- 1World Vegetable Center Korea Office, Wanju-gun, South Korea
- 2National Institute of Horticultural and Herbal Science (NIHHS), Rural Development Administration (RDA), Wanju-gun, South Korea
- 3Biological Resources and Post-harvest Division, Japan International Research Center for Agricultural Sciences (JIRCAS), Tsukuba, Japan
The effects of the climate change including an increase in the average global temperatures, and abnormal weather events such as frequent and severe heatwaves are emerging as a worldwide ecological concern due to their impacts on plant vegetation and crop productivity. In this review, the molecular processes of plants in response to heat stress—from the sensing of heat stress, the subsequent molecular cascades associated with the activation of heat shock factors and their primary targets (heat shock proteins), to the cellular responses—have been summarized with an emphasis on the classification and functions of heat shock proteins. Vegetables contain many essential vitamins, minerals, antioxidants, and fibers that provide many critical health benefits to humans. The adverse effects of heat stress on vegetable growth can be alleviated by developing vegetable crops with enhanced thermotolerance with the aid of various genetic tools. To achieve this goal, a solid understanding of the molecular and/or cellular mechanisms underlying various responses of vegetables to high temperature is imperative. Therefore, efforts to identify heat stress-responsive genes including those that code for heat shock factors and heat shock proteins, their functional roles in vegetable crops, and also their application to developing vegetables tolerant to heat stress are discussed.
Introduction
Vegetable crops mainly comprise sessile organisms. They routinely experience detrimental conditions including biotic and abiotic stresses in natural fields. The current climate changes including frequent extreme temperatures, strong storms, heavy rainfall, and harsh droughts directly threaten normal vegetable development during the entire period of vegetative and reproductive growth (Driedonks et al., 2016; Hansen et al., 2016; Bhutia et al., 2018). Global warming is one of the main issues related to global climate change and is caused by increases of greenhouse gases such as CO2, CH4, N2O, and hydrofluorocarbons (HFCs) that have been produced by urbanization and industrialization (Bhutia et al., 2018; Zandalinas et al., 2021). According to climate models (Driedonks et al., 2016) and the report from the Intergovernmental Panel on Climate Change (IPCC1), the world mean temperature will rise by 0.5 to 4°C in the twenty-first century (Hansen et al., 2016; Zandalinas et al., 2021). The changes in weather/climatic events such as temperature and rainfall are found to reduce the yield of crops. Statistical evidence shows that the temperature affects rice production in Africa. It was also found that irrigated rice yields in West Africa in the dry season would decrease by ∼45% due to reduced photosynthesis at extremely high temperatures (van Oort and Zwart, 2018). This indicates that the elevated temperature brought by climate change will result in significant losses in crop yields and production (Ortiz et al., 2008; Hansen et al., 2016). Plants have evolved to acquire the ability to induce defense mechanisms against the adverse effects of high ambient temperature on their growth (Ahuja et al., 2010; Bourgine and Guihur, 2021; Tian et al., 2021). The tolerance of plants to high ambient temperatures with no prior heat experience is known as basal thermotolerance (BTT), whereas the ability to overcome extremely high temperatures (HT) with pre-exposure to mild HT (i.e., sub-lethal temperatures) is known as acquired thermotolerance (ATT) (Ahuja et al., 2010; Bourgine and Guihur, 2021; Tian et al., 2021). The defense mechanisms against elevated temperatures in plants are tightly associated with rapid changes in gene expression in both BTT and ATT (Morimoto, 1998; Feder and Hofmann, 1999). Indeed, high ambient temperatures trigger a drastic cellular remodeling at the physiological and molecular levels in plants to maintain homeostasis, thereby allowing them to survive under adverse HT (Wang et al., 2004; Ohama et al., 2017; Tian et al., 2021). Within these mechanisms, how plants recognize HT and relay HT-induced signaling downstream to modulate transcription is a central question that plant researchers have been pondering for a long time. It has recently been reported that Ca2+ plays important roles in the perception, response, and adaptation of plants to heat stress (HS) (Mittler et al., 2012; Ohama et al., 2017; Lee and Seo, 2021). The alteration of fluidity in the plasma membrane (PM) in plants in response to HS can open cyclic