- 1Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, The Innovative Academy of Seed Design, Chinese Academy of Sciences, Harbin, China
- 2Faculty of Agriculture, Saga University, Saga, Japan
- 3Department of Biotechnology, Graduate School of Engineering, Osaka University, Suita, Japan
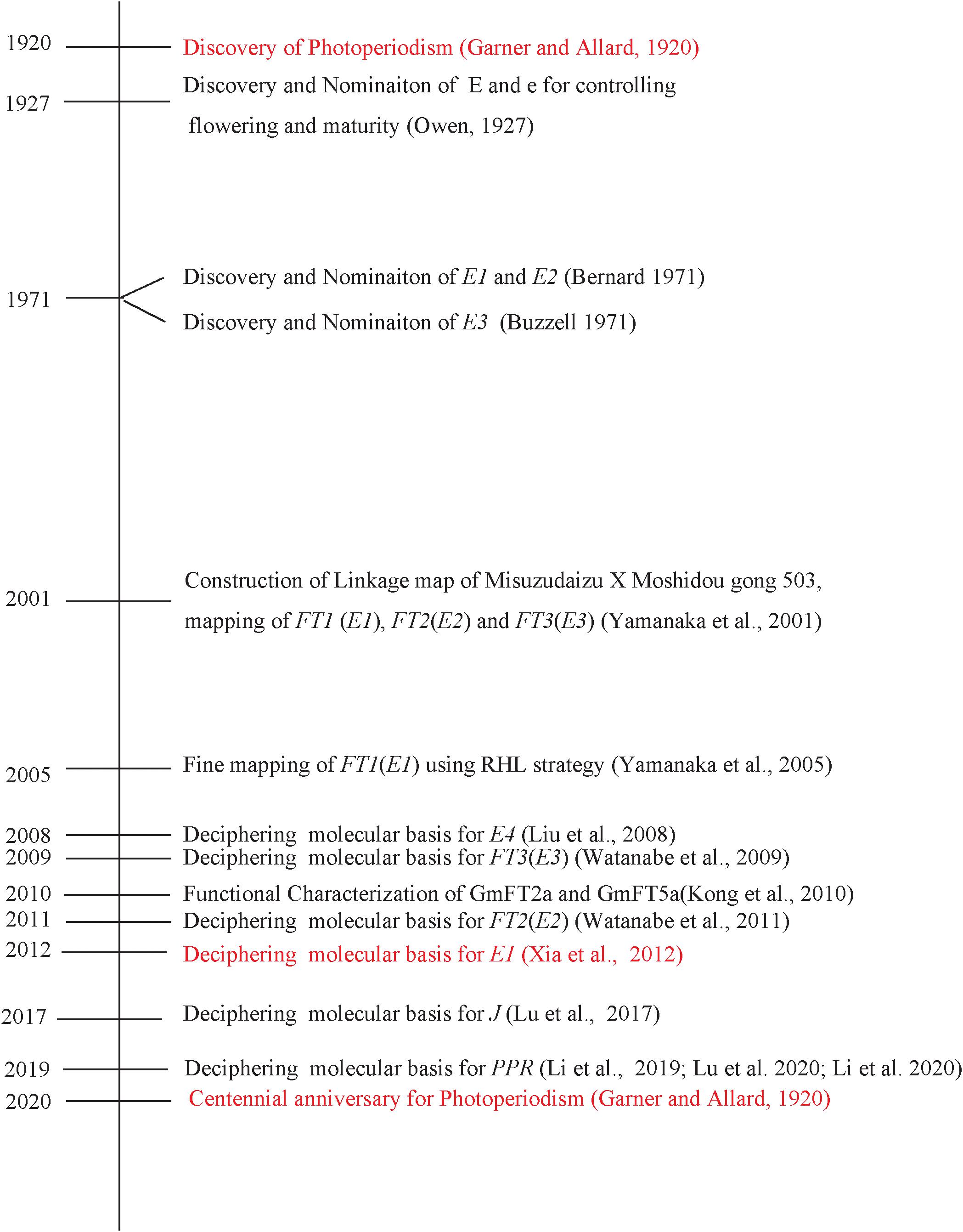
The general concept of photoperiodism, i.e., the photoperiodic induction of flowering, was established by Garner and Allard (1920). The genetic factor controlling flowering time, maturity, or photoperiodic responses was observed in soybean soon after the discovery of the photoperiodism. E1, E2, and E3 were named in 1971 and, thereafter, genetically characterized. At the centennial celebration of the discovery of photoperiodism in soybean, we recount our endeavors to successfully decipher the molecular bases for the major maturity loci E1, E2, and E3 in soybean. Through systematic efforts, we successfully cloned the E3 gene in 2009, the E2 gene in 2011, and the E1 gene in 2012. Recently, successful identification of several circadian-related genes such as PRR3a, LUX, and J has enriched the known major E1-FTs pathway. Further research progresses on the identification of new flowering and maturity-related genes as well as coordinated regulation between flowering genes will enable us to understand profoundly flowering gene network and determinants of latitudinal adaptation in soybean.
Introduction
In plants, various external cues, e.g., day length and temperature, can trigger endogenous physiological changes and lead to flowering, the critical change from vegetative growth stage to maturity stage. Garner and Allard (1920) discovered “photoperiodism” describing that day length can influence flowering time in many plant species (Garner and Allard, 1920). Along with tobacco and other plants, soybean was used as a model plant that greatly contributed to the advances of photoperiodism (Garner and Allard, 1920; Owen, 1927; Heinze et al., 1942). As the most important external cues, light is received by photoreceptors, e.g., phytochromes, cryptochromes, and phototropins. The functions of the phytochromes, the red light and far-red light absorbing photoreceptors, in initiation of flowering were extensively studied (Takimoto and Hamner, 1965). As early as in 1934, the leaf was found to sense day length (Knott, 1934). Florigen is proposed for the signal that is transmitted from leaves to the shoot apical meristem (SAM) where the flowering is initiated (Chailakhyan, 1936). Recent molecular advances have identified that FT protein, a rather small protein with a certain similarity to RAF kinase inhibitors (Kardailsky et al., 1999; Kobayashi et al., 1999), functions as Florigen, which is produced in leaves and transmitted to the SAM (Corbesier et al., 2007; Jaeger and Wigge, 2007; Tamaki et al., 2007; Notoguchi et al., 2008). The molecular mechanism of flowering has been well understood using model plants, Arabidopsis thaliana and rice (Oryza sativa). Several regulatory network pathways controlling flowering have been deciphered (Amasino, 2010; Fornara et al., 2010). In Arabidopsis, CONSTANS (CO), GIGANTEA (GI), and FLOWERING LOCUS T (FT) have been proven to be central components for initiation of flowering in long-day conditions (Koornneef et al., 1991; Kardailsky et al., 1999; Fornara et al., 2010).
In soybean, nine maturity loci, known as E-series (E1 to E8) and J conditioning flowering, have been identified and characterized genetically (Bernard, 1971; Buzzel, 1971; Buzzel and Voldeng, 1980; McBlain and Bernard, 1987; Ray et al., 1995; Bonato and Vello, 1999; Cober and Voldeng, 2001a; Cober et al., 2010). Recently, E9, E10, and E11 of E series were nominated (Kong et al., 2014; Zhai et al., 2014a; Zhao et al., 2016; Samanfar et al., 2017; Wang et al., 2019).
The E1, E3, E4, and E7 loci were proven to be photoperiod sensitive to different light quality conditions (Buzzel, 1971; Buzzel and Voldeng, 1980; McBlain et al., 1987; Cober et al., 1996a, b; Abe et al., 2003). Flowering delay under long-day for the alleles of E1, E4, and E7 was conditioned by the light quality with lower red to far-red (R:FR) quantum ratios (Cober et al., 1996a; Cober and Voldeng, 2001b). However, the E3 locus is less sensitive to light quality, which was revealed by similar flowering delays under long-day conditions with various light qualities (Cober et al., 1996a). The recessive E3 allele conditions long-day insensitivity under fluorescent light with a high R:FR ratio (Buzzel, 1971), whereas E4 needs the presence of E3 to achieve long-day insensitivity in incandescent light with a low R:FR ratio (Buzzel, 1971; Buzzel and Voldeng, 1980). Particularly, the E1 locus confers a largest effect on flowering time under various environmental conditions (Bernard, 1971; Abe et al., 2003; Stewart et al., 2003).
Characterization of isolines of E allelic combinations (Upadhyay et al., 1994a, b) revealed that each E locus exerts its influence on flowering time and maturity and also pleiotropic effects on some different developmental processes (Curtis et al., 2000), e.g., plant height and yield (Mansur et al., 1993; Chapman et al., 2003; Cober and Morrison, 2010).
Until 2000, the molecular bases for E series had not been disclosed; therefore, Professor Kyuya Harada’s research team at Chiba University, Japan had started to develop recombinant inbred line (RIL) populations for linkage maps (Yamanaka et al., 2000), and quantitative trait locus (QTL) analyses (Yamanaka et al., 2001; Watanabe et al., 2004) toward deciphering the molecular basis for the E1, E2, and E3 loci using the positional cloning strategy (Watanabe et al., 2009, 2011; Xia et al., 2012; Figure 1).

Figure 1. Linkage map construction using an F2 population derived from a cross between Misuzudaizu and Moshidou Gong 503 (adapted from Xia et al., 2007). Identified Quantitative trait loci (QTLs) of E1, E2 and E3 for flowering time were indicated by red segments. PVE, phenotypic variance explained by each QTL. Name of each linkage map is depicted on the top.
The Method and Strategy of Residual Heterozygous Lines for Positional Cloning
Mapping Population, Linkage Map, and QTL Mapping
Quantitative trait locus analysis (Tanksley, 1993) was employed to dissect the genetic factors for the quantitative trait flowering time into separate components by using RILs. The RILs were derived from a cross between Misuzudaizu, a Japanese variety, and Moshidou Gong 503, a weedy line from China.
A population of 156 RILs (F8:10) was used for QTL analysis of flowering. Three QTLs for flowering time, FT1, FT2, and FT3 were, respectively identified at LG C2 (Chr. 6), LG O (Chr. 10), and LG L (Chr. 19), which were respectively corresponding to E1, E2, and E3, according to the map positions (Yamanaka et al., 2001; Watanabe et al., 2004; Xia et al., 2007; Figure 1). All the late-flowering alleles E1, E2, and E3 were partially dominant over the early flowering alleles, e1, e2, and e3, respectively. The parent Misuzudaizu carried late-flowering allele at the E1 and E3 loci, whereas Moshidou Gong 503 had the late-flowering allele at the E2 locus.
Although near-isogenic lines (NILs) that contain a QTL in a small, defined chromosomal region are beneficial for fine mapping of the QTL, however, developing NILs is rather difficult and time and labor intensive especially in soybean. Instead, residual heterozygous lines (RHLs) were employed in our fine mapping (Yamanaka et al., 2005; Figure 2). With a set of developed molecular markers, in an RIL population, we were able to identify a given RHL or a set of given RHLs harboring a heterozygous region encompassing a given target QTL but homozygous for the most other regions of the genome, especially for the other QTL regions for the same trait. Phenotypic segregation was generally observed in the progenies of the RHL, the pattern of which depends on the effects of the target QTL (Figure 2). Similarly, heterogeneous inbred family (HIF) defined by Tuinstra et al. (1997) was successfully used to identify the QTL associated with seed weight in sorghum (Tuinstra et al., 1997).
