- 1College of Plant Science, Jilin University, Changchun, China
- 2College of Landscape Architecture and Tourism, Hebei Agricultural University, Baoding, China
- 3London Research and Development Centre, Agriculture and Agri-Food Canada, London, ON Canada
- 4Department of Biology, Western University, London, ON, Canada
- 5Department of Pain, Second Hospital of Jilin University, Changchun, China
SQUAMOSA Promoter Binding Protein (SBP) family genes act as central players to regulate plant growth and development with functional redundancy and specificity. Addressing the diversity of the SBP family in crops is of great significance to precisely utilize them to improve agronomic traits. Blueberry is an important economic berry crop. However, the SBP family has not been described in blueberry. In the present study, twenty VcSBP genes were identified through data mining against blueberry transcriptome databases. These VcSBPs could be clustered into eight groups, and the gene structures and motif compositions are divergent among the groups and similar within each group. The VcSBPs were differentially expressed in various tissues. Intriguingly, 10 VcSBPs were highly expressed at green fruit stages and dramatically decreased at the onset of fruit ripening, implying that they are important regulators during early fruit development. Computational analysis showed that 10 VcSBPs were targeted by miR156, and four of them were further verified by degradome sequencing. Moreover, their functional diversity was studied in Arabidopsis. Noticeably, three VcSBPs significantly increased chlorophyll accumulation, and qRT-PCR analysis indicated that VcSBP13a in Arabidopsis enhanced the expression of chlorophyll biosynthetic genes such as AtDVR, AtPORA, AtPORB, AtPORC, and AtCAO. Finally, the targets of VcSBPs were computationally identified in blueberry, and the Y1H assay showed that VcSBP13a could physically bind to the promoter region of the chlorophyll-associated gene VcLHCB1. Our findings provided an overall framework for individually understanding the characteristics and functions of the SBP family in blueberry.
Introduction
Blueberry (Vaccinium spp.) is a globally cultivated perennial shrub with outstanding economic value. Its fruit is not only sweet but also rich in nutrients, especially anthocyanins, which greatly promote human health such as improvement of vision, blood glucose balance, elimination of free radicals, aging delay, inhibition of obesity and hyperlipidemia, and prevention of cardiovascular diseases (Routray and Orsat, 2011). Thus, blueberry growth and development, especially the events related to fruit ripening and anthocyanin biosynthesis, have started to attract attention in recent years. To date, a few regulators have been shown to be involved in the regulation of blueberry growth and development, including transcription factor genes VcMYBs, VcSOC1-k, VcDDF1, VcFT, some miRNAs, and hormones (IAA and ABA) (Zifkin et al., 2012; Song et al., 2013; Walworth et al., 2016; Hou et al., 2017, 2020; Song and Gao, 2017; Plunkett et al., 2018). Recently, high-throughput sequencing data provided considerable information for identifying and characterizing the regulators that control blueberry growth and development (Rowland et al., 2012; Hou et al., 2017; Qi et al., 2019). However, our understanding of the regulatory network underlying blueberry growth and development are extremely limited.
SQUAMOSA Promoter Binding Proteins (SBPs) constitute a plant-specific transcription factor family featured by a highly conserved SBP domain of 76 amino acids. Generally, the SBP domain harbors three common structures: two tandem zinc fingers (C3H and C2HC) and a nuclear localization signal (NLS), partially overlapping with the second zinc finger at the C-terminal (Birkenbihl et al., 2005). It has been well known that SBP proteins can bind to a consensus DNA sequence TNCGTACAA with the GTAC as the binding core, therefore regulating the expression of their target clients (Birkenbihl et al., 2005; Kropat et al., 2005). SBP proteins play important roles in various biological and cellular processes through regulating their target clients, spanning virtually every aspect of plant growth and development as well as stress response. These include leaf morphology and leaf initiation (Preston et al., 2016), trichome formation (Yu et al., 2010), phase transition (Xu et al., 2016), shoot branching and maturation (Gao et al., 2018), regeneration of shoot and root (Barrera-Rojas et al., 2020; Ye et al., 2020), root development (Yu et al., 2015), flowering (Xie et al., 2020), male fertility (Xing et al., 2010), ovary and fruit development (Silva et al., 2014), cell number and size (Usami et al., 2009), and grain yield (Wang et al., 2017), etc. Evidently, SBP genes are a class of central players in the regulation of plant growth and development, which can be utilized for the improvement of important agronomic traits.
In 1996, the first two SBPs were identified in Antirrhinum majus, and shown to regulate the expression of the MADS-box gene SQUAMOSA directly through binding to its promoter region, therefore controlling flowering (Klein et al., 1996). With the availability of whole-genome information and transcriptome data, SBP genes have been isolated in many plant species, from the model plant Arabidopsis to economically important crops (Salinas et al., 2012; Hou et al., 2013; Bhogale et al., 2014; Li and Lu, 2014; Shalom et al., 2015). The SBP family is a relatively small group of transcription factors in plants, and the SBP family members show diverse features and evolutionary divergences. Emerging evidence indicated that SBPs exert their regulatory functions in a member-specific manner. For example, SBP-like 9 (SPL9) in Arabidopsis thaliana might serve as a negative regulator of wall ingrowth deposition in transfer cells of phloem parenchyma (Nguyen et al., 2017), whereas SPL3 cannot affect the deposition of wall ingrowth but enhance phosphate-deficient response (Lei et al., 2016). Likewise, OsSPL14 acts in controlling rice tillering growth (Luo et al., 2012), and OsSPL16 was found to be a regulator of grain size, shape, and quality in Oryza sativa (Wang et al., 2012). Nevertheless, a number of studies showed that members of the SBP family could be functionally redundant in the regulation of plant growth and development. For example, AtSPL3/4/5 redundantly promote flowering through activating the expression of LEAFY, FRUITFULL, and APETALA1 (Jung et al., 2016), while AtSPL9/15 and AtSPL2/10/11 act as regulators of plastochron and branching (Schwarz et al., 2008; Shikata et al., 2009). Additionally, a subset of SBP genes can be subjected to miR156-guided transcriptional cleavage and translational repression, for example, 11 out of the 17 SPLs in Arabidopsis and seven out of the 19 SPLs in pear, thereby being integrated into miR156/SPL modules to regulate plant growth, development, and stress response (Zhang et al., 2015; Qian et al., 2017). Clearly, the SBP family members show distinct features and perform their functions with redundancy and specificity. Thus, addressing the diversity and specificity of the SBP family in different crop species is of great significance in order to precisely utilize them to improve agronomic traits.
It has been accepted that the functional roles of SBP genes are highly conserved across plant species. However, novel functions of SBP genes have been constantly revealed in crop species with special developmental processes or organs (Bhogale et al., 2014; Silva et al., 2014; Qian et al., 2017). Fruit growth and ripening is a specific process for fruit-bearing plant species, and many fruit-specific events occur during the process. Accumulating evidence indicates that SBP genes are involved in the regulation of fruit growth and ripening. For instance, the Colorless non-ripening (CNR) locus of tomato (a homolog of AtSPL3) is crucial for fruit ripening (Manning et al., 2006), while SPL18 in grape might regulate berry development at the veraison stage in an ABA-independent manner (Xie et al., 2019). Likewise, VmTDR4 (a SQUAMOSA-class MADS-box gene) is positively involved in the regulation of anthocyanin accumulation during bilberry fruit ripening (Jaakola et al., 2010), while MaSPL16 in banana regulates carotenoid biosynthesis through promoting the expression of MaLCYBs genes (Zhu et al., 2020). These functions were not observed in non-fleshy-fruited plant species such as Arabidopsis and rice. Obviously, it is of great interest to comprehensively characterize the SBP family in crops with special developmental processes or organs and to reveal their functional roles and neo-functionalization.
Since the SBP family proteins are powerful regulators with functional diversification in plants, study of these genes will enhance understanding of the regulatory network underlying blueberry growth and development. To date, however, the characteristics and functional diversity of the SBP family have remained unexplored in blueberry. In recent years, the transcriptional profiles of blueberry leaves, flower buds, and fruits at different development stages have been investigated using high-throughput sequencing technology (Rowland et al., 2012; Gupta et al., 2015; Li et al., 2016). These transcriptome data have enabled the identification of the SBP family genes involved in blueberry growth and development. In the present study, 20 VcSBPs were identified from the blueberry transcriptome database. Gene structure, phylogeny, motif composition, miRNA target sites, and expression patterns in different tissues were systematically analyzed. Furthermore, the functional diversity of the VcSBP family genes were studied in Arabidopsis. Additionally, the targets of VcSBP proteins were investigated in blueberry. These findings lay a foundation for further studying the functional roles of the SBP genes and their regulatory mechanisms during blueberry growth and development, which will contribute to the improvement of blueberry agronomic traits.
Materials and Methods
Plant Materials
Seven-year-old blueberry trees (Vaccinium corymbosum, cv. Northland) from clonal propagation were grown at the experimental station at Jilin University (Changchun, China). Blueberry tissues were randomly harvested from six different seven-year-old blueberry plants, including new leaf, young shoot, unopened flower, opening flower, and fruit at six developmental stages [green pad (FS1), green cup I (FS2), green cup II (FS3), light green/white (FWS), pink (FPS) and blue (FMS) fruits] (Li et al., 2020), frozen in liquid nitrogen and stored at −80°C.
Arabidopsis and tobacco (Nicotiana benthamiana) plants were grown in growth chambers under long-days (16 h light/8 h dark) at 20°C with 70–80% relative humidity.
Identification of SBP Genes in Blueberry
The CDS sequences of SBP genes from Arabidopsis and grape were downloaded from the publicly available databases TAIR1 or Phytozome2, and then used as reference sequences to perform local blast searches for querying their homologs against the publicly available transcriptome databases of blueberry3 and our previously assembled transcriptome data. The conserved SBP-specific domains were confirmed using the PROSITE Server4, and all of the SBP-like genes without an SBP domain were discarded. The physicochemical properties, including molecular weight (MW), and isoelectric point (pI), of the identified SBP proteins, were predicted using the ExPASy Compute pI/Mw tool5.
Chromosomal Location and Phylogenetic Analysis of the VcSBP Family Genes
All VcSBP genes were mapped to the genome of V. corymbosum, cv. Draper, according to the approximate location information (Colle et al., 2019), and their positions were imported into the CIRCOS software to generate a circle plot (Krzywinski et al., 2009). The SBP protein sequences (17 from grape, 27 from apple, and 17 from tomato) were downloaded from Phytozome (see text footnote 2). All the SBP protein sequences from blueberry, Arabidopsis, grape, apple, and tomato were used for phylogenetic analysis, and phylogenetic trees were constructed with the MEGA7.0 software using the maximum likelihood with 1000 bootstrap replications (Kumar et al., 2016). The sequence logo was created using Weblogo online software6.
Analysis of Gene Structure and Conserved Protein Motifs
The exon/intron structure of each VcSBP gene was analyzed using the Gene Structure Display Server7 by comparing the coding sequence and genomic sequence. Potentially conserved motifs of VcSBP proteins were predicted using the online Multiple Expectation Maximization for Motif Elucidation (MEME) toolkit8, with the following parameter settings: the minimum motif width = 20, the maximum motif width = 50, and the maximum number of motifs = 20.
MicroRNA Target Prediction
To identify VcSBPs targeted by miR156/157, the coding regions and 3′ UTRs of all VcSBP sequences were analyzed at the psRNATarget server9 with blueberry miR156/157 mature sequences (Hou et al., 2017). The sequence logo of miR156/157 was created using the Weblogo online software (see text footnote 6).