nucleotide-gated calcium channels (CNGCs) controlled by nucleotide cyclases, thereby having Ca2+ move into the cytosol from the PM (Saidi et al., 2009; Finka et al., 2012; Gao et al., 2012; Mittler et al., 2012; Ohama et al., 2017). The Ca2+ ions are associated with protein calmodulin 3 (CaM3) during HS and the complex of Ca2+-CaM3 interacts with calcium/calmodulin-binding protein kinase 3 (CBK3) and phosphatase PP7 to transduce cytosol heat-stress response (HSR) signals into the nucleus by modulating phosphorytion and dephosphorylation of HSFA1, respectively (Liu et al., 2007, 2008; Mittler et al., 2012; Ohama et al., 2017). Also, the increased levels of Inositol-1,4,5-triphosphate (IP3) via the phosophoinositide-signaling pathway result in the influx of Ca2+ into cytoplasm from intracellular Ca2+ pools such as the endoplasmic reticulum (ER) and vacuole during HS (Zhang et al., 2009; Zhou et al., 2009; Mittler et al., 2012; Ohama et al., 2017). In addition, reactive oxygen species (ROS) produced by respiratory burst oxidase homolog B (RbohB), RbohD, and NADPH oxidases are other candidate sensors of HS (Königshofer et al., 2008; Miller et al., 2009; Suzuki et al., 2012). It has also been demonstrated that the ROS causes accumulation of nitric oxide (NO), which induces the activation of CaM3. The signaling cascade of CaM3 ultimately influences the association of DNA and heat shock factors (HSFs) in nucleus via the potential involvement of HSFA1 activity (Xuan et al., 2010; Wang et al., 2014; Ohama et al., 2017). Although Ca2+ and ROS are evaluated as predicted signal transducers during HS, the full activation of HSR in response of plants to HT cannot be exclusively explained by them. This indicates that there may be other signal transducers and multiple layers of signaling pathways including salicylic acid (SA), ethylene (ET), abscisic acid (ABA), and jasmonic acid (JA) signals (Fujita et al., 2006; Frank et al., 2009; Zhou et al., 2009).
The effect of HS on plants leads to diverse changes in plant cells including the state of cellular membranes, structural alterations in DNA and RNA species, and conformational changes of proteins, cytoskeleton structures, and metabolites (Ruelland and Zachowski, 2010; Mittler et al., 2012). For instance, high ambient temperature influences fluidity of cellular membranes containing primarily phospholipids, proteins, and carbohydrates with the modification of membrane rigidification (Ruelland and Zachowski, 2010). Also, high ambient temperature affects the accessibility of nucleic acids, and it has been determined that elevated temperatures induce the dissociation of the histone protein H2A.Z from nucleosomes, which promotes the chromatin accessibility to RNA polymerase II for the expression of genes for heat-shock proteins (HSP) and HSF, thus showing highly inductive and responsive gene expression dynamics (Kumar and Wigge, 2010; Zhang H. et al., 2021). RNA secondary structures can be affected by HS. It has been revealed that HT leads to a change in translation rate, resulting from the altered association of mRNAs with ribosomes (Matsuura et al., 2010). Since structured nucleic acid molecules melt as the temperature increases, it can be easily conceived that temperature changes affect the conformation of regulatory RNAs (Narberhaus et al., 2006). Indeed, the RNA secondary structure of internal ribosome entry sites (IRESs), which are translation regulatory elements of mRNAs, can be modified by HS to initiate translation in a cap-independent manner (Dinkova et al., 2005; Ruelland and Zachowski, 2010). Conversely, RNA secondary structures that mask ribosomal binding sites at optimal temperature can be modified by HS, allowing the conversion of non-functional RNA to the competent RNA species with ribosomal recruitment (Narberhaus et al., 2006). Heat stress also influences the conformational changes of proteins that act as signaling effectors in response to HT in plants (Ruelland and Zachowski, 2010). In Arabidopsis, the oligomerization of thioredoxin and/or thioredoxin-like proteins is induced by HS, causing concomitant functional switching from a disulfide reductase and foldase chaperone to a holdase chaperone (Lee et al., 2009; Park et al., 2009). It has also been reported that the elevated temperatures from 27 to 42°C in tobacco, and from 20 to 42°C in Arabidopsis cause severe damage to cytoskeletones including microtubules (Smertenko et al., 1997; Müller et al., 2007). Furthermore, tobacco BY-2 cells exposed to heat (50°C, for 5 min) exhibited depolymerization of actin microfilaments (Malerba et al., 2010), and such a defective phenotype was also observed in Arabidopsis roots (Müller et al., 2007). Based on a report demonstrating that heat triggers the accumulation of HSP70 and