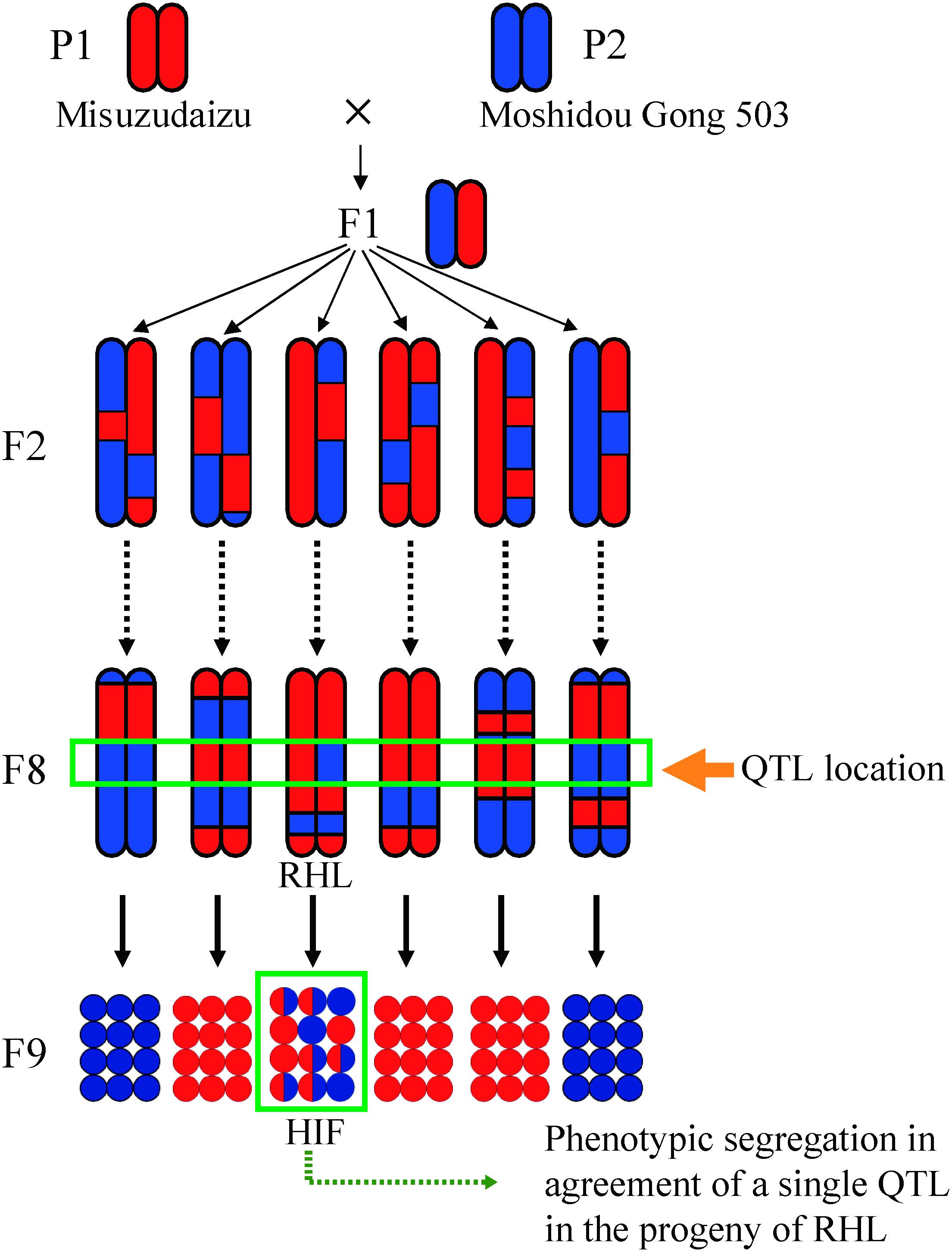
Figure 2. A schematic representation of residual heterozygous line (RHL) strategy for positional cloning. The RHL retains a heterozygous region including the target QTL region but carries homozygous regions across the genome especially for the other QTL regions detected. Meshed circles show heterozygous individuals. HIF refers to heterogeneous inbred family.
Genotypes of a given trait in recombinants identified in the progenies of RHL could be deduced from the segregation patterns in the next generation. Theoretically, the probability of successful identification of RHLs for a target QTL depends on the heterozygosity ratio and the size of the population studied (Figure 2). The formula of nCkpk (1-p)n–k can be used to calculate the possibility of the probability of successfully detecting k individuals with a heterozygous genotype at the target region, in which p is the ratio of heterozygosity of any population with given size of n. Taking an F7 generation of RILs as an example, the ratio of heterozygosity (p) is 0.0156; the probability of detecting at least one RHL in a population size of 200 is more than 0.95. In our practice, confirmed QTL analysis using the F6–F8 RIL population together with the RHL strategy is beneficial for unwinding genetic factors for an agronomic trait into each QTL (Figure 2).
Marker Development
Since cloning of E1, E2, and E3 genes started at the time before the soybean reference genome sequences of Williams 82 were available, amplified fragment length polymorphism (AFLP), simple sequence repeat (SSR), and sequence characterized amplified region (SCAR) markers were mainly used for developing new markers and genotyping a large population of the RHLs’ progenies (Xia et al., 2007; Watanabe et al., 2009).
In the given QTL region of RHL-derived population, recombinants were identified through DNA markers, whereas the genotypes of flowering time of recombinants were validated by progeny test. If the markers cosegregated with genotypes of flowering time, bacterial artificial chromosome (BAC) or transformation-competent bacterial artificial chromosome (TAC) clones compassing these markers were identified (Xia et al., 2005, 2009; Wadahama et al., 2008). Based on the fingerprinting profiles, BAC end sequencing, and relationships between BAC and markers, the BAC or TAC contig could be built. BAC clones covering the target region were selected for sequencing. The sequence data were assembled and annotated. Further functional confirmation of a candidate gene was carried out by association analysis, allelic variation, and gene disruption by induced mutation.
The Route to Successful Identification of the E3 Gene
Totally, six DNA markers, including three AFLP-derived and three PCR-based markers developed from the BAC/TAC sequences, were employed for fine mapping of the E3 locus. Through systematic fine mapping, it was strongly suggested the E3 gene had been successfully delimited to the physical region covered by TACH17D12 (Figure 3).
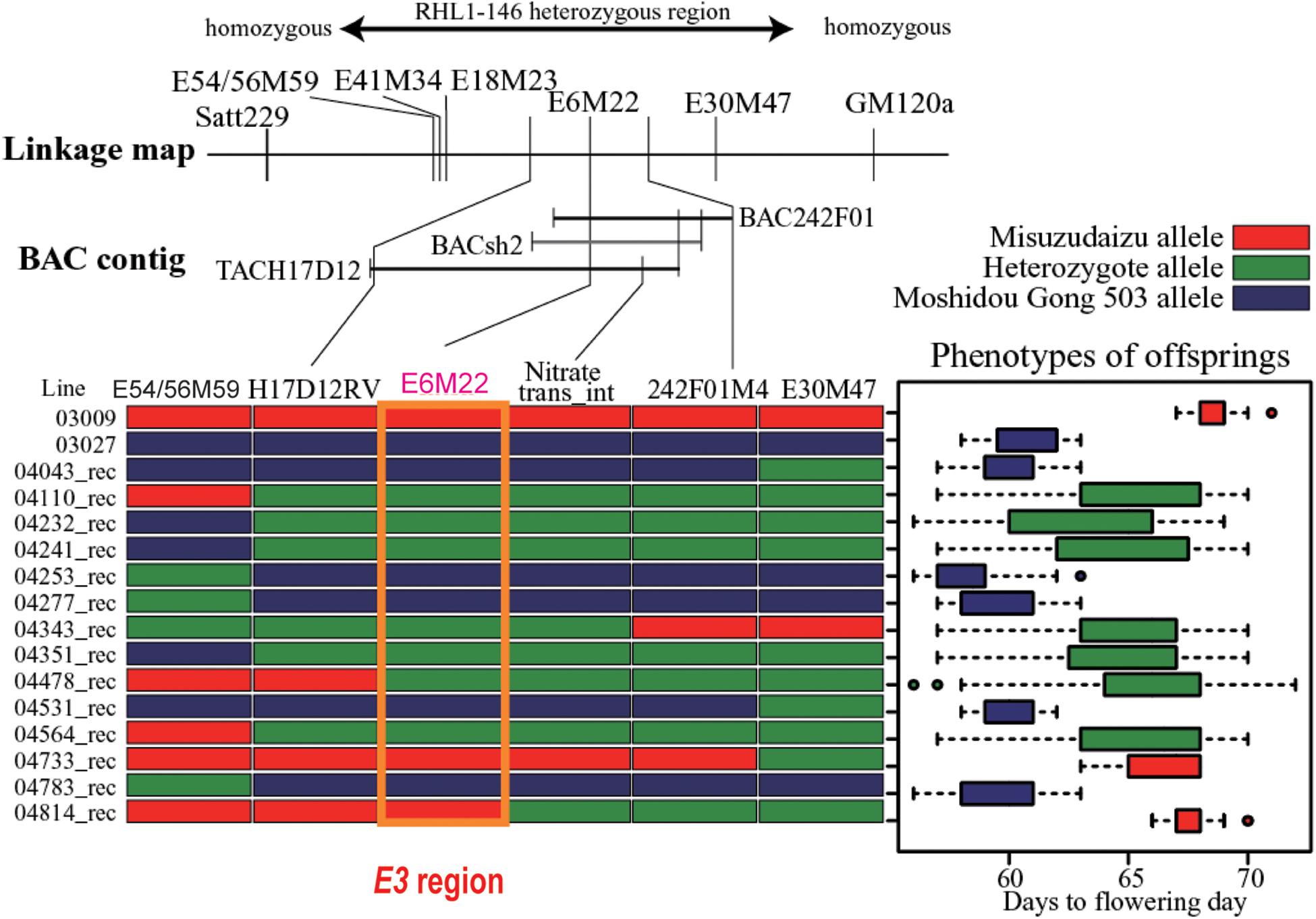
Figure 3. Fine mapping of the E3 gene. The heterozygous region is shown on the top. The linkage map of markers and bacterial artificial chromosome (BAC) contig are displayed in the middle panel. Mapping using recombinants are shown in the bottom panel: left, recombinants detected; right, phenotypic segregation patterns in the progenies. Recombination is shown by red bars representing homozygous Misuzudaizu allele, blue bars representing homozygous Moshidou Gong 503, and green bars representing the heterozygote. The phenotypic segregation is shown in boxplot format. The delimited E3 region is shown in the purple box (Adapted from Watanabe et al., 2009).
Based on the sequence of GM_TMiH_H17D12, a total of 11 genes were predicted. Considering having a large effect on flowering time under FLD conditions, a candidate for the E3 gene might be a photoreceptor (Cober et al., 1996a). The gene GmPhyA3 encoding phytochrome A was considered to be a strong candidate for E3. This E3 gene was referred to as GmPhyA3, following GmPhyA1 and GmPhyA2, that had been assigned for other phytochrome A genes when the E4 gene was cloned (Liu et al., 2008).