Expression Pattern Analysis of VcSBP Genes in Blueberry
Total RNAs were isolated from blueberry leaf, shoot, unopened flower, opening flower, fruit tissues at six developmental stages, and blueberry tissue culture seedlings as well as Arabidopsis leaf. First-strand cDNA was synthesized using the PrimeScriptTM RT reagent kit with gDNA Eraser (Takara, Japan). qRT-PCR was subsequently conducted with an ABI StepOnePlus PCR system and SYBR Premix Ex Taq (Takara, Japan). Blueberry ACTIN was set as an internal reference for data normalization. Three biological replicates with three technical replicates were performed for each sample, and data were analyzed by the software ABI StepOnePlus v2.3 and one-way ANOVA with LSD test, and p-value < 0.05 was considered to be statistically significant. Primer information is listed in Supplementary Table 1.
Vector Construction and Plant Transformation
The full-length CDS of each VcSBP was amplified using gene-specific primers (Supplementary Table 1). All purified PCR products were cloned into the Gateway entry vector pDONR207 and then transferred into the destination vector pEarleyGate101 (pEG101) through homologous recombination. All the constructed plasmids were confirmed by PCR and sequencing. The expression vectors (pEG101-VcSBPs) were individually transformed into Agrobacterium tumefaciens strain GV3101.
Arabidopsis transformation was conducted using the floral dip method described by Zhang et al. (2006). Transgenic lines were screened in the soil with 200 μg/mL glufosinate and then confirmed by PCR with gene-specific primers. Primer information is listed in Supplementary Table 1.
The VcMIR156a gene was constructed into pBI121 as described previously (Li et al., 2020) and transferred into Agrobacterium tumefaciens strain EHA105. Blueberry transformation was performed according to the method described by Song and Sink (2006). The transgenic blueberry lines were obtained and confirmed by PCR with gene-specific primers (Supplementary Table 1). The VcMIR156a-overexpressing transgenic Arabidopsis were generated as previously described (Li et al., 2020).
Prediction of VcSBP Targets in Blueberry
The genes containing the TNCGTACAA element within 2000 bp upstream were extracted against the reference genome of blueberry (V. corymbosum, cv. Draper) (Colle et al., 2019). To functionally annotate these targets, all protein sequences were analyzed using eggNOG-Mapper10. Density distribution of distance was visualized using ggplot2 in R.
Yeast One-Hybrid (Y1H) Assay
To investigate the interaction of VcSBPs and their targets, the full-length CDS of VcSBP13a was cloned and introduced into the vector pB42AD. The fragment 597-796 bp upstream of the transcription start site (TSS) of VcLHCB1 containing two TNCGTACAA elements and the fragment 1096–1548 upstream of the TSS of VcLHCB2 with five GTAC elements were cloned as promoter regions (pVcLHCB1 and pVcLHCB2) and constructed into the vector pLacZi, respectively. Three negative controls, i.e., pB42AD/pLacZi, pB42AD-VcSBP13a/pLacZi, and pB42AD/pLacZi-pVcLHCB1, pB42AD/pLacZi-pVcLHCB2, and one positive control, pB42AD-AtRVE8/placZi-AtPRR5, were also generated. Different plasmid combinations were separately co-transformed into the yeast cells (EGY48). The primers are listed in Supplementary Table 1.
Results
Identification of SBP Genes and Their Characterization in Blueberry
To identify SBP genes in blueberry, the CDS sequences of the SBP genes from both Vitis vinifera and Arabidopsis were used as queries to conduct BLASTn against the V. corymbosum GDV RefTrans V1 and our previously assembled transcriptome data (Hou et al., 2017). After removal of redundant sequences, a total of 22 SBP sequences were identified in blueberry, which are then named as VcSBP and each of them assigned a species number corresponding to their closest homolog in Arabidopsis (Supplementary Figure 1). To verify the sequences of the VcSBP genes, their full-length CDSs were amplified and sequenced, and the results showed that all the cloned SBP genes are indeed the same sequence as listed in the Genome Database for V. corymbosum cv. Draper v1.0.
The features of all the VcSBP family members were computationally characterized. As shown in Supplementary Table 2, the CDS lengths of these VcSBP genes are quite variable, ranging from 363 to 3222 bp, which is consistent with the SBP family in other plant species such as Arabidopsis, apple, grape (Hou et al., 2013; Li et al., 2013; Zhang et al., 2015). Their deduced proteins were estimated to possess the theoretical pI values from 5.96 to 10.54 and the MWs from 23.17 to 117.97 kDa. Furthermore, the SBP domains were analyzed using the online tool CD search11. As shown in Figures 1A,B, all the SBP proteins, except VcSBP6c, VcSBP14aAS, and VcSBP14cAS, contain a typical SBP domain featured by two zinc finger structures (C3H and C2HC) and an NLS motif. VcSBP14aAS harbors an SBP domain with the absence of C3H and an incomplete C2HC, while the SBP domains in VcSBP6c and VcSBP14cAS lack the NLS motif (Figures 1A,B). These results indicated that the 22 putative genes are SBP family members.
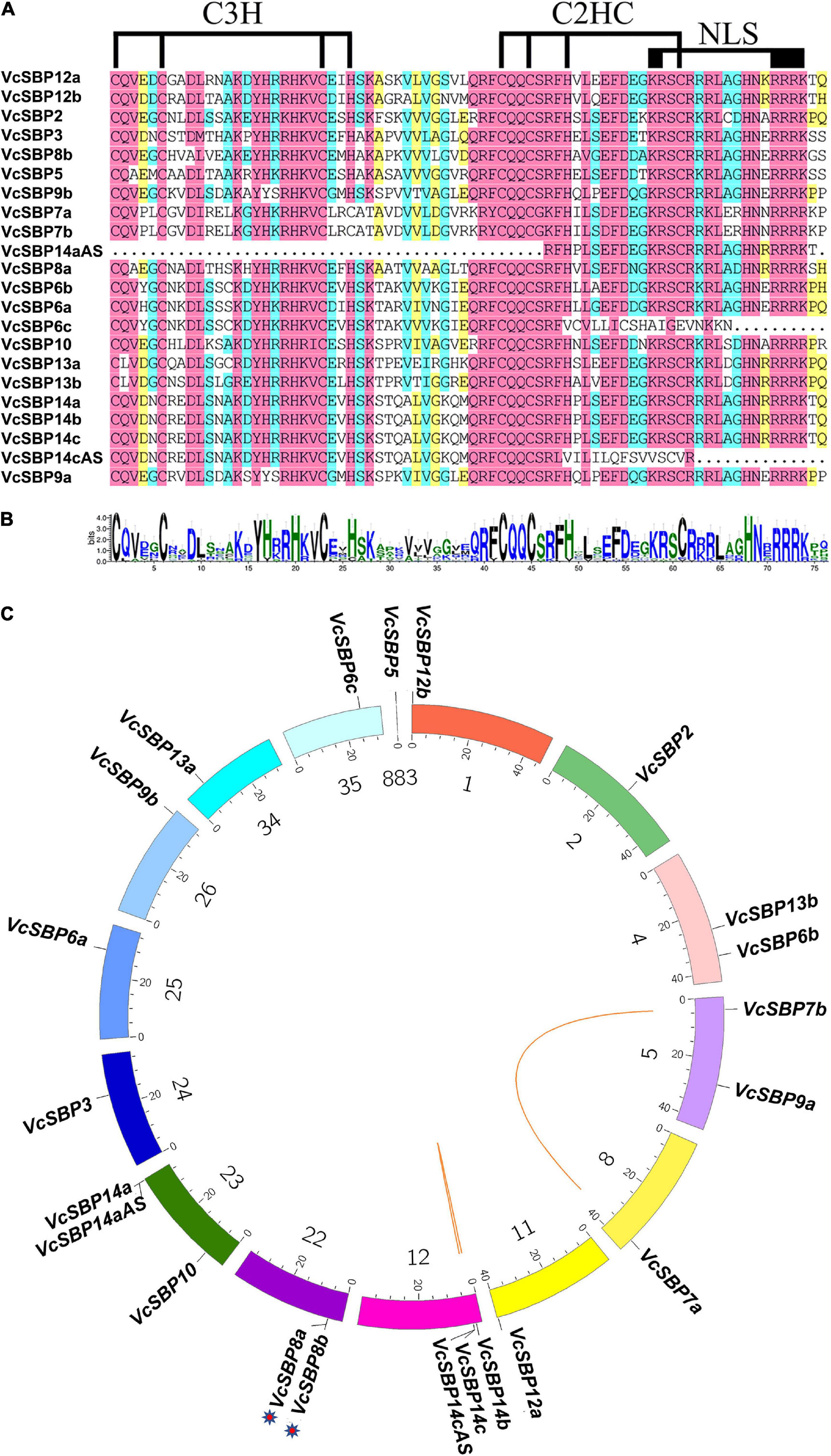
Figure 1. The SBP domains and chromosomal localization of the VcSBP family genes. (A) Multiple alignment of the SBP domains. The two conserved zinc-finger structures (C3H and C2HC) and the NLS are indicated. (B) Sequence logo of the SBP domains in VcSBPs. The total height of each stack represents the conservation degree of each position, while the height of the letters within each stack indicates the relative frequency of the corresponding amino acid. (C) Chromosomal localization and duplication of SBP genes in blueberry. Each colored box represents a scaffold. The approximate distribution of each VcSBP gene is marked on the circle with a short black line. The tandem duplication cluster is indicated with stars. Colored lines indicate the linkage group with segmental duplication.
Distribution of VcSBP Genes in the Blueberry Genome and Their Evolutionary Relationships
To date, the draft genome assembly of V. corymbosum contains 1760 scaffolds12. To map the locations of the VcSBP family genes in the draft genome, a Circos map was generated using the corresponding scaffolds where the VcSBP genes are situated. It turns out that they are unevenly distributed in 15 different scaffolds (i.e., 1, 2, 4, 5, 8, 11, 12, 22, 23, 24, 25, 26, 34, 35, 883, Figure 1C). Further observation indicated that two pairs of VcSBP genes (VcSBP14a and VcSBP14aAS, VcSBP14c, and VcSBP14cAS) were situated at the same loci with the similarity of 65.01 and 93.09%, respectively, indicating that they might be derived from different transcript splicing of the same genes. Thus, the 22 SBP sequences were likely derived from 20 SBP genes and two alternative splices. Gene family expansion can arise from gene duplication events such as tandem duplication and segmental duplication of chromosomal regions (Leister, 2004). Generally, tandem duplication refer to those closely related genes separated by the distance within 50 kb in the same chromosome. It was observed that the distance between VcSBP8a and VcSBP8b is 15,110 bp (Figure 1C and Supplementary Table 2), suggesting that they might be derived from tandem duplication. In contrast, the distances between VcSBP7a and VcSBP7b as well as VcSBP14b and VcSBP14c are relatively far from each other, and their similarities reach 99.27 and 94.26%, respectively, implying that they were possibly generated from segmental duplication.
