOS15, which is an E3 ligase, interacts with CBFs, affecting the expression of COR genes but does not participate in the regulation of CBF degradation (Park et al., 2018). Therefore, the components of the ubiquitin/26S proteasome that participate in degrading CBFs are still unknown. More proteins that are involved in the ubiquitin/26S proteasome pathway need to be identified to elucidate the mechanism of CBF stability. Our protein degradation assay in vivo and in vitro both showed that B1L affects the stability of CBF (Figures 7A–D), indicating that B1L is another regulator that participate in the regulation of CBF proteins.
The overexpression of CBFs leads to dwarfism and late-flowering (Jaglo-Ottosen, 1998; Achard et al., 2008; Zhou et al., 2017). ICE1 is a positive regulator of CBFs, and its overexpressing lines result in late-flowering, due to ICE1 directly binding to the promoter of FLC and inducing the expression of the FLC (Lee et al., 2015). Therefore, plants fine-tune CBF signaling pathway to avoid harmful effects during their development. To be specific, 14-3-3λ can be translocated to the nucleus under cold conditions, reducing the abundance of CBFs (Liu et al., 2017). Cold treatment induces the expression of B1L but does not obviously change the subcellular localization of B1L (Supplementary Figure S8). We hypothesis that the interaction between 14-3-3λ and B1L may lead to a delay in the nuclear translocation of 14-3-3λ after cold treatment. This translocation delay in turn allows plants to fine-tune the abundance of CBFs to adapt to cold stress.
Data Availability
All datasets for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
TC, J-HC, WZ, GY, and L-JY performed the experiments. TC analyzed the data. TC, HZ, and L-ZA designed the project and drafted the manuscript. D-ML, BL, and H-MS revised the manuscript critically for important intellectual content. HZ and L-ZA supervised the project and complemented the writing.
Funding
This work was supported by the Key Program National Natural Science Foundation of China (41830321); the National Natural Science Foundation of China (31770432, 31570488, 31301078); the National Basic Research Program of China (973 Program) (2013CB429904); the Science and Technology Partnership Program, Ministry of Science and Technology of China (KY201501008); the National Science and Technology Major Project in Gansu (17ZD2WA017); and the Ministry of Agriculture of the People’s Republic of China (2016ZX08009-003-002).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jia Li and Xiao-Ping Gou (Lanzhou University) as well as Shu-Hua Yang (China Agricultural University) for kindly providing Arabidopsis seeds. We thank Sui-Wen Hou (Lanzhou University) for valuable comments on the manuscript. We are grateful to Xiu-Le Yue and the Core Facility for Life Science Research (Lanzhou University) for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00807/full#supplementary-material
Footnotes
References
Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., and Genschik, P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. doi: 10.1105/tpc.108.058941
Agarwal, M., Hao, Y., Kapoor, A., Dong, C. H., Fujii, H., Zheng, X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. doi: 10.1074/jbc.M605895200
Catala, R., Lopez-Cobollo, R., Mar Castellano, M., Angosto, T., Alonso, J. M., Ecker, J. R., et al. (2014). The Arabidopsis 14-3-3 protein rare cold inducible 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26, 3326–3342. doi: 10.1105/tpc.114.127605
Chen, L., Guan, L., Qian, P., Xu, F., Wu, Z., Wu, Y., et al. (2016). NRPB3, the third largest subunit of RNA polymerase II, is essential for stomatal patterning and differentiation in Arabidopsis. Development 143, 1600–1611. doi: 10.1242/dev.129098
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Cotelle, V., Meek, S. E., Provan, F., Milne, F. C., Morrice, N., and MacKintosh, C. (2000). 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J. 19, 2869–2876. doi: 10.1093/emboj/19.12.2869
de Boer, A. H., van Kleeff, P. J. M. M., and Gao, J. (2013). Plant 14-3-3 proteins as spiders in a web of phosphorylation. Protoplasma 250, 425–440. doi: 10.1007/s00709-012-0437-z
Ding, Y., Jia, Y., Shi, Y., Zhang, X., Song, C., Gong, Z., et al. (2018). OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 37:e98228. doi: 10.15252/embj.201798228
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Dong, M. A., Farre, E. M., and Thomashow, M. F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 7241–7246. doi: 10.1073/pnas.1103741108
Gampala, S. S., Kim, T. W., He, J. X., Tang, W., Deng, Z., Bai, M. Y., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13, 177–189. doi: 10.1016/j.devcel.2007.06.009
Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M., and Thomashow, M. F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. doi: 10.1046/j.1365-313x.1998.00310.x
Guy, C. L. (1990). Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 187–223.