GmPhyA3 from Misuzudaizu (GmPhyA3-Mi) encodes a 1130 amino acid protein. GmPhyA3-Mi carries normal conserved domains for phytochrome A type protein, including two Per/Arnt/Sim (PAS) domains, a histidine kinase domain, and a chromophore-attached domain. GmPhyA3-Mo from Moshidou Gong 503 carries a large insertion in the fourth intron and one functional single-nucleotide polymorphism (SNP) (glycine to arginine) in the third exon. Amazingly, this SNP was captured by AFLP technique as marker E6M22 (Figure 3). The insertion sequence is approximately 2.5 kb of the non-long-terminal-repeat (non-LTR) retrotransposon reverse transcriptase element, a portion of which is highly homogeneous to the Ty1/copia or Ty1/gypsy sequences in the E4 allele (Liu et al., 2008).
E3 gene sequences from Harosoy and Harosoy-e3 were, respectively designated as GmPhyA3-E3 and GmPhyA3-e3 (Figure 4). In addition, a retrotransposon-like insertion sequence was also identified in GmPhyA3-E3, as well as in GmPhyA3-Mo (Figure 4). However, the amino acid sequences encoded by GmPhyA3-Mi and GmPhyA3-E3 were identical.
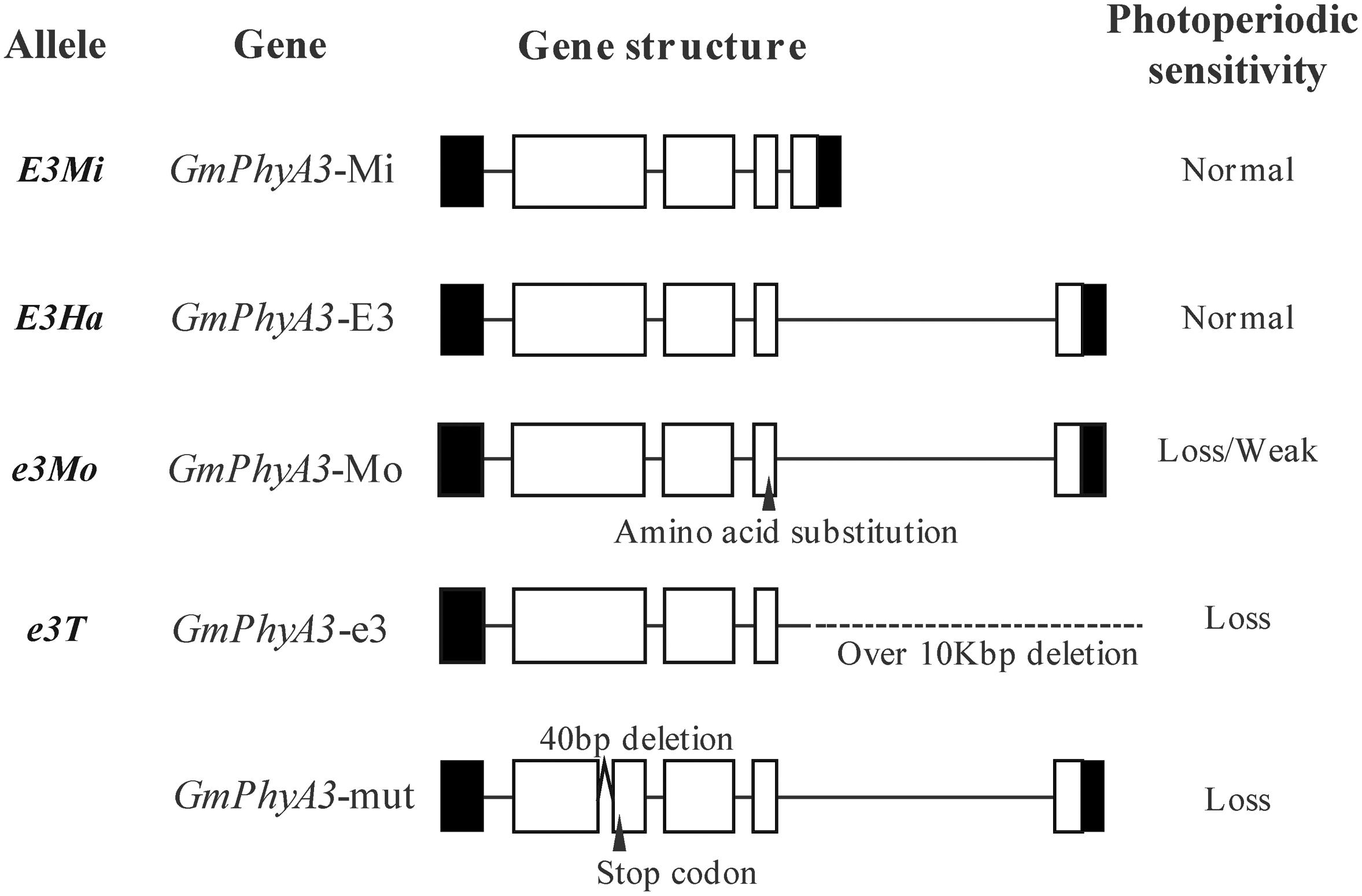
Figure 4. The allelic variation of the E3 gene. Open boxes, shaded boxes, and horizontal lines, respectively indicate exons, untranslated regions (UTRs), and introns of protein structure. Variations such as deletion, insertion, and presence of the stop codon are indicated. On the right side are shown their photoperiod sensitivity.
Additionally, a large deletion of 13.33 kb occurred at the beginning of the third exon in GmPhyA3-e3. Furthermore, a mutant (GmPhyA3-mut), with a 40-bp deletion in GmPhyA3 gene, was identified from the mutant libraries of Bay using targeting-induced local lesions in genomes (TILLING) (Figure 4; Watanabe et al., 2009).
Genetic analysis revealed that F2 population derived from a cross between Harosoy and 6-22-ft3 showed a significant difference on flowering time in agreement with E3 genetic effect, indicating the E3 and FT3 alleles are eventually identical.
In addition, large retrotransposon sequences inserted into GmPhyA3-E3 and GmPhyA3-Mo might exert no noticeable effect on the phenotype, whereas the single AA substitution that occurred in the GmPhyA-Mo might have a weak effect on the E3 allele (Figure 4; Watanabe et al., 2009).
Considering that a large effect under FLD had been reported for the E3 allele (Cober et al., 1996b), the sensitivities of the three NILs (Harosoy and -E3, 6-22-FT3 and -ft3, 1-146-FT3 and -ft3) and the mutant line for the GmPhyA3 gene to FLD conditions were evaluated. The result showed that the effect of the E3 allele was promoted under FLD conditions in all the NILs, although different genetic backgrounds also can determine the basal line of flowering days. The GmPhyA3-mut mutant flowered 15 days earlier than the wild-type cultivar Bay under FLD mimic condition, in which sunlight was extended with a mercury-vapor lamp with high red/far-red (R/FR) ratio (Watanabe et al., 2009). Refer to the formal publication on the positional cloning of the E3 gene (Watanabe et al., 2009) for the detailed cloning procedure.
Recently, Liu Y. et al. (2020) systematically illustrated the dynamic allelic variations in the E3 gene based on pan-genome information of wild and cultivated soybean. In addition, the existence of a read-through type gene fusion between E3 and its neighboring genes including SoyZH13_19G210600 was demonstrated.
The Route to Successful Identification of the E2 Gene
The strategy that has been employed for cloning of the E3 gene was used for cloning of the E2 gene. The FT2 locus corresponded to the maturity locus E2 (Yamanaka et al., 2001). In the RIL population, the line RIL6-8 was identified to carry heterozygous region covering the E2 locus; therefore, this line is hereafter referred to as RHL6-8 (Figure 5; Watanabe et al., 2011).
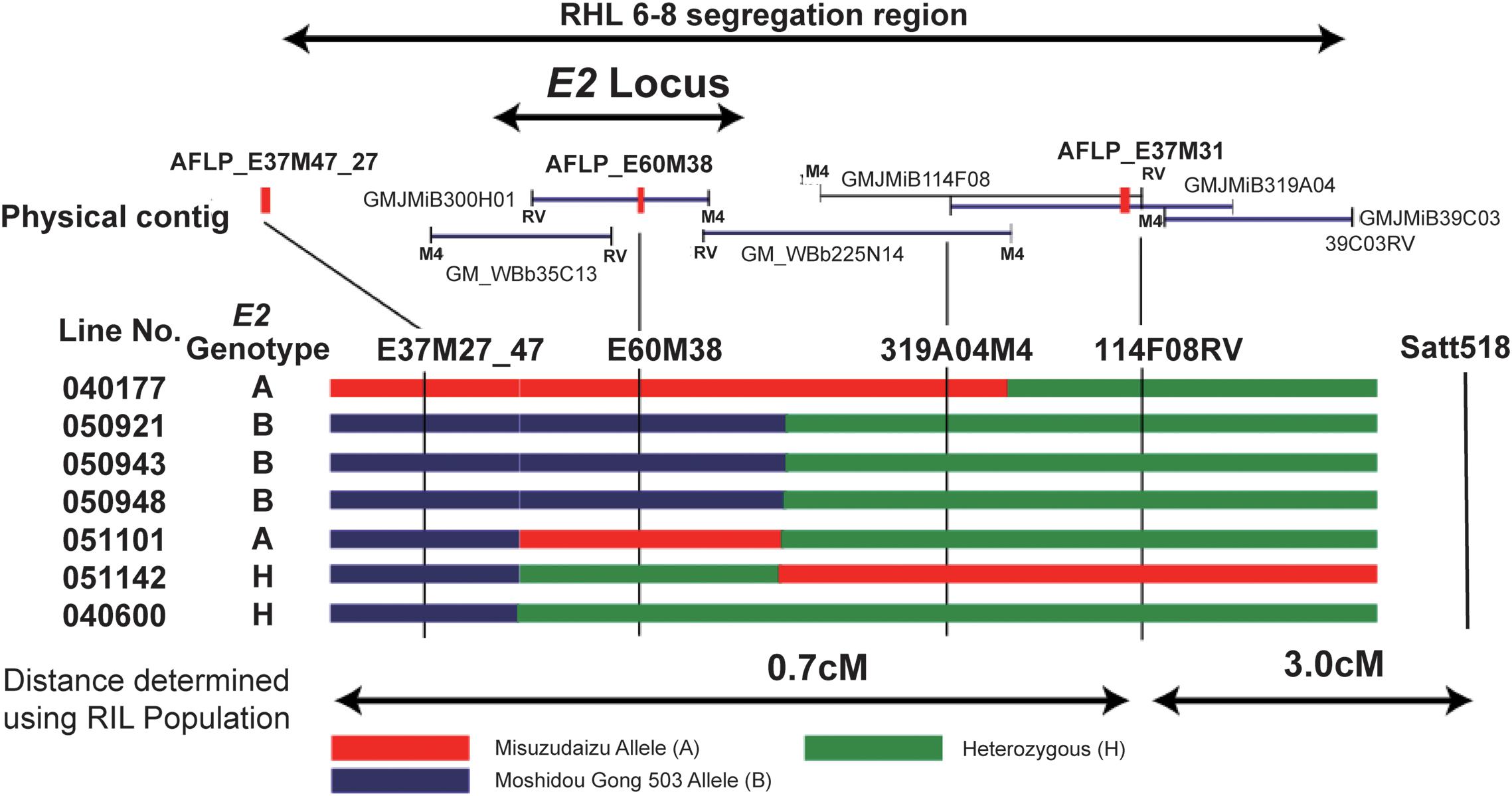
Figure 5. Fine mapping of the E2 gene using residual heterozygous line (RHL) strategy. The segregation region of the line of RHL 6–8 are shown on the top. The physical contig are shown in the middle panel, in which the BAC or TAC clones and the developed markers are placed in the relative physical position. The M4 end or RV end is also indicated. Mapping using recombinants are shown in the bottom panel. Left: Recombinant line detected. Recombination is shown by black bars representing homozygous Misuzudaizu allele, white bars representing homozygous Moshidou Gong 503, and dotted bars representing the heterozygote. Genotypes of E2 are judged based on the phenotypic segregation in the next generation. The delimited E2 region is shown. Right: phenotypic segregation patterns in the following year in the progenies (Adapted from Watanabe et al., 2011).