To explore the evolutionary relationships among the SBP family proteins, a phylogenetic tree was generated using the protein sequences of VcSBPs and the SBPs from apple, grape, tomato, and Arabidopsis. As shown in Supplementary Figure 2, all the SBP proteins were classified into six different groups (G1-G6), and VcSBPs were separately distributed to the 6 groups, suggesting that the VcSBP family might have experienced evolutionary diversification similar to those in the other four plant species. For example, seven small VcSBP proteins with no more than 254 aa (VcSBP3, VcSBP5, VcSBP6c, VcSBP9a, VcSBP10, VcSBP14aAS, and VcSBP14cAS) were separately distributed into the six groups, while the large proteins with more than 800 aa (VcSBP14a, VcSBP12a, and VcSPB7a/7b) were clustered into G5 and G6, respectively. Further observation indicated that VcSBPs were closer to their homologs from apple, grape, Arabidopsis and/or tomato in the phylogenetic tree. For instance, VcSBP3 was grouped together with SlySBP3, AtSBP3, VvSBP9, CNR, and while VcSBP2 was distributed into the subgroup of SBP2/10/11 with the inclusion of VvSBP2, SlySBP2, and AtSBP2.
VcSBP Family Shows Diverse Gene Structures and Motif Compositions
To understand the structural diversity of VcSBP family genes, the exon/intron structures were generated according to the gene coding and genomic sequences. Consistent with previous reports in other plant species (Hou et al., 2013; Li et al., 2013; Zhang et al., 2015), VcSBP genes showed a high variation in the number of exons. As indicated in Figure 2A, four VcSBP genes (VcSBP7a, VcSBP7b, VcSBP12a, and VcSBP14a) comprise 10 exons with intron intervals. In contrast, VcSBP6c, VcSBP14aAS, and VcSBP14cAS harbor only one exon without intron. The remaining VcSBPs have 2–4 exons. Furthermore, integration analysis of exon/intron structures with phylogenetic relationship and sequence identity was conducted. It turns out that the pairs of VcSBPs in the same clade basically display similar exon/intron structures (Figure 2A). Two pairs of duplicated genes (VcSBP7a and VcSBP7b, VcSBP14b, and VcSBP14c) show not only similar exon/intron structure but also high similarity with the values of 99.27 and 94.21%, respectively (Supplementary Table 2), supporting that they might undergo similar exon/intron gain or loss events with less functional diversification. However, the remaining VcSBP pairs with similar exon/intron structure in the same clade displayed relatively low similarities ranging from 8–48% (Figure 2A and Supplementary Table 2), implying the diversity of their functional roles.
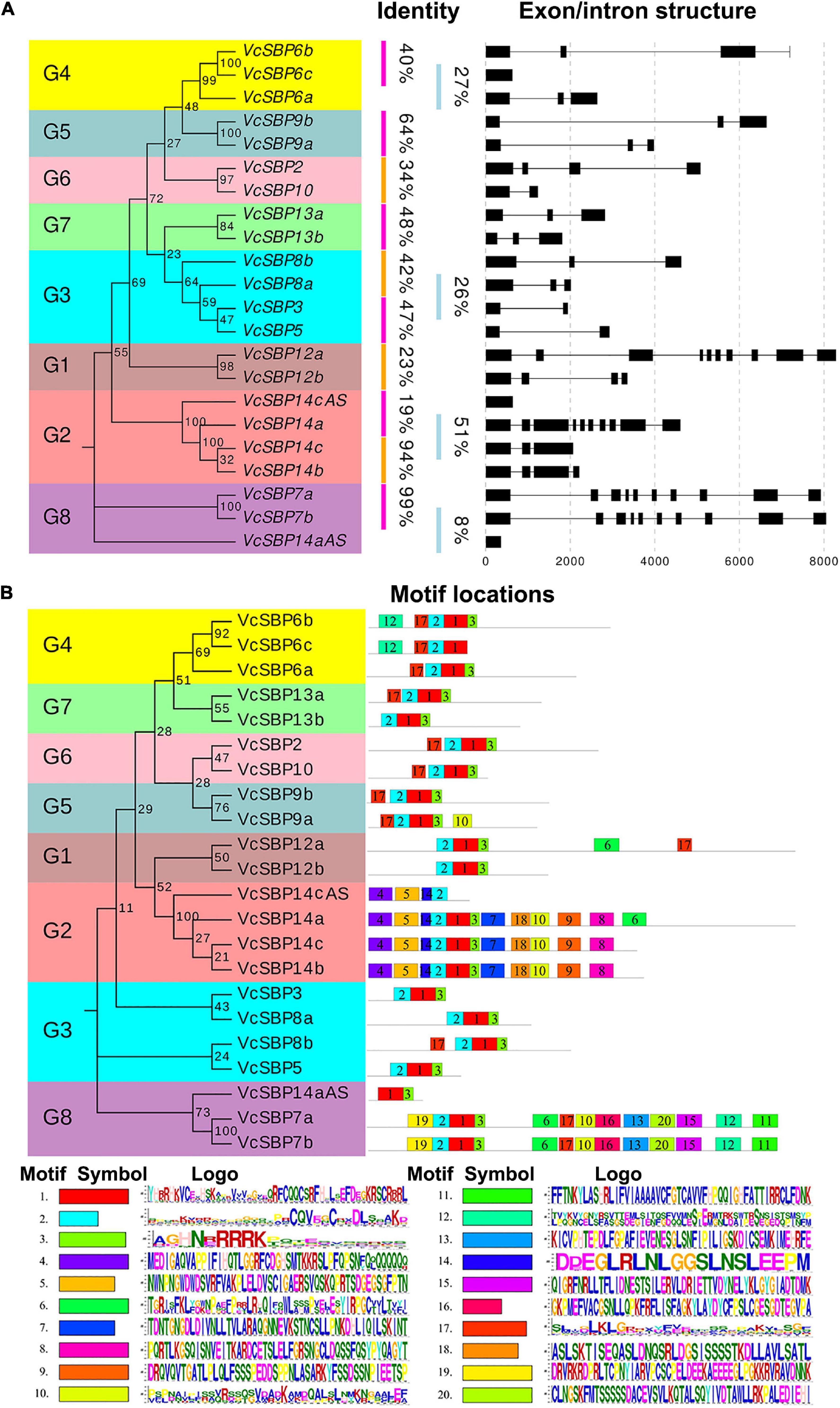
Figure 2. Gene structures and protein motif compositions of the VcSBP family. (A) The exon/intron structures of VcSBP genes. The left panel is the phylogenetic tree of VcSBP genes. Eight groups are clustered (G1–G8), and the percent similarity between the gene pair is listed. The right panel shows the intron-exon structures where the exons are shown by rectangular, and the introns are represented by thin lines. (B) Motif analysis of VcSBP proteins. The left panel is the phylogenetic tree of VcSBP proteins, and eight groups are clustered (G1–G8). The right panel shows the motif compositions of VcSBP proteins. The motifs were identified using the program MEME, represented with boxes of different colors labeled by number (1–20). The sequence logos of 20 motifs are listed, and the height of the letters within each stack indicates the relative frequency. The symbol and the ID number are corresponding to the colored box and the number in the right panel.
To provide clues about the functional diversity of VcSBP family, conserved motifs in each of the VcSBP proteins were predicted using the online tool ScanProsite. As shown in Figure 2B, twenty conserved motifs were identified in VcSBPs, and two motifs (the motifs 1 and 2) constitute the SBP domain. Five VcSBPs (VcSBP7a/7b and VcSBP14a/14b/14c) harbor 11–13 motifs, while the remaining VcSBPs contain 2–5 motifs. Although most of the 20 motifs are functionally unknown, the existence of multiple motif compositions implied the functional diversity of the SBP family in blueberry. The 20 VcSBPs and two alternatively spliced species were clustered into eight groups in the phylogenetic tree. It was observed that the VcSBP proteins in the same group in the phylogenetic tree basically show similar motif composition, suggesting possible functional redundancy within the same group.
VcSBPs Are Differentially Expressed in Different Tissues and Throughout Fruit Development
To obtain clues about the functional roles of VcSBP genes, their expression patterns in five tissues (new leaf, young shoot, opening, and unopened flower, and mature fruit; Figure 3A) were examined using qRT-PCR. Since high sequence similarity exists within each of the three VcSBP groups (VcSBP7a/b, VcSBP6b/c, and VcSBP14a/b/c/cAS), only one gene was chosen as representative for each group (VcSBP6b, VcSBP7a, and VcSBP14a). The examined VcSBP genes were found to be differentially expressed in the five tissues. As shown in Figures 3B,C, 10 SBP genes showed the highest expression in shoot, especially VcSBP13b and VcSBP9a, with 7.46–509.52 and 7.12–118.31-fold increase as compared to the other four tissues. Meanwhile, three VcSBP genes (VcSBP8a, VcSBP8b, and VcSBP12a) were highly expressed in opening flower (Figure 3D), and three VcSBP genes (VcSBP5, VcSBP6b, and VcSBP13a) in unopened flower and shoot (Figure 3E). In mature fruit, all the VcSBP genes were expressed at relatively low levels except VcSBP9b, VcSBP12b, VcSBP14a, and VcSBP14aAS (Figure 3F). These results suggested that the VcSBP family might perform functions in an organ-specific manner.
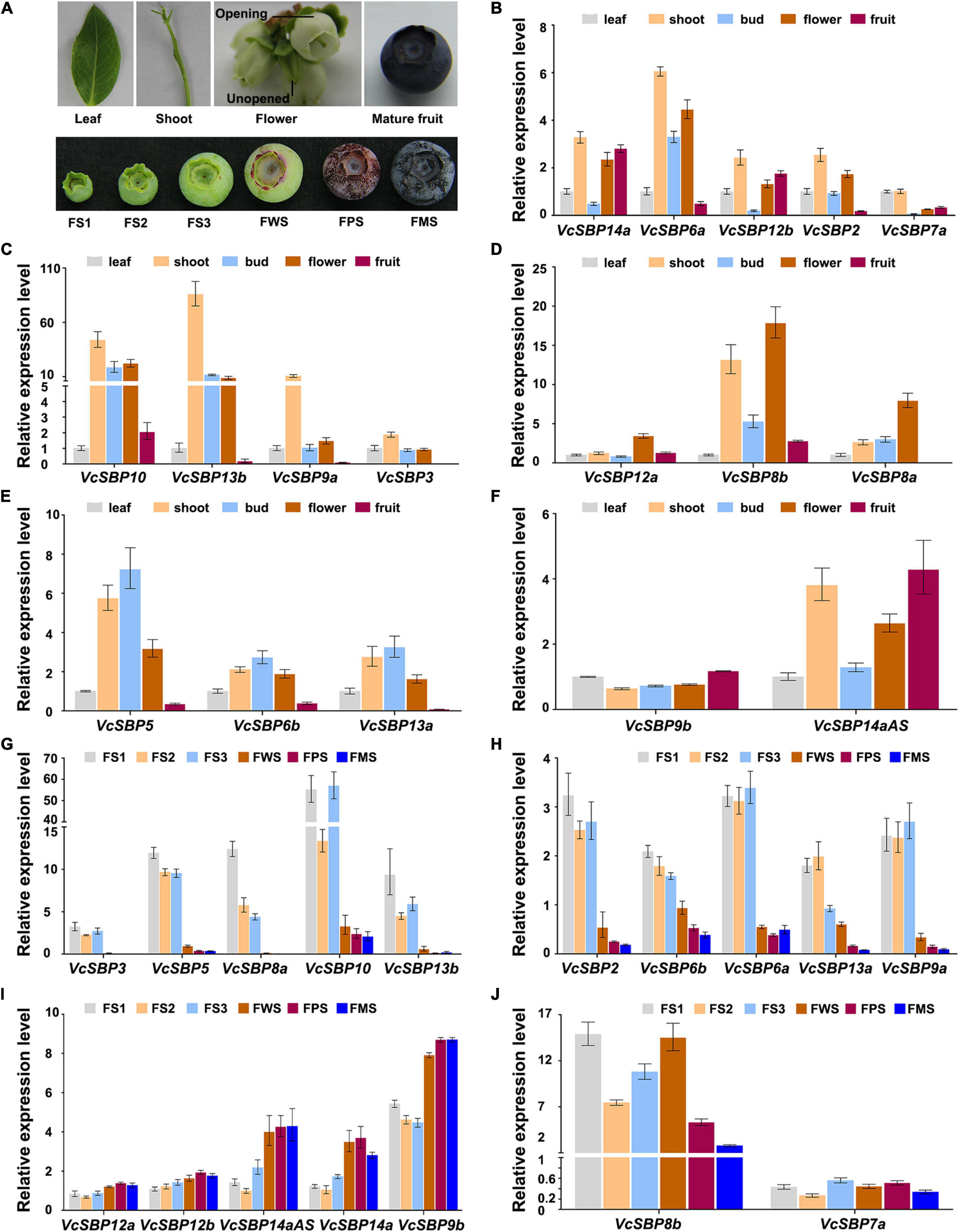
Figure 3. Expression patterns of VcSBP genes in different tissues and during fruit development. (A) Photograph of different tissues and fruits at six developmental stages. Different tissues include blueberry new leaf, young shoot, unopened flower, opening flower, and mature fruit, while FS1, FS2, FS3, FWS, FPS, and FMS refer to green pad, green cup I, green cup II, light green/white, pink, and blue stages, respectively. (B–F) Expression pattern of VcSBP genes in different blueberry tissues. (G–J) Expression pattern of VcSBP genes in fruits at six developmental stages. Total RNAs extracted from the above different tissues and fruits at six developmental stages were used for qRT-PCR analysis. Three biological replicates and three technical replicates for each biological replicate were performed. Error bars indicate standard errors. The data were normalized against the gene VcACTIN.