Huang, X., Zhang, Q., Jiang, Y., Yang, C., Wang, Q., and Li, L. (2018). Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. elife 7, 470–476. doi: 10.7554/eLife.31636
Jaglo-Ottosen, K. R. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. doi: 10.1126/science.280.5360.104
Jaspert, N., Throm, C., and Oecking, C. (2011). Arabidopsis 14-3-3 proteins: fascinating and less fascinating aspects. Front. Plant Sci. 2:96. doi: 10.3389/fpls.2011.00096
Jia, Y., Ding, Y., Shi, Y., Zhang, X., Gong, Z., and Yang, S. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212, 345–353. doi: 10.1111/nph.14088
Jiang, B., Shi, Y., Zhang, X., Xin, X., Qi, L., Guo, H., et al. (2017). PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 114, E6695–E6702. doi: 10.1073/pnas.1706226114
Kaundal, A., Ramu, V. S., Oh, S., Lee, S., Pant, B. D., Lee, H.-K., et al. (2017). General control nonrepressible4 (GCN4) degrades 14-3-3 and the RIN4 complex to regulate stomatal aperture with implications on nonhost disease resistance and drought tolerance. Plant Cell 29, 2233–2248. doi: 10.1105/tpc.17.00070
Keicher, J., Jaspert, N., Weckermann, K., Möller, C., Throm, C., Kintzi, A., et al. (2017). Arabidopsis 14-3-3 epsilon members contribute to polarity of PIN auxin carrier and auxin transport-related development. elife 6, 1–21. doi: 10.7554/eLife.24336.001
Kulich, I., Cole, R., Drdová, E., Cvrčková, F., Soukup, A., Fowler, J., et al. (2010). Arabidopsis exocyst subunits SEC8 and EXO70A1 and exocyst interactor ROH1 are involved in the localized deposition of seed coat pectin. New Phytol. 188, 615–625. doi: 10.1111/j.1469-8137.2010.03372.x
Lee, J. H., Jung, J. H., and Park, C. M. (2015). Inducer of CBF expression 1 integrates cold signals into flowering locus C-mediated flowering pathways in Arabidopsis. Plant J. 84, 29–40. doi: 10.1111/tpj.12956
Lee, D.-K., Parrott, D. L., Adhikari, E., Fraser, N., and Sieburth, L. E. (2016). The mobile bypass signal arrests shoot growth by disrupting shoot apical meristem maintenance, cytokinin signaling, and WUS transcription factor expression. Plant Physiol. 171, 2178–2190. doi: 10.1104/pp.16.00474
Li, H., Ding, Y., Shi, Y., Zhang, X., Zhang, S., Gong, Z., et al. (2017a). MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 43, 630–642. doi: 10.1016/j.devcel.2017.09.025
Li, H., Ye, K., Shi, Y., Cheng, J., Zhang, X., and Yang, S. (2017b). BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 10, 545–559. doi: 10.1016/j.molp.2017.01.004
Liu, Z., Jia, Y., Ding, Y., Shi, Y., Li, Z., Guo, Y., et al. (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66, 117–128.e5. doi: 10.1016/j.molcel.2017.02.016
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Liu, L., Zhang, Y., Tang, S., Zhao, Q., Zhang, Z., Zhang, H., et al. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61, 893–903. doi: 10.1111/j.1365-313X.2009.04109.x
Ma, S., and Bohnert, H. J. (2007). Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol. 8:R49. doi: 10.1186/gb-2007-8-4-r49
Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V., Miura, T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414. doi: 10.1105/tpc.106.048397
Park, J., Lim, C. J., Shen, M., Park, H. J., Cha, J.-Y., Iniesto, E., et al. (2018). Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. USA 115, E5400–E5409. doi: 10.1073/pnas.1721241115