Three SCAR markers that had been successfully developed from these five polymorphic products were used to screen two independent BAC libraries, and a total of 10 BAC clones were acquired and a contig of approximately 430 kb was built (Watanabe et al., 2011). Three molecular markers, one AFLP-derived and two BAC-sequence-derived markers, were employed for the fine mapping to delimit the E2 locus (Watanabe et al., 2011). The E2 locus could explain 87.9% of the total variance in flowering time, indicating that a single QTL or gene controls this trait observed in this population. The marker 2 (E60M38) cosegregated with E2 judging from the flowering time, indicating that this marker was physically close to E2 (Watanabe et al., 2011). Judging from the phenotypes and genotyping data of recombinants as well as the positions where recombination events occurred, the E2 locus could be delimited into the single BAC clone, MiB300H01 (Watanabe et al., 2011; Figure 5). The whole sequence of the BAC clone, MiB300H01, was determined using shotgun sequencing. Among the nine genes annotated for the 94-kb sequence of MiB300H01, Glyma10g36600 was considered to be the strongest candidate for the E2 locus based on the functional annotation in junction with the functional interpretations in previous genetic studies (Buzzel, 1971; Buzzel and Voldeng, 1980; Cober et al., 1996a).
The candidate E2 gene was referred to as GmGIa. The coding sequence of GmGIb, the closest homolog of GmGIa in the genome, was also predicted.
The coding sequence of GmGIa-Mo from Moshidou Gong 503 containing 14 exons is prolonged to a 20-kb genomic region. Interestingly, the marker 2 derived from AFLP polymorphic band E60M38 was located in the fifth intron and cosegregated with E2 (Watanabe et al., 2011). Four SNPs were detected in the coding sequence of GmGIa-Mi, the Misuzudaizu early flowering allele, in comparison with GmGIa-Mo. Especially, an SNP in the 10th exon resulted in a premature stop codon mutation leading to a truncated 521 AA GI protein in GmGIa-Mi. Considering this stop codon mutation is functional in GmGIa, a derived amplified polymorphic sequence (dCAPs) marker was developed to genotype other corresponding NILs of Harosoy (e2/e2). The genotypes of the E2 in all NILs tested were completely consistent with the genotypes of this dCAPs marker. This result further verified the candidacy of GmGIa for the E2 loci and that this conserved stop codon mutation was a causal factor for the early flowering phenotype (Watanabe et al., 2011). To further validate whether mutations in the GmGIa can cause profound impact on flowering time and maturity, we identified a mutant line from X-ray-irradiated and ethyl methanesulfonate (EMS)-derived libraries by TILLING (McCallum et al., 2000). In comparison with wild-type cultivar Bay carrying the E2 allele, the mutant line whose E2 gene had a deletion in the 10th exon leading to a truncated protein (735 amino acids) showed a significant earlier (8 days) flowering phenotype under natural day-length conditions (Watanabe et al., 2011).
Taken together, GmGIa is the responsible gene for the E2 locus. Refer to the formal publication on the positional cloning of the E2 gene (Watanabe et al., 2011) for the detailed cloning procedure.
Three GmGIa haplotypes (H1, H2, and H3) were identified amid cultivated cultivars and their wild relatives in soybean. Interestingly, additional 44 haplotypes occur in wild soybeans (Wang et al., 2016). In cultivated as well as wild-type soybeans, H2 often occur in the southern part of China, while H3 was constrained to areas adjacent to the northeast region of China. H1, a domesticated haplotype, is the variant of H2, which was found to be profoundly distributed among cultivated soybeans. Intriguingly, the ortholog of H1 was present only at a low frequency in wild populations from Yellow River (Wang et al., 2016).
The Route to Successful Identification of the E1 Gene
The RHL1-156 line with a heterozygous segment (approximately 17 cM) comprising the E1 locus was screened out from the RILs population derived from a cross between Misuzudaizu and Moshidou Gong 503. Importantly, all other flowering-time-related QTL loci (except for the E1 locus) anchoring segments were homozygous in this line. Upon segregation, a population of 1,006 individuals was derived from the RHL1-156. The E1 locus could be mapped between Satt365 and GM169, at the distances of about 0.1 and 0.4 cM. The E1 locus is located in the pericentromeric region of chromosome 6 in soybean1, with a high ratio of physical to genetic distance. Accordingly, no polymorphic AFLP bands had been detected between bulks of E1 and e1, thus fine mapping halted due to the lack of molecular marker. It was difficult to develop new molecular markers in the era before the genome information publically available. Therefore, we shifted the cloning strategy and generated a mapping population of Harosoy-E1 (E1e2E3E4e5) × Harosoy(e1) (e1e2E3E4e5), both of which carry identical genetic background except the E1 locus. Flowering times of Harosoy-E1, F1 plant, and Harosoy (e1) were 45.0 ± 0.78 days (mean ± SD), 41.5 ± 1.16 days, and 34.9 ± 0.83 days, respectively, at Matsudo, Japan (35°78′N, 139°90′E), in 2005. The results indicated that the effects of the E1 locus were about 10 days, and the E1 allele is partially dominant over e1. For the F2 population (117 plants), E1 was initially mapped between markers Satt365 and Satt289 by means of QTL analysis of flowering time at Matsudo in 2005, and the closest marker was Satt557. Among an F2:3 population of 1442 plants derived from 51 F2 plants that were heterozygous at Satt557, seven recombinants between markers Satt365 and Satt289 were identified (Figure 6).
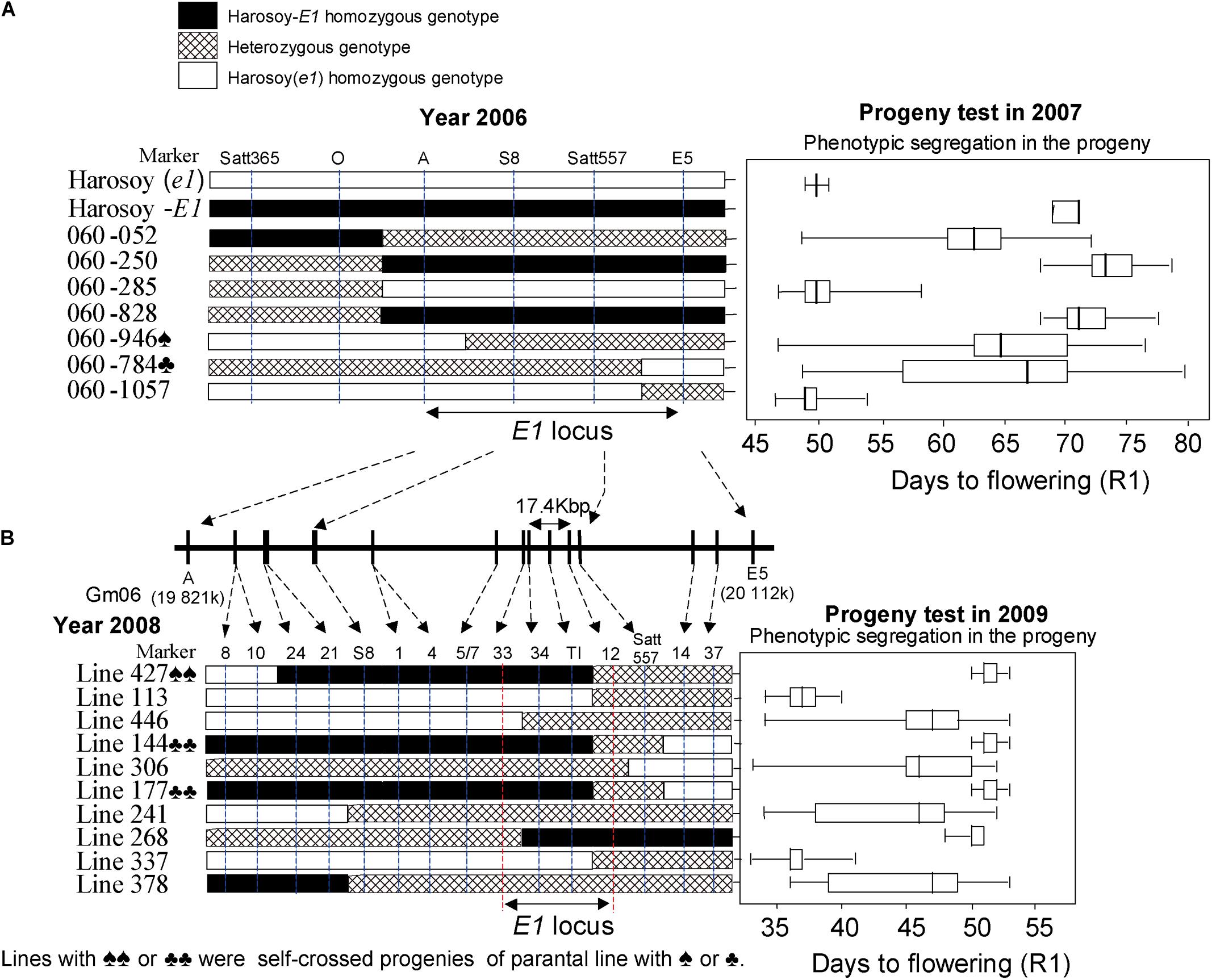
Figure 6. Fine mapping of the E1 gene. Graphical genotypes of soybean recombinants carrying recombination in the E1 region in the 2006–2009 experiments. Left: Recombinants detected. Right: Phenotypic segregation patterns in the progenies. Recombination is shown by white bars representing Harosoy (e1), black bars representing Harosoy-E1, and cross-hatched bars representing the heterozygote. The phenotypic segregation is presented in boxplot format. The box, the bold vertical line, and the horizontal line, respectively represent the interquartile region, median, and range of flowering time. In 2006–2007 (A), with seven recombinants, we were able to delimit the E1 to a 289-kb region. In 2008–2009 (B), with 10 recombinants, we further delimited the E1 region to a 17.4-kb region (adapted from Xia et al., 2012).
The segregation patterns of flowering time among its progeny in 2007 at Tsukuba, Japan (36°03′N, 140°04′E) were used to accurately estimate the E1 genotype for each recombinant. Despite a physical distance of 133 kb, we could not detect any recombination event occurring between the markers S8 and Satt557, which might be ascribed to a low recombination rate occurring in the pericentromeric region.
Therefore, the E1 region was only located to an interval of ∼289 kb between markers A and marker E5 (Figure 6). According to the prediction using RiceGAAS (Sakata et al., 2002), more than 40 genes were annotated for this 289-kb region (Figure 6). Therefore, a new round of fine mapping became necessary to further delimit the region of E1.