Blueberry fruit development can be generally divided into three phases: fruit growth, a transition from growth to maturation, and maturation (Zifkin et al., 2012). To explore the functional roles of SBP family during fruit development, the expression patterns of VcSBPs were investigated in fruits at six developmental stages (green pad, green cup I, green cup II, light green/white, pink, and blue fruit, Figure 3A). The three early developmental stages represent the growth phase; the light green/white stage corresponds to the transition stage; the pink and blue stages refer to the maturation phase. As shown in Figures 3G,H, 10 VcSBP genes were highly expressed at the three early developmental stages (green pad, green cup I, green cup II), and dramatically decreased at the light green stage (especially VcSBP3, VcSBP5, VcSBP9b, VcSBP10, and VcSBP13b with more than 10-fold changes as compared to the ones at green cup II), and then remained at a low level until fruit maturation. Conversely, the expression levels of some VcSBP genes (VcSBP9b, VcSBP12a, VcSBP12b, VcSBP14a, and VcSBP14aAS) were relatively low at the three early developmental stages, but then increased from the light green stage until fruit maturation (Figure 3I). Also, it was observed that VcSBP8b was gradually increased from the green cup I stage to the light green stage, and then remarkably decreased at the maturation stage (Figure 3J). These results suggested that VcSBP family might play different, even opposite, roles during blueberry fruit development.
A Subset of SBP Genes Are Targeted by miR156 in Blueberry
It is well acknowledged that most SBP family members can be regulated through miR156/157-mediated mRNA cleavage or translational repression in plants (Wang and Wang, 2015). Previously we identified six MIR156/MIR157 genes in blueberry (Hou et al., 2017). To computationally identify the SBP family members targeted by miR156/157 in blueberry, the miRNA responsive elements (MREs) were searched using the complementary sequences of six miR156/157s against the 20 VcSBPs and the 2 alternatively spliced variants (Supplementary Figure 3). It was found that 10 of the VcSBPs harbor one or two MRE(s) for miR156/157 (Figure 4A), suggesting that they have the potentials to be targeted by miR156/157. Further examination indicated that the MREs were located in the coding region of 8 VcSBPs (VcSBP2, VcSBP6a, VcSBP6b, VcSBP8b, VcSBP9a, VcSBP9b, VcSBP13a, and VcSBP13b) and 3′-UTR region of two VcSBPs (VcSBP3 and VcSBP5). Noticeably, two MREs for miR156/157 were observed in the 3′-UTR region of VcSBP3. It is worth mentioning that, previously, VcSBP2/SPL12 was experimentally verified to be targeted by miR156/157 (Li et al., 2020). Here, further mining of our degradome data revealed four additional miR156/157-guided cleavages of VcSBP transcripts, including the assembled sequences Comp14442, Comp42467, Comp25517, and Comp34004 (Figure 4B), which correspond to the cDNA sequences of VcSBP9b, VcSBP6b, VcSBP5, and VcSBP3, respectively. These data suggest that these VcSBPs might be targeted by miR156 in vivo.
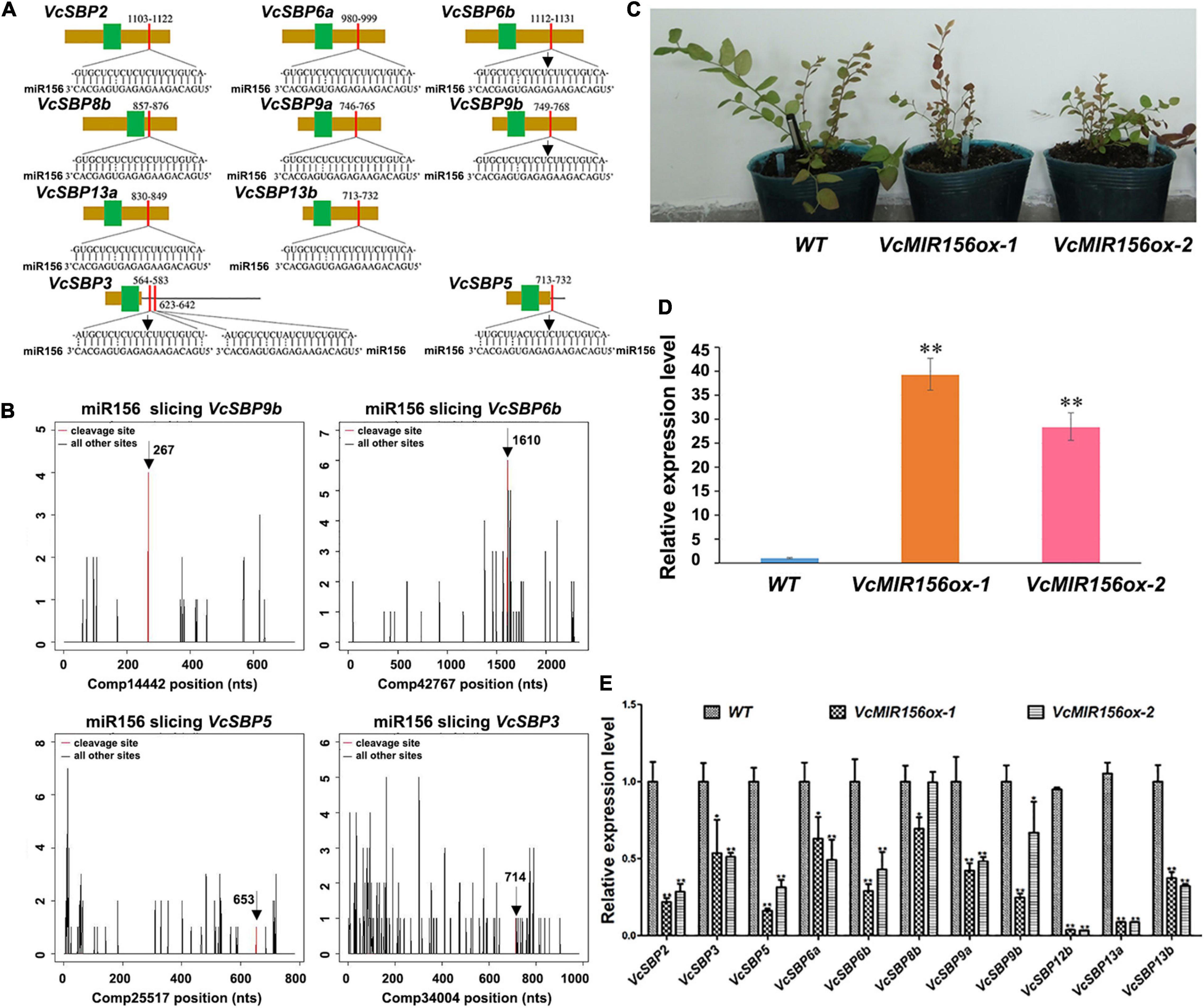
Figure 4. miR156-targeted SBP family members in blueberry. (A) The diagrams of VcSBP sequences targeted by miR156. The brown box, black line, green box, and red box represent CDS sequence, UTR regions, SBP domains, and miR156 responsive elements, respectively. (B) Target plots (t-plots) of the miR156-targeted VcSBP transcripts (VcSBP9b, VcSBP6b, VcSBP5, and VcSBP3). The vertical lines represent the peak of different sliced VcSBP transcripts, and the red line denotes the peak of miR156-mediated transcript slices. The exact cleavage sites are indicated by vertical arrows in the (A,B). (C) The photograph of transgenic blueberry lines with VcMIR156a overexpression and untransformed control (WT). (D) Expression analysis of VcMIR156a in transgenic blueberry lines. (E) The expression pattern of VcSBP genes in WT and transgenic blueberry lines overexpressing VcMIR156a. Total RNAs extracted from blueberry leaf was used for qRT-PCR analysis. Three biological replicates and three technical replicates for each biological replicate were performed. Error bars indicate standard errors. The data were normalized against the gene VcACTIN, and significant differences are denoted by asterisks: ∗P < 0.05, ∗∗P < 0.01.
To verify the SBP family members targeted by miR156/157s in vivo, genetic transformation was performed to obtain transgenic blueberry lines overexpressing VcMIR156a (Figure 4C). qPCR analysis indicated that the expression of VcMIR156a was indeed increased in the two transgenic blueberry lines (Figure 4D). Furthermore, the expressions of the above 10 SBPs and VcSBP12b were examined in the transgenic blueberry lines and untransformed control. As shown in Figure 4E, all the examined VcSBP genes were significantly repressed by the VcMIR156a overexpression, especially VcSBP2, VcSBP5, VcSBP6b, VcSBP9a, VcSBP12b, VcSBP13a, and VcSBP13b with more than two-fold decreases, suggesting that these eleven SBP genes can be regulated through miR156/157-mediated mRNA cleavage in vivo.
VcSBP Family Plays Diverse Roles in Arabidopsis and Affects Chlorophyll Accumulation
To investigate the functional roles of VcSBP genes, transgenic Arabidopsis lines were generated for the VcSBP genes. Phenotypic analysis indicated that the VcSBP family genes performs diverse functions in Arabidopsis, mainly involved in four aspects of biological or developmental processes: flowering, leaf development, trichome formation, and chlorophyll accumulation. Overexpression of seven VcSBPs (VcSBP7a/7b, VcSBP14a/14b, VcSBP3, VcSBP5, and VcSBP13a) led to early flowering (Figure 5A and Supplementary Figures 4A,B), whereas VcSBP8b repressed plant flowering and trichome formation in Arabidopsis (Figure 5A and Supplementary Figures 4C,D). It was also observed that curling leaf could be arisen from overexpression of each of the three VcSBP genes, VcSBP10, VcSBP13a, or VcSBP13b (Figure 5A and Supplementary Figure 4E), while the transgenic lines overexpressing VcSBP13a or VcSBP8b showed narrow leaf (Figures 5A,B). Additionally, serrated leaf was observed in the transgenic lines overexpressing VcSBP12b or VcSBP13a (Figure 5A and Supplementary Figure 4F). Clearly, the VcSBPs in the same phylogenetic clade cannot always generate similar morphological characters (Figure 5A).