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24, 2578–2595. doi: 10.1105/tpc.112.098640
Stockinger, E. J., Gilmour, S. J., and Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Van Kleeff, P. J., Jaspert, N., Li, K. W., Rauch, S., Oecking, C., and de Boer, A. H. (2014). Higher order Arabidopsis 14-3-3 mutants show 14-3-3 involvement in primary root growth both under control and abiotic stress conditions. J. Exp. Bot. 65, 5877–5888. doi: 10.1093/jxb/eru338
Van Norman, J. M., Frederick, R. L., and Sieburth, L. E. (2004). BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr. Biol. 14, 1739–1746. doi: 10.1016/j.cub.2004.09.045
Van Norman, J. M., Murphy, C., and Sieburth, L. E. (2011). BYPASS1: synthesis of the mobile root-derived signal requires active root growth and arrests early leaf development. BMC Plant Biol. 11:28. doi: 10.1186/1471-2229-11-28
Visscher, A. M., Belfield, E. J., Vlad, D., Irani, N., Moore, I., and Harberd, N. P. (2015). Overexpressing the multiple-stress responsive gene At1g74450 reduces plant height and male fertility in Arabidopsis thaliana. PLoS One 10:e0140368. doi: 10.1371/journal.pone.0140368
Wang, H., Yang, C., Zhang, C., Wang, N., Lu, D., Wang, J., et al. (2011). Dual role of BKI1 and 14-3-3 s in Brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21, 825–834. doi: 10.1016/j.devcel.2011.08.018
Wang, F., Zhu, D., Huang, X., Li, S., Gong, Y., Yao, Q., et al. (2009). Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell Online 21, 2378–2390. doi: 10.1105/tpc.108.065433
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-Acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Yoon, G. M., and Kieber, J. J. (2013). 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 25, 1016–1028. doi: 10.1105/tpc.113.110106
Zhao, C., Wang, P., Si, T., Hsu, C.-C., Wang, L., Zayed, O., et al. (2017). MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 43, 1–12. doi: 10.1016/j.devcel.2017.09.024
Zhou, M., Chen, H., Wei, D., Ma, H., and Lin, J. (2017). Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci. Rep. 7:39819. doi: 10.1038/srep39819
Zhou, H., Lin, H., Chen, S., Becker, K., Yang, Y., Zhao, J., et al. (2014). Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26, 1166–1182. doi: 10.1105/tpc.113.117069
Keywords: 14-3-3 proteins, Arabidopsis, CBF pathway, cold stress, DUF793, protein degradation
Citation: Chen T, Chen J-H, Zhang W, Yang G, Yu L-J, Li D-M, Li B, Sheng H-M, Zhang H and An L-Z (2019) BYPASS1-LIKE, A DUF793 Family Protein, Participates in Freezing Tolerance via the CBF Pathway in Arabidopsis. Front. Plant Sci. 10:807. doi: 10.3389/fpls.2019.00807
Edited by:
William Walter Adams III, University of Colorado Boulder, United StatesReviewed by:
Jian-Kang Zhu, Purdue University, United StatesTimothy Artlip, Appalachian Fruit Research Station (ARS-USDA), United States
Copyright © 2019 Chen, Chen, Zhang, Yang, Yu, Li, Li, Sheng, Zhang and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Zhang, zhanghua@lzu.edu.cn; Li-Zhe An, lizhean@lzu.edu.cn
 Tao Chen
Tao Chen Jia-Hui Chen1
Jia-Hui Chen1 Hong-Mei Sheng
Hong-Mei Sheng Hua Zhang
Hua Zhang Li-Zhe An
Li-Zhe An