With the aid of a simple seed genotyping developed in the lab, 13,761 F2:5 seeds having a heterozygous E1 background and 10 recombinants carrying crossovers within the 289-kb region were successfully screened out. Similarly, the phenotypic segregation pattern of the progeny was evaluated at Tsukuba in 2009 to judge the E1 genotype of each recombinant (Figure 6).
The E1 gene was delimited to the region between markers 12 and 33, judging by the fact that the phenotypes cosegregated with markers 34 and TI among these recombinants (Figure 6). Molecular markers of E1 region were used to screen in two independent BAC libraries of Misuzudaizu and Williams 82. In order to construct the BAC contigs, BACs were selected for shotgun sequencing based on the presence of molecular markers including BAC end sequencing-derived makers and the fingerprinting profiling of each BAC clone digested with HindIII (Xia et al., 2005, 2009).
Sequences yield from single BAC were assembled individually, and two physical contigs were successfully built for Misuzudaizu and Williams 82, respectively. The delimited E1 region corresponds to 17,372 bp in Misuzudaizu (dominant E1) and 22,876 bp in Williams 82 (recessive E1). In the 17,372 bp from Misuzudaizu and Harosoy-e1, a single intron-free gene (AB552962, 525 bp, 174 aa) was consistently annotated by various software, such as GenScan (Burge and Karlin, 1997), and was designated as the E1 gene. In recessive e1 cultivars of Williams 82 and Harosoy (e1), a single missense point mutation was detected in the coding region of E1, resulting in a change from threonine to arginine at AA 15. This recessive allele was referred as to e1-as (AB552963). In Sakamotowase and its derived NILs, a 1-bp deletion in codon 17 at the E1 locus resulted in a premature stop, designated as e1-fs (AB552971).
In some early flowering cultivars such as Fiskeby V, Yukihomare, Toyosuzu, Toyomusume, Hejian 1, and Heihe 2, there was approximately 130 kb deletion (including the entire E1 gene) and was designated e1-nl. Both in the growth chamber and in the field, cultivars with the e1-as genotype generally flowered and matured intermediate between the E1 and e1-fs genotypes, demonstrating that e1-as is a leaky allele and retains partial E1 function. The function of E1 in delaying flowering was confirmed by the EMS-derived E1 mutants showing early flowering phenotype.
The E1 gene encodes a protein that contains a putative bipartite nuclear localization signal (NLS) and a B3 domain, suggesting that this protein is a transcription factor. This mutation from E1 to e1-as occurs in the first basic domain (amino acid motif KKRK) of the putative bipartite NLS, which might affect nuclear targeting. Through analysis of transformed Arabidopsis protoplasts and onion cells, the E1 protein was mainly localized in the nucleus, whereas the e1-as was found in the nucleus as well as in the cytoplasm. E1 expression was highly repressed under both short- and long-day conditions in cultivars carrying e3e3/e4e4.
The E1 expression level was negatively correlated with the transcriptional abundance of FT2a and FT5a, two homologs of Arabidopsis FT that promote flowering (Kong et al., 2010) under the regulation of the E3 and E4 loci (Xia et al., 2012). Refer to the formal publication on the positional cloning of the E1 gene (Xia et al., 2012) for the detailed cloning procedure.
The molecular identification of E1 for the repression of flowering at the E1 locus represents a significant step forward in photoperiodic flowering and thus has implications in breeding programs and cultivation practices. The expression level of functional E1 gene was strongly associated with flowering time (Zhai et al., 2015).
The soybean genome has two E1 homologs, E1La (Glyma04g24640, Glyma.04G156400.1) and E1Lb (Glyma18g22670, renamed as Glyma.04G143300.1).
Under long-day conditions, the expressions of all three genes of Harosoy peaked before dusk and after dawn the next day. The transition between light and dark phases and night–break experiments revealed that E1 family genes were expressed solely during light periods (Xu et al., 2015). In the cultivar “Toyomusume,” which lacks the E1 gene, silencing of E1La and E1Lb resulted in the upregulation of the expression of FT2a and FT5a and early flowering phenotype. Thus, E1La and E1Lb might have similar function to E1 in flowering (Xu et al., 2015). E1Lb suppresses flowering under long-day conditions by blocking the expression of FT2a and FT5a in a fashion independent of E1 (Zhu et al., 2019). Regulation of E1 and E1L expression by light is dominated by E3 and E4, and regulation of FT2a and FT5a expression is controlled by E1 and E1L (Xia et al., 2012; Xu et al., 2015). This module may be a major regulator in photoperiodic flowering of soybean (Xia et al., 2012; Xu et al., 2015), which is different from CO/FT module in Arabidopsis (Samach et al., 2000) and rice (Kojima et al., 2002).
The E1 homolog Phvul.009G204600 (PvE1L) from common bean, a short-day leguminous species, was proven to delay the onset of flowering in soybean (Zhang et al., 2016). However, Medtr2g058520, the E1 homolog from long-day leguminous species, promotes flowering (Zhang et al., 2016). Thus, the functional conservation and diversification of E1 family genes from legumes may be associated with lineage specification (Zhang et al., 2016).
Although both FT2a and FT5a are under the control of E1, and collectively regulate flowering time, the function of FT2a is more prominent in SD. However, FT5a functions more prominently in LD, which affects adaptability of soybean to high latitude (Kong et al., 2014; Takeshima et al., 2016). The ef allele at FT5a is a rare haplotype, conferring an adaptive option at latitudes when early flowering is needed (Cai et al., 2020). FT4 and FT1a were proven to be repressing flowering, which are antagonistic to FT2a and FT5a. Both genes are expressed at higher levels under LD compared SD, indicating that both are induced by E1 (Zhai et al., 2014a; Liu et al., 2018).
Soybean genome has 12 FT-like genes, which scattered in six homologous pairs, FT1a/b, FT2a/b, FT2c/d, FT3a/b, FT5a/b, and FT4/6 (Wu et al., 2017). Evolutionary trajectories of duplicated FT homologs and their functional roles in soybean domestication were reported (Wang et al., 2015; Wu et al., 2017). The FT2c allele having a transposon insertion is widely spread in soybean landraces but not in domesticated soybean, indicating that this allele spreads at the beginning of soybean domestication (Wu et al., 2017). FT2a was identified to be responsible for E9 (Zhao et al., 2016). Studies on the expression levels of different alleles among NILs and photoperiodic-insensitive cultivars indicated that the SORE-1 (a Ty1/copia-like retrotransposon) insertion in E9 diminished FT2a expression (Zhao et al., 2016).
Allelic Combinations of the E1 to E4 Loci Primarily Determine Latitudinal Distribution
GmPhyA2, another phytochrome A gene, was proven to be the causal gene for the E4 locus by using a candidate gene approach (Liu et al., 2008). At the recessive allele (E4-SORE-1), the insertion of a Ty1/copia-like retrotransposon into exon 1 of the E4 gene weakens the function of the E4 gene on repressing flowering (Liu et al., 2008; Figure 7).
Natural variations in E1–E4 genes were determined for 62 cultivars or landraces and a wild soybean accession (Tsubokura et al., 2014). Allelic combinations at the E1–E4 loci are associated with ecological types, and 62–66% variation in flowering time could be explained by these loci (Tsubokura et al., 2014). The association of maturity group of soybean varieties and the adaptation to diverse ecological or latitudinal regions with allelic variation in E1–E4 were also performed in China (Jiang et al., 2014; Zhai et al., 2014b), the United States (Langewisch et al., 2014, 2017; Wolfgang and Charles, 2017), and Europe (Kurasch et al., 2017).
Liu L. P. et al. (2020) reported that the allele combinations of e1-as/e2-ns/e3-tr/E4, E1/e2-ns/E3/E4, and E1/E2/E3/E4 are dominant genotypes in the Northeast China, Huang-Huai-Hai (HHH) Rivers Valley, and South China regions, respectively. Notably, E1 and E2, especially E2, affected flowering and variation maturity time of soybean significantly.
Among the soybean population at Novi Sad, Serbia, e1-as/E2/E3/E4 was the most dominant genotype and presented the best performance in terms of yield. This allelic combination is putatively the optimal one suitable for the environments of Central-Eastern Europe (Miladinovic et al., 2018).
A total of 15 multilocus genotypes at the E1–E4 loci were identified from 53 photoperiod-insensitive accessions. At either the E3 or E4 locus, a recessive allele was observed for all of the 53 accessions. A loss-of-function of e1-fs or e1-nl or hypomorphic e1-as allele at the E1 locus always occurred when a dominant allele is present at the other loci (Xu et al., 2013).
Soybean RIL lines with various allele combinations at the E1, E2, E3, and J loci were field tested for days to flowering (DTF) and days to maturity (DTM) in short-day tropical environments in Ghana. The alleles of these genes interacted with each other for DTF but not for DTM. The mutant allele J and E1 had profound impact on DTF and DTM (Miranda et al., 2020).
“Enrei” (E1/e2/e3/E4) is one of the leading cultivars in Japan. In order to expand the adaptability of “Enrei,” NILs for E2 and E3 were developed, and their flowering, maturity, seed productivity, and seed-quality traits were evaluated in five different locations (Yamada et al., 2012). The dominant alleles E2 and E3 were introduced from “Sachiyutaka” (E1/E2/e3/E4) and “Fukuyutaka” (E1/E2/E3/E4), respectively, by recurrent backcrosses based on the functional DNA markers. The modification of genotypes at maturity loci provides new varieties that are adaptive to environments of different latitudes while retaining almost the same seed quality as that of the original cultivar. Modification of maturity loci is underway for several other cultivars. E1 and E1La/b were simultaneously silenced via RNA interference, and a super-early maturity line was developed that will adapt to high-latitude short-season regions (Liu L. et al., 2020). In addition, targeted mutations of soybean flowering genes by CRISPR/Cas9 technology to modify flowering and maturity have been reported for FT2a (Cai et al., 2018), for FT2a and FT5a (Cai et al., 2020), and for E1 (Han et al., 2019).
Identification of New Genes Controlling Flowering Time
A potential candidate gene for E10 was proposed as FT4 (Samanfar et al., 2017). FT4, a homolog of FT, is positively regulated by E1 and was proven to function as a flowering repressor (Zhai et al., 2014a).
E11 is a recently reported locus that influences both flowering time and maturity, and the most likely candidate is reported to be a soybean homolog of LATE ELONGATED HYPOCOTYL (LHY) (Wang et al., 2019). A homolog of EARLYFLOWERING 3 (GmELF3) was identified as a gene for J locus (Lu et al., 2017; Yue et al., 2017). J protein physically associates with E1 promoter and downregulates its transcription (Lu et al., 2017). The GmFLC-like protein can directly suppress the expression of FT2a by physically interacting with its promoter region. GmFLC-like might be involved in long-term low temperature-triggered late flowering by repressing FT gene expression. The result of treatments with various low temperature durations showed that GmFLC-like acts as a floral repressor (Lyu et al., 2020).
GmAGL1 was proven to promote flowering possibly in a fashion of photoperiodic regulation. Overexpression of GmAGL1 leads to early maturity, but no reduction occurs in seed traits or oil and protein contents (Zeng et al., 2018).