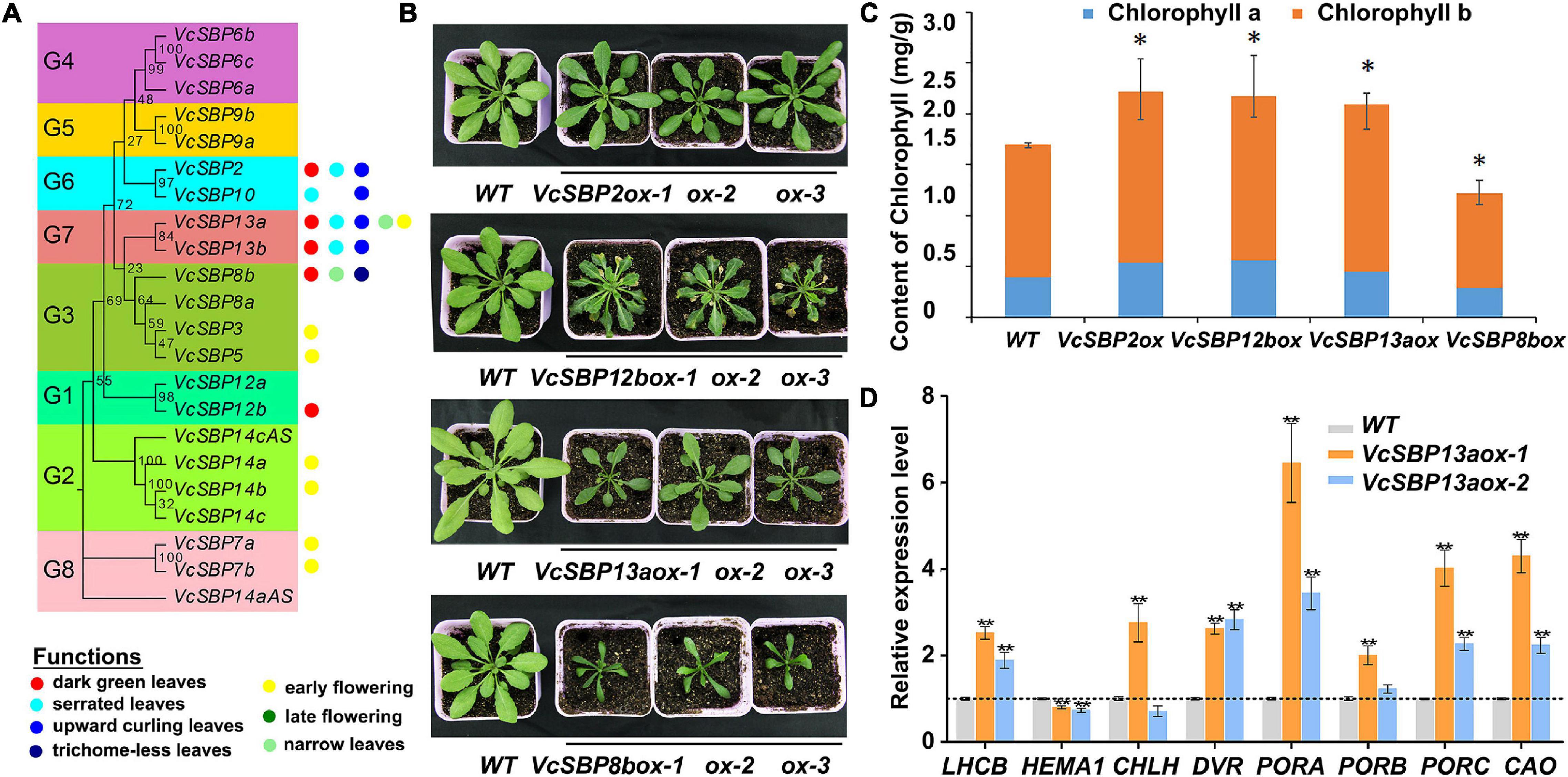
Figure 5. VcSBP family play diverse roles in Arabidopsis and affects chlorophyll accumulation. (A) Summery of functional roles of the VcSBP family genes in Arabidopsis. (B) Photographs of transgenic Arabidopsis lines overexpressing VcSBP genes (VcSBP2, VcSBP12b, VcSBP13a, and VcSBP8b). (C) Chlorophyll contents of wild type and transgenic lines overexpressing VcSBP2, VcSBP12b, VcSBP13a, or VcSBP8b. (D) Expression patterns of 8 chlorophyll-associated genes (AtHEMA1, AtDVR, AtPORA, AtPORB, AtPORC, AtCAO, AtCHLH, and AtLHCB) in wild type and transgenic lines overexpressing VcSBP13a. Total RNAs were exacted from 14-day-old transgenic seedlings and wild type. Values were normalized against the gene AtACTIN8. Error bars in (C,D) indicate standard errors of three biological and technical replicates, and significant differences are denoted by asterisks: *P < 0.05, **P < 0.01.
Previously, we reported that overexpression of VcSBP2/SPL12 enhanced chlorophyll accumulation in Arabidopsis (Li et al., 2020). Here, we noticed that transgenic plants overexpressing each of the 4 VcSBP genes (VcSBP2, VcSBP12b, VcSBP13a, and VcSBP8b) clearly showed dark green leaves (Figure 5B). Also, a little succulence was observed for the leaves of the VcSBP8b-overexpressing transgenic lines (Supplementary Figures 4C,D). Chlorophyll contents were then determined in the transgenic Arabidopsis lines. As shown in Figure 5C, total chlorophyll contents in the transgenic lines overexpressing VcSBP2, VcSBP12b, or VcSBP13a were 1.28, 1.31, and 1.24 times higher, respectively, than that in WT. Consistently, both chlorophyll a and b were increased by 1.15–1.43 and 1.24–1.29-folds as compared to WT, respectively (Figure 5C). However, chlorophyll content was decreased in the VcSBP8b-overexpressing transgenic lines, which might be due to the succulent leaves. Furthermore, the expressions of eight chlorophyll-associated genes were examined in the VcSBP13a-overexpressing transgenic lines, including 7 chlorophyll biosynthetic genes (AtHEMA1, AtDVR, AtPORA, AtPORB, AtPORC, AtCAO, and AtCHLH) and one chlorophyll-binding protein gene (AtLHCB). Consequently, all the genes were significantly upregulated by overexpression of VcSBP13a except AtHEMA1 that showed a slight decrease (Figure 5D). Especially, AtPORA was remarkably increased by 10-folds (Figure 5D). These results indicated that VcSBPs affect chlorophyll accumulation via regulating the expression of chlorophyll-associated genes in Arabidopsis. Since SBP family is transcriptionally regulated by miR156, the expressions of the above eight chlorophyll biosynthetic genes were examined in the VcMIR156a-overexpressing transgenic Arabidopsis. Consistently, AtDVR and AtPORC were significantly repressed by VcMIR156a overexpression (Supplementary Figure 5).
SBP Family Might Affect the Expression of VcLHCB1 via Targeting Its Promoter in Blueberry
Increasing evidence indicated that SBPs are able to bind to the consensus sequence TNCGTACAA with GTAC as its essential core (Kropat et al., 2005). To provide some clues for understanding the targets of SBPs in blueberry, the potential genome-wide binding sites were searched using the consensus sequence against the V. corymbosum cv. Draper v1.0 genome. Consequently, 2568 genes were found to harbor the potential binding motif of SBP proteins in their promoter regions (Supplementary Table 4), suggesting that they are possible targets of SBP proteins in blueberry. The potential targets were classified into five groups based on their functional roles, including transcription, DNA-or-RNA-related; metabolism defense or protein binding; cellular process; synthesis, catalysis or modification; biological process unknown (Figure 6A). Further examination indicated that the distribution of the potential binding sites in the promoter regions varied among the five groups. The density of the binding sites over the target genes in the groups G5, G3, and G2 peaked around ∼900, 1100, and 1200 bp upstream of their TSSs, while no obvious peak was found for the target genes in the groups G4 and G1 (Figure 6B).
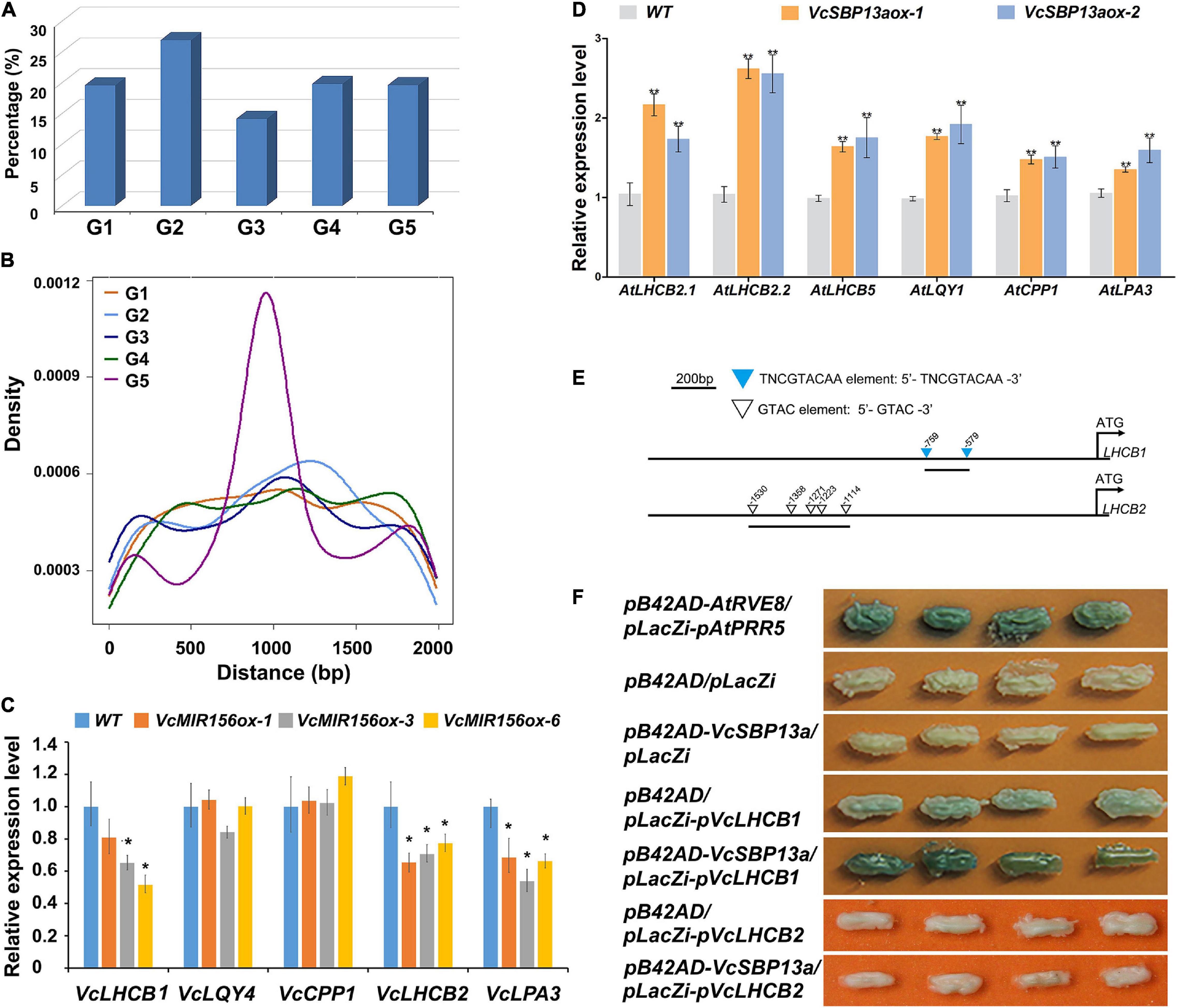
Figure 6. Computational and experimental analysis of the target client(s) of SBP(s) in blueberry. (A) The categories of putative target clients. Five categories were classified in terms of their functional roles, including transcription, DNA-or-RNA-related (G1); metabolism defense or protein binding (G2); cellular process (G3); synthesis, catalysis, or modification (G4); and biological process unknown (G5). (B) The density of SBP binding sites in the promoter regions of each group of target clients. (C) The expression patterns of chlorophyll-associated genes (VcLHCB1, VcLQY4, VcCPP1, VcLHCB2, and VcLPA3) in WT and transgenic blueberry lines overexpressing VcMIR156a. (D) The expression patterns of chlorophyll-associated genes (AtLHCB2.1, AtLHCB2.2, AtLHCB5, AtLQY4, AtCPP1, and AtLPA3) in WT and transgenic Arabidopsis lines overexpressing VcSBP13a. Values were normalized against the gene VcACTIN (C) or AtACTIN8 (D). Error bars indicate standard errors of three biological and technical replicates, and significant differences are denoted by asterisks: *P < 0.05, **P < 0.01. (E) The schematic diagrams of the promoters of VcLHCB1 and VcLHCB2. The downward triangles indicate the elements of TNCGTACAA and GTAC, and the number shows the start site of each element. (F) Interaction assay between VcSBP13a and the promoters of VcLHCB1 and VcLHCB2 using the yeast one-hybrid assay. pB42AD/pLacZi-pVcLHCB1, pB42AD/pLacZi-pVcLHCB2, pB42AD/pLacZi, and pB42AD-VcSBP13a/pLacZi were set as negative controls; and pB42AD-AtRVE8/placZi-AtPRR5 as positive control.