Analysis of variations in coding and non-coding regions of the GmGBP1 genes in 278 soybean accessions showed that the shorter growth period might be largely ascribed to higher GmGBP1 expression. In addition, RNA-interference-mediated downregulation of GmGBP1 resulted in a longer growth period under different day lengths. It was showed that GmGBP1 can act as a positive regulator of FT2a and FT5a to promote the expression of GmFULc, leading to early flowering under short-days (Zhao et al., 2018).
Two pairs of homologs COL1a/b and COL2a/b and other 22 CO-like genes have been identified in the soybean genome. Although the RNAi-mediated downregulation of COL1a/b could lead to the downregulation of E1 (Wu et al., 2019), the function of COL genes in soybean has not been well understood. The mutant lacking COL2b putatively weakens the repression of flowering by cool temperature, in which the expressions of E1, FT2a, and FT5a have been altered (Zhang et al., 2020a, b).
Recently, a great progress has been made on connection of clock genes with E1-FTs, the major flowering pathway in soybean (Lu et al., 2017, 2020; Li Y. et al., 2019; Bu et al., 2021).
The QTLs, qFT12-1/Gp12/Tof12 or Gp11/Tof11, in chromosomes 11 and 12 have been identified to be GmPRR3a and GmPRR3b, two homologs of Arabidopsis PSEUDO-RESPONSE REGULATOR (PRR) 3 (Li M. W. et al., 2019; Li Y. et al., 2019; Li et al., 2020; Lu et al., 2020). Through the LHY homologs, both GmPRR3a and GmPRR3b function to promote E1 expression and thus delay flowering under long-days (Lu et al., 2020). The allelic variation in GmPRR3b has been widely chosen through modern breeding (Li Y. et al., 2019; Li et al., 2020). The causal SNP (Chr12:5520945) likely confers GmPRR3b a suitable level of activity, resulting in early flowering and vigorous growth. This functional variation is preferentially retained during breeding or improvement of landraces or cultivars. This gene, showing rhythmic and photoperiod-dependent expression, is specifically induced in LD and appears to act as a transcriptional repressor of GmCCA1a, which directly moderates J/GmELF3a to control flowering time (Li et al., 2020).
Overexpression of GmPRR37 noticeably repressed the flowering of transgenic soybean in LD but not in SD (Wang et al., 2020). GmPRR37 downregulated the expression of FT2a and FT5a, the flowering-promoting FT homologs, and upregulated FT1a expression, flowering-repressing FT homolog under long-day conditions (Wang et al., 2020).
The long-juvenile (LJ) trait can increase the vegetative phase under short-day conditions, ensuing higher yield and enabling expansion of cultivation in tropical regions. J locus, the major classical locus conferring the LJ trait, was identified as the ortholog of A. thaliana EARLY FLOWERING 3 (ELF3), which depends genetically on the legume-specific flowering repressor E1 (Lu et al., 2017; Yue et al., 2017). J protein physically associates with the E1 promoter to downregulate its transcription, alleviating suppression of two important FT genes and promoting flowering under short-days (Lu et al., 2017).
Evening complex (EC) can be formed by both LUX1 and LUX2 by interacting with J, which promotes flowering redundantly. The EC represses the expression of E1 and its homologs by binding to the LBS (a specific LUX binding site) of their promoters. Thus, FT2a and FT5a were abundantly produced to induce flowering in SD (Bu et al., 2021).
Conclusion and Future Perspective
To mark the centennial of photoperiodism, we reviewed our efforts toward successful cloning of responsible genes at the major maturity loci E1, E2, and E3. Indeed, international efforts have been made including the discovery of the genetic factor controlling flowering and maturity, nomination, development of NIL, construction of linkage maps and BAC libraries, QTL mapping, fine mapping, and positional cloning using RHL and NIL. Since the successful identification of molecular basis of E1, E2, and E3 genes, great progress has been made in identification of new genes that control or regulate flowering time and maturity and in flowering time gene networks especially related to circadian clock (Figures 7, 8). The central role of E1 gene in photoperiodic flowering has been recently understood at molecular level. Both E3 and E4 genes mediate flowering responses under high ratio of R and FR light. Under LD, the E3 and E4 genes induce the expression of E1 and E1Lb. PRR3a and PRR3b inhibit the expression of GmLHY/GmCCA1 by binding to their promoters. Furthermore, GmLHY and GmCCA1 can bind to the E1 promoter and thus suppress its expression. E1 can essentially repress the expression of flowering-inducing factors FT2a and FT5a and promote the expression of flowering-inhibitory factors FT4 and FT1a. As a result, flowering is delayed under LD. Under SD, the functions of E3 and E4 are greatly weakened, leading to a suppressed expression of the E1. Meanwhile, J can inhibit E1 expression. Consequently, the E1 expression is strongly repressed in SD. The repressing effect of FT2a and FT5a by E1 is strongly alleviated; in contrast, the expression of FT1a and FT4 is suppressed (Figure 8). Therefore, flowering is strongly promoted in SD.
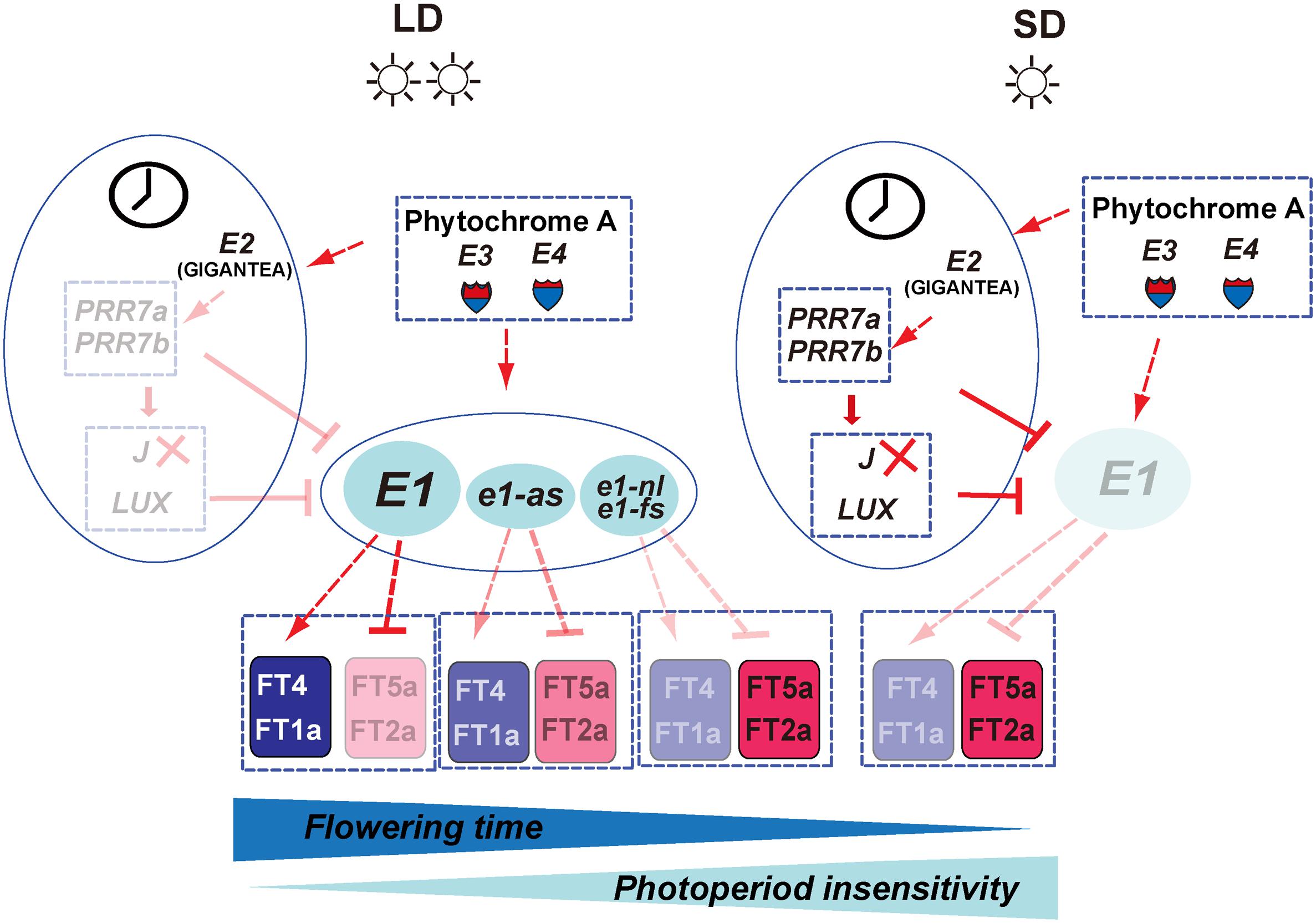
Figure 8. The putative flowering time gene network controlling the photoperiodic sensitivity in soybean. On the left panel, under long-day conditions, the expression of the E1 gene is predominately promoted by the E3 and E4 genes. The elevated E1 expression promotes the FT4a and FT1a expression and represses the FT2a and FT5a, leading to late flowering and higher photoperiod sensitivity. However, leaky allele e1-as displays partial function of the E1 gene, and the non-functional allele, e1-nl or e1-fs, totally loses the promotion activity for the expression of FT4a and FT1a as well as the suppression activity for the expression of FT2a and FT5a. In addition, circadian clock genes such as E2 as well as several downstream components such as PRR3/7a, PRR3/7b, LUX, and J are proven to participate in the control of E1 expression. Under short-day condition, E1 is strongly suppressed and leads to promoted expression of the FT2a/FT5a and early flowering time. The solid and dotted lines, respectively represent direct and indirect regulations. The arrow and T shape represent positive and negative regulation, respectively.
To date, the draft flowering time gene network of Phytochrome-clock-related gene E1-FTs has been built. However, the detailed regulatory mechanism remains poorly understood. Although the E1 gene stands as a key hub gene in the regulation of flowering time in soybean, its pleiotropic function on other agronomic or phenotypic traits has not been well exploited. We also needed to clarify the functions of large numbers of flowering time gene homologs present in soybean genome, as well as their functional diversification and evolution in relation to domestication and modern breeding. Further identification of important components of E1 pathway and studies on the detailed and coordinate regulation of flowering time gene network starting from the light reception to the full maturity will enable us to understand the nature of photoperiodism at molecular level in soybean.
Author Contributions
ZX, SW, and KH: conceptualization and writing (review and editing). HZ, HW, and KX: investigation and visualization. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Project (2016YFD0100201 and 2016YFD0101900) from the Ministry of Science and Technology of China; Key Deployment Project (ZDRW-ZS-2019-2); Strategic Priority Research Program of XDA24010105-4 from the Chinese Academy of Sciences; and Programs (31771869, 31771818, and 31971970) from the National Natural Science Foundation of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Abe, J., Xu, D., Miyano, A., Komatsu, K., Kanazawa, A., and Shimamoto, Y. (2003). Photoperiod-insensitive Japanese soybean landraces differ at two maturity loci. Crop Sci. 43, 1300–1304. doi: 10.2135/cropsci2003.1300
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013. doi: 10.1111/j.1365-313x.2010.04148.x
Bernard, R. I. (1971). 2 Major genes for time to flowering and maturity in soybeans. Crop Sci. 11, 242–247. doi: 10.2135/cropsci1971.0011183x001100020022x
Bonato, E. R., and Vello, N. A. (1999). E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet. Mol. Biol. 22, 229–232. doi: 10.1590/s1415-47571999000200016
Bu, T., Lu, S., Wang, K., Dong, L., Li, S., Xie, Q., et al. (2021). A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Natl. Acad. Sci. U S A. 118:e2010241118. doi: 10.1073/pnas.2010241118
Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. doi: 10.1006/jmbi.1997.0951
Buzzel, R. I. (1971). Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can. J. Genet. Cytol. 13, 703–707. doi: 10.1139/g71-100
Buzzel, R. I., and Voldeng, H. D. (1980). Inheritance of insensitivity to long daylength. Soybean Genet. Newslett. 7, 26–29.