Consistent with the above results that VcSBPs affect chlorophyll accumulation in Arabidopsis, nine chlorophyll-associated genes were found to harbor the potential binding site of SBP proteins, including three LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEINs (VcLHCBs), two LOW QUANTUM YIELD OF PHOTOSYSTEM II1 (VcLQY1), 3 CHAPERONE-LIKE PROTEIN OF POR1-like (VcCPP1), and one LOW PSII ACCUMULATION 3 (VcLPA3). Subsequently, five genes were chosen as representatives to examine their expression patterns in transgenic blueberry plants overexpressing VcMIR156a where 11 VcSBPs were transcriptionally repressed (Figure 4E). As shown in Figure 6C, the expressions of VcLPA3 and two VcLHCBs were significantly downregulated by MIR156a overexpression in blueberry, whereas no significant change was observed for VcLQY1 and VcCPP1. Furthermore, the expression patterns of their corresponding Arabidopsis homologs (such as AtLHCB2.1, AtLHCB2.2, AtLHCB5, AtLQY1, AtCPP1, and AtLPA3) were investigated in transgenic Arabidopsis with VcSBP13a overexpression. Consequently, all these homologous genes were significantly promoted by VcSBP13a overexpression (Figure 6D).
To examine if the SBP proteins bind to these genes, VcSBP13a and two VcLHCBs (VcLHCB1 and VcLHCB2) were separately chosen as representatives of baits and preys to perform Y1H analysis. Sequence analysis indicated that the promoter region of VcLHCB1 contained two typical binding sites (TNCGTACAA element), whereas only GTAC elements were observed in the promoter region of VcLHCB2 (Figure 6E). As shown in Figure 6F, like the positive control (pB42AD-AtRVE8/placZi-AtPRR5), strong blue colonies were observed when VcSBP13a acts as bait and the fragment of VcLHCB1 promoter as prey. In contrast, very light blue appeared in the colonies containing pB42AD as bait and pLacZi-pVcLHCB1 as prey, and no blue color was shown for the other two negative controls (pB42AD/pLacZi and pB42AD-VcSBP13a/pLacZi). These results indicated that physical interaction occurred between VcSBP13a and the VcLHCB1 promoter. However, no blue color was observed in the colonies when pB42AD-VcSBP13a acted as bait and pLacZi-pVcLHCB2 as prey.
Discussion
SBP genes belong to a small family of plant-specific transcription factors. In the present study, 20 SBP genes were identified in blueberry, and the number of VcSBP family members is similar to the ones in Petunia (21), Tartary buckwheat (24), grape (17), and pear (19) (Hou et al., 2013; Li et al., 2013; Zhou et al., 2018; Liu et al., 2019), supporting the notion that the number of SBP genes in different plant species is relatively stable during evolution (Liu et al., 2019). Gene family generally arises from gene duplication during evolution, therefore leading to the acquisition of neofunctionalizations and subfunctionalizations as well as the emergence of backup or redundant genes (Preston and Hileman, 2013; Wang and Wang, 2015). Among the 20 identified VcSBPs, only two gene pairs (VcSPB7a and VcSBP7b, VcSBP14b, and VcSBP14c) might have been derived from segmental duplication, and one pair (VcSBP8a and VcSBP8b) from tandem amplification (Figure 1C). Noticeably, the VcSBP genes in the same group in the phylogenetic tree showed relatively low identity (8–48%) except the two segmental duplication pairs (VcSPB7a and VcSBP7b, VcSBP14b, and VcSBP14c, Figures 1C, 2A), suggesting that most VcSBPs might be single-copy genes with functional specificity. However, it was estimated that at least three rounds of whole-genome duplication occurred during the evolution of blueberry species (Wang et al., 2020), which are supposed to facilitate the generation of multiple copy genes. It can be explained by at least two reasons: (1) it is still possible that the number of VcSBP genes might have been underestimated since the identification of SBP genes was conducted on the basis of available transcriptome data; (2) VcSBPs might belong to the duplication-resistant genes, which generally return to single-copy status through the duplication-resistant system or genetic drift after suffering duplication events (Wang et al., 2020).
SBP family performs diverse functions during plant growth and development. Here, we presented four aspects of evidence to show the functional diversity of the SBP family in blueberry. Firstly, it has been proposed that the diversification of gene structures and conservation of motifs may be tightly associated with the functional evolution of SBP genes (Salinas et al., 2012; Hou et al., 2013; Li et al., 2013; Li and Lu, 2014; Shalom et al., 2015; Liu et al., 2019). In the present study, it was revealed that the majority of VcSBP family members belonging to the same phylogenetic group showed similar motif compositions and gene structures, while the diversity in motif compositions and gene structures was observed between the SBP family members in different phylogenetic groups (Figure 2). These observations support a scenario that the SBP family underwent functional conservation and diversification during evolution (Preston and Hileman, 2013; Wang and Wang, 2015; Zhang et al., 2015). Secondly, the spatio-temporal expression is generally thought as key contributors to functional specificity for a gene family (Paul et al., 2012; Hasan et al., 2017). VcSBP genes displayed tissue-specific and fruit development stage-specific expression patterns (Figure 3), implying that the VcSBP family might exert diverse functions in blueberry. Thirdly, the SBP family can be regulated via miR156-guided transcript cleavage or translational repression, and a subset of SBPs have been proved to be targeted by miR156 through recognizing MREs on their transcripts, for example, 11 out of the 17 SBPs in Arabidopsis, 15 out of the 27 SBPs in apple and 7 out of the 19 SBPs in pear (Li et al., 2013; Zhang et al., 2015; Qian et al., 2017). In the present study, several members of the VcSBP family were computationally and experimentally demonstrated to be targets of miR156 (Figure 4), implying that a subset of VcSBPs are able to form a regulatory hub with miR156, therefore exerting vital functions during blueberry growth and development. Lastly, overexpression of VcSBPs in Arabidopsis gave rise to multiple morphological phenotypes (Figure 5 and Supplementary Figure 4). Thus, our results provided an overall framework for understanding the functional diversity of VcSBP genes, which will contribute to the genetic improvement of the agronomic traits of blueberry.
The SBP family acts as pivotal regulators of diverse biological and physiological processes in plants. In the present study, functional analysis in Arabidopsis indicated that VcSBP genes might be involved in multiple developmental processes such as leaf shape regulation (serrated leaf formation, VcSBP12b/13a; narrow leaf, VcSBP8b/13a), trichome formation (VcSBP8b), and flowering time control (VcSBP7a/7b/14a/14b/3/5/13a, Figure 5 and Supplementary Figure 4). These observations are consistent with previous reports in Arabidopsis. For example, AtSPL10 overexpression causes narrow leaf in Arabidopsis (Gao et al., 2018); AtSPL3/4/5 exert important functions in regulating Arabidopsis flowering and developmental transition (Jung et al., 2016; Xu et al., 2016); AtSPL3/4/5/8/9/10/13 affect trichome formation (Yu et al., 2010); and loss-of-function mutation of AtSPL14 increases the number of leaf hydathodes and enhances leaf margin serration (Stone et al., 2005). Thus, our results support the notion that the functionality of the SBP family proteins is highly conserved among distinct plant species (Preston and Hileman, 2013). However, not all the VcSBPs display the same functional roles as their counterparts in other plant species. For instance, overexpression of VcSBP8b leads to a very narrow leaf in Arabidopsis (Figure 5B), whereas its Arabidopsis counterpart AtSPL8 fails to generate similar leaf morphology, and it is AtSPL10 instead that was reported to modulate leaf morphology (Gao et al., 2018). Previous studies also indicated that mutation of LG1, the closest homolog of Arabidopsis AtSPL8, in maize, rice, and barley gave rise to the lack of ligules and auricles (Lee et al., 2007; Wang and Wang, 2015), whereas in Arabidopsis mutation of AtSPL8 fails to cause a similar structure of ligules. Thus, it seems that it is not always possible to foretell the functional roles of individual SBP genes based on homology, although the SBP family as a whole shows functional conservation across diverse plant species. More interestingly, overexpression of three VcSBP genes (VcSBP10/13a/13b) in Arabidopsis causes the formation of curling leaves (Supplementary Figure 4E). Previous report indicated that mutation in rSPL13 led to an up-curled leaf phenotype in alfalfa (Gao et al., 2018). Nevertheless, no evidence shows the formation of curling leaves by being members of the SBP family in Arabidopsis. Thus, it appears that SBP family might show species-dependent functions or novel function in some specific plant species.
Several studies indicated that the SBP family plays important roles during fruit ripening. For example, VmTDR4 (a SQUAMOSA-class MADS-box gene) is positively involved in the regulation of anthocyanin accumulation during bilberry fruit ripening (Jaakola et al., 2010), while MaSPL16 in banana regulates carotenoid biosynthesis through promoting the expression of MaLCYBs (Zhu et al., 2020). Likewise, SlSPL-CNR, an SBP transcription factor in tomato, is mainly expressed in ripening fruits and serves as a positive player in the regulation of fruit ripening and cell death (Lai et al., 2020). In the present study, five VcSBP genes were found to be expressed at relatively low levels at three early stages of fruit development and significantly increased during fruit ripening (Figure 3I), suggesting that they might act as regulatory hubs to control fruit ripening in blueberry. In contrast, 10 VcSBP genes were highly expressed at three early stages of fruit development and dramatically decreased to a low level when fruit initiates ripening (Figure 3G,H), which is consistent with previous reports that the expressions of VvSPL6/10/13 were gradually decreased as grape berry develops and ripens (Cui et al., 2018), while the FvSPLs were transcriptionally decreased during strawberry fruit ripening (Xiong et al., 2018). These results suggest that the 10 VcSBPs might be required for fruit development and suppressed during fruit ripening in blueberry.
Generally, the development and ripening of fleshy fruits are accompanied by a wide range of changes at cellular, molecular and metabolic levels, including fruit enlargement, degreening, accumulation of pigments, softening, etc. Previously, we revealed that VcSBP2/SPL12 affects the accumulation of chlorophylls in Arabidopsis (Li et al., 2020). In the present study, the contents of chlorophyll a and b were found to be increased by the overexpression of at least three SBP genes (VcSBP2, VcSBP12a, and VcSBP13a) in Arabidopsis (Figure 5). Moreover, the chlorophyll biosynthetic genes in Arabidopsis were indeed elevated by VcSBP13a overexpression (Figure 5D). These observations indicated that a subset of VcSBPs might be involved in the regulation of chlorophyll accumulation. Previous studies have revealed that SBP family proteins can directly interact with their clients (for example, AtFUL, AtSOC1, AtDFR, AtAP1, MdWRKY100, MaLCYB1.1, and MaLCYB1.2, MADS5, and MADS32), thereby regulating diverse biological processes in plants such as flowering, inflorescence formation, biosynthesis of secondary metabolites, root regeneration, and response to stress (Yamaguchi et al., 2009; Gou et al., 2019; Ma et al., 2020; Zhu et al., 2020). However, the targets of SBP proteins associated with chlorophyll accumulation have remained to be found. Our Y1H assay showed that VcSBP13a could physically bind to the promoter region of an LHCB gene in blueberry (Figure 6). Thus, we proposed that VcSBPs are able to positively regulate the expressions of chlorophyll-associated genes (at least VcLHCB1) directly through binding to their promoter regions to affect chlorophyll accumulation in blueberry.
In conclusion, the SBP family was systematically identified and functionally characterized in blueberry, and they show conservation and divergence in characteristics and functional roles across plant species. Based on the targets and functional roles of VcSBPs as well as their expression patterns, we propose that a subset of VcSBPs might be involved in the regulation of chlorophyll accumulation directly through targeting to the chlorophyll-associated genes such as VcLHCB1. These findings provide the first comprehensive understandings of the features and functional diversity of the SBP family in blueberry, which will facilitate their utilization in the improvement of the agronomic traits of blueberry.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SB and XL designed the experiments. XX, SY, BS, HL, and PY performed the experiments. JW and SL performed the data analyzes. SB, XL, and YC wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [Grant number 31872075, 2019–2022].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.703994/full#supplementary-material
Supplementary Figure 1 | Phylogenetic analysis of SBP genes in blueberry and Arabidopsis. The CDS sequences of AtSBPs were downloaded from the TAIR website (www.arabidopsis.org). A neighbor-joining tree was generated with the MEGA7 software using the CDS sequences of the SBP genes in blueberry and Arabidopsis.
Supplementary Figure 2 | Phylogenetic relationship of VcSBP proteins with the SBPs in other plant species. A maximum likelihood tree was generated with the MEGA X software using the putative amino acid sequences of 101 SBP proteins, which were clustered into eight groups (G1-G6). The SBP proteins in the same species are represented with the same symbol: blue check, V. corymbosum; orange circle, Vitis vinifera; yellow square, Malus domestica; pink star, Solanum lycopersicum; gray triangle, Arabidopsis thaliana.
Supplementary Figure 3 | Alignment of miR156 complementary sequences within VcSBP genes. (A) Complementary sequences are within coding regions and the 3′UTR. Reverse complement sequences of the mature vco-miR156a, vco-miR156b-5p, vco-miR156c-5p, vco-miR156e-5p, vco-miR156g-5p, and vco-miR157a genes are shown below the alignment for comparison. (B) The sequence logo of miR156 responsive elements in VcSBP genes. The total height of each stack represents the conservation degree of each position, while the height of the letters within each stack indicates the relative frequency of the corresponding amino acid.
Supplementary Figure 4 | Phenotype of the transgenic Arabidopsis individually overexpressing the VcSBP family genes. Early (A, B) – and – late (C) flowering phenotypes of the transgenic Arabidopsis overexpressing the gene VcSBP7a, VcSBP7b, VcSBP14a, VcSBP14b, VcSBP3, VcSBP5, VcSBP13a, or VcSBP8b. (D) Less trichome phenotype of the transgenic Arabidopsis overexpressing the gene VcSBP7a. (E) Curled leaf phenotype of the transgenic Arabidopsis overexpressing the gene VcSBP10, VcSBP13a, or VcSBP13b. (F) Serrated leaf phenotype of the transgenic Arabidopsis overexpressing the gene VcSBP13a or VcSBP12b.
Supplementary Figure 5 | Expression patterns of 8 chlorophyll-associated genes (AtLHCB, AtHEMA1, AtCHLH, AtDVR, AtPORA, AtPORB, AtPORC, and AtCAO) in wild type and transgenic lines overexpressing VcMIR156a. Total RNAs were exacted from 7-day-old transgenic seedlings and wild type. Values were normalized against the gene AtACTIN8. Error bars indicate standard errors of three biological and technical replicates, and significant differences are denoted by asterisks: *P < 0.05, **P < 0.01.
Supplementary Table 1 | Primers used in the study.
Supplementary Table 2 | Characterization of the SBP family in blueberry.
Supplementary Table 3 | Similarity and identity between different VcSBP family members.
Supplementary Table 4 | The predicted targets of the VcSBP family proteins in blueberry.
Footnotes
- ^ https://www.arabidopsis.org/
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ www.vaccinium.org
- ^ https://prosite.expasy.org/
- ^ https://web.expasy.org/compute_pi/
- ^ http://weblogo.threeplusone.com/
- ^ https://gsds.cbi.pku.edu.cn/
- ^ http://meme-suite.org/tools/meme
- ^ http://plantgrn.noble.org/psRNATarget/
- ^ http://eggnog-mapper.embl.de/
- ^ www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ https://www.vaccinium.org/analysis/49
References
Barrera-Rojas, C. H., Rocha, G. H. B., Polverari, L., Brito, D. A. P., Batista, D. S., Notini, M. M., et al. (2020). miR156-targeted SPL10 controls Arabidopsis root meristem activity and root-derived de novo shoot regeneration via cytokinin responses. J. Exp. Bot. 71, 934–950. doi: 10.1093/jxb/erz475
Bhogale, S., Mahajan, A. S., Natarajan, B., Rajabhoj, M., Thulasiram, H. V., and Banerjee, A. K. (2014). MicroRNA156: a potential graft-transmissible MicroRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp andigena. Plant Physiol. 164, 1011–1027. doi: 10.1104/pp.113.230714
Birkenbihl, R. P., Jach, G., Saedler, H., and Huijser, P. (2005). Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352, 585–596. doi: 10.1016/j.jmb.2005.07.013
Colle, M., Leisner, C. P., Wai, C. M., Ou, S., Bird, K. A., Wang, J., et al. (2019). Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. Gigascience 8:giz012. doi: 10.1093/gigascience/giz012
Cui, M., Wang, C., Zhang, W., Pervaiz, T., Haider, M. S., Tang, W., et al. (2018). Characterization of Vv-miR156: Vv-SPL pairs involved in the modulation of grape berry development and ripening. Mol. Genet. Genomics 293, 1333–1354. doi: 10.1007/s00438-018-1462-1
Gao, R., Gruber, M. Y., Amyot, L., and Hannoufa, A. (2018). SPL13 regulates shoot branching and flowering time in Medicago sativa. Plant Mol. Biol. 96, 119–133. doi: 10.1007/s11103-017-0683-8
Gou, J., Tang, C., Chen, N., Wang, H., Debnath, S., Sun, L., et al. (2019). SPL7 and SPL8 represent a novel flowering regulation mechanism in switchgrass. New Phytol. 222, 1610–1623. doi: 10.1111/nph.15712
Gupta, V., Estrada, A. D., Blakley, I., Reid, R., Patel, K., Meyer, M. D., et al. (2015). RNA-Seq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. Gigascience 4:5. doi: 10.1186/s13742-015-0046-9
Hasan, M. A., Ahmad, S., and Molla, M. K. (2017). Protein subcellular localization prediction using multiple kernel learning based support vector machine. Mol. Biosyst. 13, 785–795. doi: 10.1039/c6mb00860g
Hou, H., Li, J., Gao, M., Singer, S. D., Wang, H., Mao, L., et al. (2013). Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS One 8:e59358. doi: 10.1371/journal.pone.0059358
Hou, Y., Zhai, L., Li, X., Xue, Y., Wang, J., Yang, P., et al. (2017). Comparative analysis of fruit ripening-related miRNAs and their targets in blueberry using small RNA and degradome sequencing. Int. J. Mol. Sci. 18:2767. doi: 10.3390/ijms18122767
Hou, Y. M., Li, H. X., Zhai, L. L., Xie, X., Li, X. Y., and Bian, S. M. (2020). Identification and functional characterization of the Aux/IAA gene VcIAA27 in blueberry. Plant Signal. Behav. 15:1700327. doi: 10.1080/15592324.2019.1700327
Jaakola, L., Poole, M., Jones, M. O., Kamarainen-Karppinen, T., Koskimaki, J. J., Hohtola, A., et al. (2010). A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 153, 1619–1629. doi: 10.1104/pp.110.158279
Jung, J. H., Lee, H. J., Ryu, J. Y., and Park, C. M. (2016). SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 9, 1647–1659. doi: 10.1016/j.molp.2016.10.014
Klein, J., Saedler, H., and Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250, 7–16. doi: 10.1007/BF02191820
Kropat, J., Tottey, S., Birkenbihl, R. P., Depege, N., Huijser, P., and Merchant, S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. U. S. A. 102, 18730–18735. doi: 10.1073/pnas.0507693102