Cai, Y., Chen, L., Liu, X., Guo, C., Sun, S., Wu, C., et al. (2018). CRISPP/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotech. J. 16, 176–185. doi: 10.1111/pbi.12758
Cai, Y., Wang, L., Chen, L., Wu, T., Liu, L., Sun, S., et al. (2020). Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotech. J. 18, 298–309. doi: 10.1111/pbi.13199
Chailakhyan, M. K. (1936). New facts in support of the hormonal theory of plant development. Doklady Akademii Nauk SSSR 13, 79–83.
Chapman, A., Pantalone, V. R., Ustun, A., Allen, F. L., Landau-Ellis, D., Trigiano, R. N., et al. (2003). Quantitative trait loci for agronomic and seed quality traits in an F(2) and F(4 : 6) soybean population. Euphytica 129, 387–393.
Cober, E. R., and Morrison, M. J. (2010). Regulation of seed yield and agronomic characters by photoperiod sensitivity and growth habit genes in soybean. Theor. Appl. Genet. 120, 1005–1012. doi: 10.1007/s00122-009-1228-6
Cober, E. R., and Voldeng, H. D. (2001a). A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci. 41, 698–701. doi: 10.2135/cropsci2001.413698x
Cober, E. R., and Voldeng, H. D. (2001b). Low R:FR light quality delays flowering of E7E7 soybean lines. Crop Sci. 41, 1823–1826. doi: 10.2135/cropsci2001.1823
Cober, E. R., Molnar, S. J., Charette, M., and Voldeng, H. D. (2010). A new locus for early maturity in soybean. Crop Sci. 50, 524–527. doi: 10.2135/cropsci2009.04.0174
Cober, E. R., Tanner, J. W., and Voldeng, H. D. (1996a). Genetic control of photoperiod response in early-maturing, near-isogenic soybean lines. Crop Sci. 36, 601–605. doi: 10.2135/cropsci1996.0011183x003600030013x
Cober, E. R., Tanner, J. W., and Voldeng, H. D. (1996b). Soybean photoperiod-sensitivity loci respond differentially to light quality. Crop Sci. 36, 606–610. doi: 10.2135/cropsci1996.0011183x003600030014x
Corbesier, L., Vincent, C., Jang, S., Fornata, F., Fan, O., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Curtis, D. F., Tanner, J. W., Luzzi, B. M., and Hume, D. J. (2000). Agronomic and phonological differences of soybean isolines differing in maturity and growth habit. Crop Sci. 40, 1624–1629. doi: 10.2135/cropsci2000.4061624x
Fornara, F., and Montaigu Ade, and Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550–e2.
Garner, W. W., and Allard, H. A. (1920). Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J. Agri. Res. 18, 553–606.
Han, J. N., Guo, B. F., Guo, Y., Zhang, B., Wang, X. B., and Qiu, L. J. (2019). Creation of early flowering germplasm of soybean by CRISPR/Case9 technology. Front. Plant Sci. 10:1446. doi: 10.3389/fpls.2019.01446
Heinze, P. H., Parker, M. W., and Borthwick, H. A. (1942). Floral initiation in biloxi soybean as influenced by grafting. Bot Gazette 103, 518–530. doi: 10.1086/335066
Jaeger, K. E., and Wigge, P. A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17, 1050–1054. doi: 10.1016/j.cub.2007.05.008
Jiang, B., Nan, H., Gao, Y., Tang, L., Yue, Y., Lu, S., et al. (2014). Allelic Combinations of soybean maturity Loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS One 9:e106042. doi: 10.1371/journal.pone.0106042
Kardailsky, L., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. doi: 10.1126/science.286.5446.1962
Knott, J. E. (1934). Effect of a localized photoperiod on spinach. Proc. Natl. Acad. Sci. U S A. 31, 152–154.
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pairs of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1015. doi: 10.1093/pcp/pcf156
Kong, F., Liu, B., Xia, Z., Sato, S., Kim, B. M., Watanabe, S., et al. (2010). Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231. doi: 10.1104/pp.110.160796
Kong, F., Nan, H., Cao, D., Li, Y., Wu, F., Wang, J., et al. (2014). A new dominant gene conditions early flowering and maturity in soybean. Crop Sci. 54, 2529–2535. doi: 10.2135/cropsci2014.03.0228
Koornneef, M., Hanhart, C. J., and van der Veen, J. H. (1991). A genetic and physiological analysis of late flowering mutations in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. doi: 10.1007/bf00264213
Kurasch, A. K., Hahn, V., Vollmann, J., Schori, A., Betrix, C. A., Mayr, B., et al. (2017). Identification of mega-environments in Europe and effect of allelic variation at maturity E loci on adaptation of European soybean Plant. Cell Environ. 40, 765–778. doi: 10.1111/pce.12896
Langewisch, T., Lenis, J., Jiang, G. L., Wang, D., Pantalone, V., and Bilyeu, K. (2017). The development and use of a molecular model for soybean maturity groups. BMC Plant Biol. 17:91. doi: 10.1186/s12870-017-1040-4
Langewisch, T., Zhang, H., Vincent, R., Joshi, T., Xu, D., and Bilyeu, K. (2014). Major soybean maturity gene haplotypes revealed by SNPViz analysis of 72 sequenced soybean genomes. PLoS One 9:e94150. doi: 10.1371/journal.pone.0094150
Li, C., Li, Y., Li, Y., Lu, H., Hong, H., Yu, T., et al. (2020). A domestication-associated gene GmPRR3b regulates circadian clock and flowering time in soybean. Mol. Plant 13, 745–759. doi: 10.1016/j.molp.2020.01.014
Li, M. W., Liu, W., Lam, H. M., and Gendron, J. M. (2019). Characterization of two growth period QTLs reveals modification of PRR3 genes during soybean domestication. Plant Cell Physiol. 60, 407–420. doi: 10.1093/pcp/pcy215
Li, Y., Dong, Y., Wu, H., Hu, B., Zhai, H., Yang, J., et al. (2019). Positional cloning of the flowering time QTL qFT12-1 reveals the link between the clock related PRR homolog with photoperiodic response in soybeans. Front. Plant Sci. 10:1303. doi: 10.3389/fpls.2019.01303
Liu, B., Kanazawa, A., Matsumura, H., Takahashi, R., Harada, K., and Abe, J. (2008). Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome. Gene Genet. 180, 995–1007. doi: 10.1534/genetics.108.092742
Liu, L. P., Song, W. W., Wang, L. W., Sun, X. G., Qi, Y. P., Wu, T. T., et al. (2020). Allele combinations of maturity genes E1-E4 affect adaptation of soybean to diverse geographic regions and farming systems in China. PLoS One 15:e0235397. doi: 10.1371/journal.pone.0235397
Liu, L., Gao, L., Zhang, L., Cai, Y., Song, W., Chen, L., et al. (2020). Co-silencing E1 and its homologs in an extremely late-maturing soybean cultivar confers super-early maturity and adaptation to high-latitude short-season regions. J. Integ. Agri. 19:2011.
Liu, W., Jiang, B., Ma, L., Zhang, S., Zhai, H., Xu, X., et al. (2018). Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. N. Phytol. 217, 1335–1345. doi: 10.1111/nph.14884
Liu, Y., Du, H., Li, P., Shen, Y., Peng, H., Liu, S., et al. (2020). Pan-genome of wild and cultivated soybeans. Cell 182:162. doi: 10.1016/j.cell.2020.05.023
Lu, S., Dong, L., Fang, C., Liu, S., Kong, L., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436. doi: 10.1038/s41588-020-0604-7
Lu, S., Zhao, X., Hu, Y., Liu, S., Nan, H., Li, X., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779. doi: 10.1038/ng.3819
Lyu, J., Cai, Z. D., Li, Y. H., Suo, H. C., Yi, R., Zhang, S., et al. (2020). The floral repressor GmFLC-like is involved in regulating flowering time mediated by low temperature in soybean. Int. J. Mol. Sci. 21:1322. doi: 10.3390/ijms21041322
Mansur, L. M., Lark, K. G., Kross, H., and Oliveira, A. (1993). Interval mapping of quantitative trait loci for reproductive, morphological, and seed traits of soybean (Glycine max L). Theor. Appl. Genet. 86, 907–913. doi: 10.1007/bf00211040
McBlain, B. A., and Bernard, R. L. (1987). A new gene affecting the time of flowering and maturity in soybean. J. Hered. 178, 68–70.
McBlain, B. A., Bernard, R. L., Cremeens, C. R., and Korczak, J. F. (1987). A rocedure to identify genes affecting maturity using soybean soline testers. Crop Sci. 27, 1127–1132. doi: 10.2135/cropsci1987.0011183x002700060008x
McCallum, C. M., Comai, L., Greene, E. A., and Henikoff, S. (2000). Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442. doi: 10.1104/Pp.123.2.439
Miladinovic, J., Ceran, M., Dordevic, V., Balesevic-Tubic, S., Petrovic, K., Dukic, V., et al. (2018). Allelic variation and distribution of the major maturity genes in different soybean collections. Front. Plant Sci. 9:1286. doi: 10.3389/fpls.2018.01286
Miranda, C., Scaboo, A., Cober, E., Denwar, N., and Bilyeu, K. (2020). The effects and interaction of soybean maturity gene alleles controlling flowering time, maturity, and adaptation in tropical environments. BMC Plant Biol. 20:65. doi: 10.1186/s12870-020-2276-y
Notoguchi, M., Abe, M., Kimura, T., Daumon, Y., Kobayashi, T., Yamaguchi, A., et al. (2008). Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49, 1645–1658. doi: 10.1093/pcp/pcn154
Owen, F. V. (1927). Inheritance studies in soybeans II Glabrousness, color of pubescence, time of maturity, and linkage relations. Genetics 12, 519–523. doi: 10.1093/genetics/12.6.519
Ray, J. D., Hinson, K., Mankono, J. E. B., and Malo, M. F. (1995). Genetic control of a long-juvenile trait in soybean. Crop Sci. 35, 1001–1006. doi: 10.2135/cropsci1995.0011183x003500040012x
Sakata, K., Nagamura, Y., Numa, H., Antonio, B. A., Nagasaki, H., Idonuma, A., et al. (2002). RiceGAAS: An automated annotation system and database for rice genome sequence. Nucleic Acids Res. 30, 98–102. doi: 10.1093/nar/30.1.98
Samach, A., Onouch, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 613–616.
Samanfar, B., Molnar, S. J., Charette, M., Schoenrock, A., Dehne, F., Golshani, A., et al. (2017). Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. Theor. Appl. Genet. 130, 377–390. doi: 10.1007/s00122-016-2819-7
Stewart, D. W., Cober, E. R., and Bernard, R. L. (2003). Modeling genetic effects on the photothermal response of soybean phenological development. Agronomy J. 95, 65–70. doi: 10.2134/agronj2003.0065
Takeshima, R., Hayashi, T., Zhu, J., Zhao, C., Xu, M., Yamaguchi, N., et al. (2016). A soybean quantitative trait locus that promotes flowering under long days is identified as FT5a, a FLOWERING LOCUS T ortholog. J. Exp. Bot. 67, 5247–5258. doi: 10.1093/jxb/erw283
Takimoto, A., and Hamner, K. C. (1965). Effect of far-red light and its interaction with red light in photoperiodic response of Pharbitis Nil. Plant Physiol. 40:859. doi: 10.1104/pp.40.5.859
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Tanksley, S. D. (1993). Mapping polygenes. Ann. Rev. Genet. 27, 205–233. doi: 10.1146/annurev.ge.27.120193.001225
Tsubokura, Y., Watanabe, S., Xia, Z., Kanamori, H., Yam, H., Kaga, A., et al. (2014). Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann. Bot. 113, 429–441. doi: 10.1093/aob/mct269
Tuinstra, M. R., Ejeta, G., and Goldsbrough, P. B. (1997). Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative traits loci. Theor. Appl. Genet. 95, 1005–1011. doi: 10.1007/s001220050654
Upadhyay, A. P., Ellis, R. H., Summerfield, R. J., Roberts, E. H., and Qi, A. (1994a). Characterization of photothermal flowering responses in maturity isolines of soya bean [Glycine max (L) Merrill] cv Clark. Ann. Bot. 74, 87–96. doi: 10.1093/aob/74.1.87
Upadhyay, A. P., Ellis, R. H., Summerfield, R. J., Roberts, E. H., and Qi, A. (1994b). Variation in the duration of the photoperiod-sensitive and photoperiod-insensitive phase of development to flowering among 8 maturity isoline of soybean [Glycine max (L) Merrill]. Ann. Bot. 74, 97–101. doi: 10.1093/aob/74.1.97
Wadahama, H., Kamauchi, S., Nakamoto, Y., Nishizawa, K., Ishimoto, M., Kawada, T., et al. (2008). A novel plant protein disulfide isomerase family homologous to animal P5-molecular cloning and characterization as a functional protein for folding of soybean seed-storage proteins”. FEBS J. 275, 399–410. doi: 10.1111/j.1742-4658.2007.06199.x
Wang, F., Nan, H., Chen, L., Fang, C., Zhang, H., Su, T., et al. (2019). A new dominant locus, E11, controls early flowering time and maturity in soybean. Mol. Breed 39:70.
Wang, L., Sun, S., Wu, T., Liu, L., Sun, X., Cai, Y., et al. (2020). Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotech. J. 18, 1869–1881. doi: 10.1111/pbi.13346
Wang, Y., Gu, Y., Gao, H., Qiu, L., Chang, R., Chen, S., et al. (2016). Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol. Biol. 16:79. doi: 10.1186/s12862-016-0653-9
Wang, Z., Zhou, Z., Liu, Y., Liu, T., Li, Q., Ji, Y., et al. (2015). Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell 27, 323–336. doi: 10.1105/tpc.114.135103
Watanabe, S., Hideshima, R., Xia, Z., Tsubokura, Y., Sato, S., Nakamoto, Y., et al. (2009). Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182, 1251–1262. doi: 10.1534/genetics.108.098772
Watanabe, S., Tajuddin, T., Yamanaka, N., Hayashi, M., and Harada, K. (2004). Analysis of QTLs for reproductive development and seed quality traits in soybean using recombinant inbred lines. Breed Sci. 54, 399–407. doi: 10.1270/jsbbs.54.399
Watanabe, S., Xia, Z., Hideshima, R., Tsubokura, Y., Sato, S., Yamanaka, N., et al. (2011). A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188, 395–407. doi: 10.1534/genetics.110.125062
Wolfgang, G., and Charles, Y. Q. (2017). Genetic separation of southern and northern soybean breeding programs in North America and their associated allelic variation at four maturity loci. Mol. Breed 37:8.
Wu, F., Kang, X., Wang, M., Haider, W., Price, W. B., Hajek, B., et al. (2019). Transcriptome-enabled network inference revealed the GmCOL1 feed-forward loop and its roles in photoperiodic flowering of soybean. Front. Plant Sci. 10:1221. doi: 10.3389/fpls.2019.01221
Wu, F., Sedivy, E. J., Price, W. B., Haider, W., and Hanzawa, Y. (2017). Evolutionary trajectories of duplicated FT homologues and their roles in soybean domestication. Plant J. 90, 941–953. doi: 10.1111/tpj.13521
Xia, Z., Sato, S., Watanabe, S., Yamanaka, N., Kawasaki, S., and Harada, K. (2005). Construction and characterization of a BAC library of soybean. Euphytica 141, 129–137. doi: 10.1007/s10681-005-6380-8
Xia, Z., Tsubokura, Y., Hoshi, M., Hanawa, M., Yano, C., Okamura, K., et al. (2007). An integrated high-density linkage map of soybean with RFLP, SSR, STS, and AFLP markers using a single F2 population. DNA Res. 14, 257–269. doi: 10.1093/dnares/dsm027
Xia, Z., Watanabe, S., Chen, Q., Sato, S., and Harada, K. (2009). A novel manual pooling system for preparing three-dimensional pools of a deep coverage soybean bacterial artificial chromosome library. Mol. Ecol. Resour. 9, 516–524. doi: 10.1111/j.1755-0998.2008.02503.x
Xia, Z., Watanabe, S., Yamada, T., Tsubokura, Y., Nakashima, H., Zhai, H., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. U S A. 109, E2155–E2164.
Xu, M., Xu, Z., Liu, B., Kong, F., Tsubokura, Y., Watanabe, S., et al. (2013). Genetic variation in four maturity genes affects photoperiod insensitivity and phyA-regulated post-flowering responses of soybean. BMC Plant Biol. 13:91. doi: 10.1186/1471-2229-13-91
Xu, M., Yamagishi, N., Zhao, C., Takeshima, R., Kasai, M., Watanabe, S., et al. (2015). The soybean-specific maturity gene E1 family of floral repressors controls night-break responses through down-regulation of FLOWERING LOCUS T orthologs. Plant Physiol. 168, 1735–1746. doi: 10.1104/pp.15.00763
Yamada, T., Hajika, M., Yamada, N., Hirata, K., Okabe, A., Oki, N., et al. (2012). Effects on flowering and seed yield of dominant alleles at maturity loci E2 and E3 in a Japanese cultivar, Enrei. Breed Sci. 61, 653–660. doi: 10.1270/jsbbs.61.653
Yamanaka, N., Nagamura, Y., Tsubokura, Y., Yamamoto, K., Takahashi, R., Kouchi, H., et al. (2000). Quantitative trait locus analysis of flowering time in soybean using a RFLP linkage map. Breed Sci. 50, 109–115. doi: 10.1270/jsbbs.50.109
Yamanaka, N., Ninomiya, S., Hoshi, M., Tsubokura, Y., Yano, M., Nagamura, Y., et al. (2001). An informative linkage map of soybean reveals QTLs for flowering time, leaflet morphology and regions of segregation distortion. DNA Res. 8, 61–72. doi: 10.1093/dnares/8.2.61
Yamanaka, N., Watanabe, S., Toda, K., Hayashi, M., Fuchigami, H., Takahashi, R., et al. (2005). Fine mapping of the FT1 locus for soybean flowering time using a residual heterozygous line derived from a recombinant inbred line. Theor. Appl. Genet. 110, 634–639. doi: 10.1007/s00122-004-1886-3
Yue, Y., Liu, N., Jiang, B., Li, M., Wang, H., Jiang, Z., et al. (2017). A single nucleotide deletion in J encoding GmELF3 confers long juvenility and is associated with adaption of tropic soybean. Mol. Plant 10, 656–658. doi: 10.1016/j.molp.2016.12.004
Zeng, X. R., Liu, H. L., Du, H. Y., Wang, S. J., Yang, W. M., Chi, Y. J., et al. (2018). Soybean MADS-box gene GmAGL1 promotes flowering via the photoperiod pathway. BMC Genomics 19:51. doi: 10.1186/s12864-017-4402-2
Zhai, H., Lü, S., Liang, S., Wu, H., Zhang, X., Liu, B., et al. (2014a). GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS One 9:e89030. doi: 10.1371/journal.pone.0089030
Zhai, H., Lü, S., Wang, Y., Chen, X., Ren, H., Yang, J., et al. (2014b). Allelic variations at four major maturity E genes and transcriptional abundance of the E1 gene are associated with flowering time and maturity of soybean cultivars. PLoS One 9:e97636. doi: 10.1371/journal.pone.0097636
Zhai, H., Lu, S., Wu, H., Zhang, Y., Zhang, X., Yang, J., et al. (2015). Diurnal expression pattern, allelic variation, and association analysis reveal functional features of the E1 gene in control of photoperiodic flowering in soybean. PLoS One 10:e0135909. doi: 10.1371/journal.pone.0135909
Zhang, J., Xu, M., Dwiyanti, M. S., Watanabe, S., Yamada, T., Hase, Y., et al. (2020a). A soybean deletion mutant that moderates the repression of flowering by cool temperatures. Front. Plant Sci. 11:429. doi: 10.3389/fpls.2020.00429
Zhang, L., Liu, W., Tsegaw, M., Xu, X., Qi, Y., Sapey, E., et al. (2020b). Principles and practices of the photo-thermal adaptability improvement in soybean. J. Integr. Agric. 19, 295–310. doi: 10.1016/s2095-3119(19)62850-9
Zhang, X., Zhai, H., Wang, Y., Tian, X., Zhang, Y., Wu, H., et al. (2016). Functional conservation and diversification of the soybean maturity gene E1 and its homologs in legumes. Sci. Rep. 6:29548.
Zhao, C., Takeshima, R., Zhu, J., Xu, M., Sato, M., Watanabe, S., et al. (2016). A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 16:20. doi: 10.1186/s12870-016-0704-9
Zhao, L., Li, M. M., Xu, C. J., Yang, X., Li, D. M., Zhao, X., et al. (2018). Natural variation in GmGBP1 promoter affects photoperiod control of flowering time and maturity in soybean. Plant J. 96, 147–162. doi: 10.1111/tpj.14025
Keywords: soybean, flowering time, maturity, photoperiodic response, positional cloning, E1
Citation: Xia Z, Zhai H, Wu H, Xu K, Watanabe S and Harada K (2021) The Synchronized Efforts to Decipher the Molecular Basis for Soybean Maturity Loci E1, E2, and E3 That Regulate Flowering and Maturity. Front. Plant Sci. 12:632754. doi: 10.3389/fpls.2021.632754
Received: 24 November 2020; Accepted: 02 March 2021;
Published: 28 April 2021.
Edited by:
George Coupland, Max Planck Institute for Plant Breeding Research, GermanyReviewed by:
Karen A. Hudson, Crop Production and Pest Control Research, United States Department of Agriculture, Agricultural Research Service, United StatesTakeshi Kurokura, Utsunomiya University, Japan
Copyright © 2021 Xia, Zhai, Wu, Xu, Watanabe and Harada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengjun Xia, xiazhj@iga.ac.cn; Kyuya Harada, haradaq@nifty.com
 Zhengjun Xia
Zhengjun Xia Hong Zhai1
Hong Zhai1