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lai, T., Wang, X., Ye, B., Jin, M., Chen, W., Wang, Y., et al. (2020). Molecular and functional characterization of the SBP-box transcription factor SPL-CNR in tomato fruit ripening and cell death. J. Exp. Bot. 71, 2995–3011. doi: 10.1093/jxb/eraa067
Lee, J., Park, J. J., Kim, S. L., Yim, J., and An, G. (2007). Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 65, 487–499. doi: 10.1007/s11103-007-9196-1
Lei, K. J., Lin, Y. M., Ren, J., Bai, L., Miao, Y. C., An, G. Y., et al. (2016). Modulation of the phosphate-deficient responses by MicroRNA156 and its targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 in Arabidopsis. Plant Cell Physiol. 57, 192–203. doi: 10.1093/pcp/pcv197
Leister, D. (2004). Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20, 116–122. doi: 10.1016/j.tig.2004.01.007
Li, C., and Lu, S. (2014). Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol. 14:131. doi: 10.1186/1471-2229-14-131
Li, J., Hou, H., Li, X., Xiang, J., Yin, X., Gao, H., et al. (2013). Genome-wide identification and analysis of the SBP-box family genes in apple (Malus x domestica Borkh.). Plant Physiol. Biochem. 70, 100–114. doi: 10.1016/j.plaphy.2013.05.021
Li, L., Zhang, H., Liu, Z., Cui, X., Zhang, T., Li, Y., et al. (2016). Comparative transcriptome sequencing and de novo analysis of Vaccinium corymbosum during fruit and color development. BMC Plant Biol. 16:223. doi: 10.1186/s12870-016-0866-5
Li, X., Hou, Y., Xie, X., Li, H., Li, X., Zhu, Y., et al. (2020). A blueberry MIR156a-SPL12 module coordinates the accumulation of chlorophylls and anthocyanins during fruit ripening. J. Exp. Bot. 71, 5976–5989. doi: 10.1093/jxb/eraa327
Liu, M., Sun, W., Ma, Z., Huang, L., Wu, Q., Tang, Z., et al. (2019). Genome-wide identification of the SPL gene family in Tartary Buckwheat (Fagopyrum tataricum) and expression analysis during fruit development stages. BMC Plant Biol. 19:299. doi: 10.1186/s12870-019-1916-6
Luo, L., Li, W., Miura, K., Ashikari, M., and Kyozuka, J. (2012). Control of tiller growth of rice by OsSPL14 and Strigolactones, which work in two independent pathways. Plant Cell Physiol. 53, 1793–1801. doi: 10.1093/pcp/pcs122
Ma, Y., Xue, H., Zhang, F., Jiang, Q., Yang, S., Yue, P., et al. (2020). The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 19, 311–323. doi: 10.1111/pbi.13464
Manning, K., Tor, M., Poole, M., Hong, Y., Thompson, A. J., King, G. J., et al. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38, 948–952. doi: 10.1038/ng1841
Nguyen, S. T., Greaves, T., and Mccurdy, D. W. (2017). Heteroblastic development of transfer cells is controlled by the microRNA miR156/SPL module. Plant Physiol. 173, 1676–1691. doi: 10.1104/pp.16.01741
Paul, A. L., Denison, F. C., Schultz, E. R., Zupanska, A. K., and Ferl, R. J. (2012). 14-3-3 phosphoprotein interaction networks – does isoform diversity present functional interaction specification? Front. Plant Sci. 3:190. doi: 10.3389/fpls.2012.00190
Plunkett, B. J., Espley, R. V., Dare, A. P., Warren, B. A. W., Grierson, E. R. P., Cordiner, S., et al. (2018). MYBA from blueberry (Vaccinium Section Cyanococcus) is a subgroup 6 type R2R3MYB transcription factor that activates anthocyanin production. Front. Plant Sci. 9:1300. doi: 10.3389/fpls.2018.01300
Preston, J. C., and Hileman, L. C. (2013). Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 4:80. doi: 10.3389/fpls.2013.00080
Preston, J. C., Jorgensen, S. A., Orozco, R., and Hileman, L. C. (2016). Paralogous SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes differentially regulate leaf initiation and reproductive phase change in petunia. Planta 243, 429–440. doi: 10.1007/s00425-015-2413-2
Qi, X., Ogden, E. L., Ehlenfeldt, M. K., and Rowland, L. J. (2019). Dataset of de novo assembly and functional annotation of the transcriptome of blueberry (Vaccinium spp.). Data Brief. 25:104390. doi: 10.1016/j.dib.2019.104390
Qian, M., Ni, J., Niu, Q., Bai, S., Bao, L., Li, J., et al. (2017). Response of miR156-SPL module during the red peel coloration of bagging-treated chinese sand pear (Pyrus pyrifolia Nakai). Front. Physiol. 8:550. doi: 10.3389/fphys.2017.00550
Routray, W., and Orsat, V. (2011). Blueberries and their anthocyanins: factors affecting biosynthesis and properties. Compr. Rev. Food Sci. F. 10, 303–320. doi: 10.1111/j.1541-4337.2011.00164.x
Rowland, L. J., Alkharouf, N., Darwish, O., Ogden, E. L., Polashock, J. J., Bassil, N. V., et al. (2012). Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol. 12:46. doi: 10.1186/1471-2229-12-46
Salinas, M., Xing, S., Hohmann, S., Berndtgen, R., and Huijser, P. (2012). Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 235, 1171–1184. doi: 10.1007/s00425-011-1565-y
Schwarz, S., Grande, A. V., Bujdoso, N., Saedler, H., and Huijser, P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67, 183–195. doi: 10.1007/s11103-008-9310-z
Shalom, L., Shlizerman, L., Zur, N., Doron-Faigenboim, A., Blumwald, E., and Sadka, A. (2015). Molecular characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family from Citrus and the effect of fruit load on their expression. Front. Plant Sci. 6:389. doi: 10.3389/fpls.2015.00389
Shikata, M., Koyama, T., Mitsuda, N., and Ohme-Takagi, M. (2009). Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 50, 2133–2145. doi: 10.1093/pcp/pcp148
Silva, G. F. F., Silva, E. M., M da Azevedo, S., Guivin, M. A., Ramiro, D. A., et al. (2014). microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 78, 604–618. doi: 10.1111/tpj.12493
Song, G. Q., and Gao, X. (2017). Transcriptomic changes reveal gene networks responding to the overexpression of a blueberry DWARF AND DELAYED FLOWERING 1 gene in transgenic blueberry plants. BMC Plant Biol. 17:106. doi: 10.1186/s12870-017-1053-z
Song, G. Q., and Sink, K. C. (2006). Blueberry (Vaccinium corymbosum L.). Methods Mol. Biol. 344, 263–272. doi: 10.1385/1-59745-131-2:263
Song, G. Q., Walworth, A., Zhao, D., Hildebrandt, B., and Leasia, M. (2013). Constitutive expression of the K-domain of a Vaccinium corymbosum SOC1-like (VcSOC1-K) MADS-box gene is sufficient to promote flowering in tobacco. Plant Cell Rep. 32, 1819–1826. doi: 10.1007/s00299-013-1495-1
Stone, J. M., Liang, X., Nekl, E. R., and Stiers, J. J. (2005). Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 41, 744–754. doi: 10.1111/j.1365-313X.2005.02334.x
Usami, T., Horiguchi, G., Yano, S., and Tsukaya, H. (2009). The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136, 955–964. doi: 10.1242/dev.028613
Walworth, A. E., Chai, B., and Song, G. Q. (2016). Transcript profile of flowering regulatory genes in VcFT-overexpressing blueberry plants. PLoS One 11:e0156993. doi: 10.1371/journal.pone.0156993
Wang, H., and Wang, H. Y. (2015). The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant 8, 677–688. doi: 10.1016/j.molp.2015.01.008
Wang, S., Wu, K., Qian, Q., Liu, Q., Li, Q., Pan, Y., et al. (2017). Non-canonical regulation of SPL transcription factors by a human OTUB1-like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res. 27, 1142–1156. doi: 10.1038/cr.2017.98
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, Y., Nie, F., Shahid, M. Q., and Baloch, F. S. (2020). Molecular footprints of selection effects and whole genome duplication (WGD) events in three blueberry species: detected by transcriptome dataset. BMC Plant Biol. 20:250. doi: 10.1186/s12870-020-02461-w
Xie, Y., Zhou, Q., Zhao, Y., Li, Q., Liu, Y., Ma, M., et al. (2020). FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate arabidopsis flowering. Mol. Plant 13, 483–498. doi: 10.1016/j.molp.2020.01.013
Xie, Z., Su, Z., Wang, W., Guan, L., Bai, Y., Zhu, X., et al. (2019). Characterization of VvSPL18 and its expression in response to exogenous hormones during grape berry development and ripening. Cytogenet. Genome Res. 159, 97–108. doi: 10.1159/000503912
Xing, S., Salinas, M., Hohmann, S., Berndtgen, R., and Huijser, P. (2010). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22, 3935–3950. doi: 10.1105/tpc.110.079343
Xiong, J. S., Zheng, D., Zhu, H. Y., Chen, J. Q., Na, R., and Cheng, Z. M. (2018). Genome-wide identification and expression analysis of the SPL gene family in woodland strawberry Fragaria vesca. Genome 61, 675–683. doi: 10.1139/gen-2018-0014
Xu, M., Hu, T., Zhao, J., Park, M. Y., Earley, K. W., Wu, G., et al. (2016). Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 12:e1006263. doi: 10.1371/journal.pgen.1006263
Yamaguchi, A., Wu, M. F., Yang, L., Wu, G., Poethig, R. S., and Wagner, D. (2009). The MicroRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17, 268–278. doi: 10.1016/j.devcel.2009.06.007
Ye, B. B., Shang, G. D., Pan, Y., Xu, Z. G., Zhou, C. M., Mao, Y. B., et al. (2020). AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 32, 226–241. doi: 10.1105/tpc.19.00378
Yu, N., Cai, W. J., Wang, S., Shan, C. M., Wang, L. J., and Chen, X. Y. (2010). Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22, 2322–2335. doi: 10.1105/tpc.109.072579
Yu, N., Niu, Q. W., Ng, K. H., and Chua, N. H. (2015). The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 83, 673–685. doi: 10.1111/tpj.12919
Zhang, S. D., Ling, L. Z., and Yi, T. S. (2015). Evolution and divergence of SBP-box genes in land plants. BMC Genomics 16:787. doi: 10.1186/s12864-015-1998-y
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhou, Q., Zhang, S., Chen, F., Liu, B., Wu, L., Li, F., et al. (2018). Genome-wide identification and characterization of the SBP-box gene family in Petunia. BMC Genomics 19:193. doi: 10.1186/s12864-018-4537-9
Zhu, L. S., Liang, S. M., Chen, L. L., Wu, C. J., Wei, W., Shan, W., et al. (2020). Banana MaSPL16 modulates carotenoid biosynthesis during fruit ripening through activating the transcription of lycopene beta-cyclase genes. J. Agr. Food Chem. 68, 1286–1296. doi: 10.1021/acs.jafc.9b07134
Zifkin, M., Jin, A., Ozga, J. A., Zaharia, L. I., Schernthaner, J. P., Gesell, A., et al. (2012). Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 158, 200–224. doi: 10.1104/pp.111.180950
Keywords: blueberry, SBP gene, miR156, chlorophyll accumulation, SBP targets
Citation: Xie X, Yue S, Shi B, Li H, Cui Y, Wang J, Yang P, Li S, Li X and Bian S (2021) Comprehensive Analysis of the SBP Family in Blueberry and Their Regulatory Mechanism Controlling Chlorophyll Accumulation. Front. Plant Sci. 12:703994. doi: 10.3389/fpls.2021.703994
Received: 01 May 2021; Accepted: 09 June 2021;
Published: 01 July 2021.
Edited by:
Gang Wu, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Jiaqiang Sun, Chinese Academy of Agricultural Sciences (CAAS), ChinaJiyuan Shen, South China Agricultural University, China
Copyright © 2021 Xie, Yue, Shi, Li, Cui, Wang, Yang, Li, Li and Bian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaomin Bian, shmbian@jlu.edu.cn; Xuyan Li, xuyanli@jlu.edu.cn
†These authors have contributed equally to this work
 Xin Xie1†
Xin Xie1† Shaokang Yue
Shaokang Yue Yuhai Cui
Yuhai Cui Xuyan Li
Xuyan Li Shaomin Bian
Shaomin Bian