the heat-activated MAP kinase (HAMK), both HSP70 and HAMK are likely to be necessary to disassemble the cytoskeleton under HS (Suri and Dhindsa, 2008). Altered enzymatic activities such as the catalytic rate, and the un- or mis-folding of enzymes can also be affected by HS, resulting in the imbalance of cellular metabolism in plants (McClung and Davis, 2010; Ruelland and Zachowski, 2010; Suzuki et al., 2012). The steady-state efflux and influx of metabolites such as sucrose, prolines, glycine-betaine, ascorbate, glutathione, and ROS play an important role in heat response and tolerance (Wang et al., 2004; Al-Whaibi, 2011; Mittler et al., 2012). Reactive oxygen species were initially regarded as a toxic by-product of aerobic metabolism. However, it is now apparent that ROS such as superoxide and hydrogen peroxide are able to function as signal molecules to induce the HSR (Miller et al., 2007, 2009; McClung and Davis, 2010; Ruelland and Zachowski, 2010; Suzuki et al., 2012). In particular, the levels of ROS are influenced by the participation of ROS-generating enzymes in plant response to HT (Königshofer et al., 2008). The acquisition of plant heat tolerance is closely associated with the synthesis of chaperone proteins and the levels of non-enzymatic antioxidants in response to HT (Kotak et al., 2007; Wahid et al., 2007; Frank et al., 2009; Rampino et al., 2009). Many reports have been published showing that HS influences protein conformation which can drive a protein to be denatured, aggregated, and un- or mis-folded, thereby being directly recognized by several HSPs (Yamada et al., 2007; Scharf et al., 2012; Ohama et al., 2017). Notably, plant HSPs play a crucial role in conferring plant tolerance to HS, and they help facilitate proper folding of target proteins by hindering denaturation and aggregation of the proteins as molecular chaperones (Ahuja et al., 2010; Jacob et al., 2017). For instance, under normal temperature conditions, HSFs regulate the HSR and form inactive multiprotein complexes with HSPs. On the other hand, under HS, HSFs dissociate from the complex and form phosphorylated trimers, thereby allowing their nuclear translocation and binding to heat-shock element (HSE) to induce transcription of target genes (Kotak et al., 2007; Ahuja et al., 2010; Scharf et al., 2012; Jacob et al., 2017; Ohama et al., 2017). Indeed, transcriptomic and proteomic analyses revealed that the abrupt changes in gene expression in response to high ambient temperatures enhance a selected regulatory response and synthesis of proteins linked to HSPs, HSFs, and HSR (Al-Whaibi, 2011; Jacob et al., 2017; Zandalinas et al., 2021). However, the players and their mode of action in heat perception, HS-signaling pathways and HSR still remain elusive in vegetable crops.
In this review, we give an overview of the HSPs with focus on vegetable crops. Heat shock proteins play an essential role in the regulation of HSFs and subsequently, the expression of heat responsive genes. Moreover, a better understanding of HSPs will enable us to widen our knowledge of interconnected mechanisms underlying the complex regulatory networks of HSFs and heat responsive genes at the physiological and molecular levels during the adaptation of plants against HS. We also discuss the potential applications of biotechnology for efficient development of crops with enhanced thermotolerance to cope with climate change.
Heat Shock Proteins Involved in Heat Stress
In nature, plants are often exposed to various kinds of abiotic stresses including low or high temperature, deficiency or excess of water, high salinity, heavy metals and ultraviolet radiation (Rucińiska-Sobkowiak, 2010; Bita and Gerats, 2013; Osakabe et al., 2013; He et al., 2018). Among these, HS has significant effects on plant growth, metabolism, and productivity (Rodríguez et al., 2015). HS causes protein misfolding and/or denaturation, leading to protein aggregation in plant cells by interactions between exposed hydrophobic amino acid residues of affected proteins (Nakajima and Suzuki, 2013). In response to HS, plants synthesize molecular chaperones including HSPs that recognize hydrophobic amino acid residues of non-native proteins and promote folding and refolding of denatured proteins (Figure 1). They are also responsible for assembling of multi-protein complexes, transporting, and sorting of proteins into correct compartments, controlling cell cycle and signal-transduction under various stress conditions. The different classes of HSPs play complementary and sometimes overlapping roles in protein stabilization under thermal stress. The HSPs are generally grouped into five major families based on their molecular weight: HSP100, 90, 70, 60 and the small HSPs (sHSPs) (Figure 2 and Table 1).
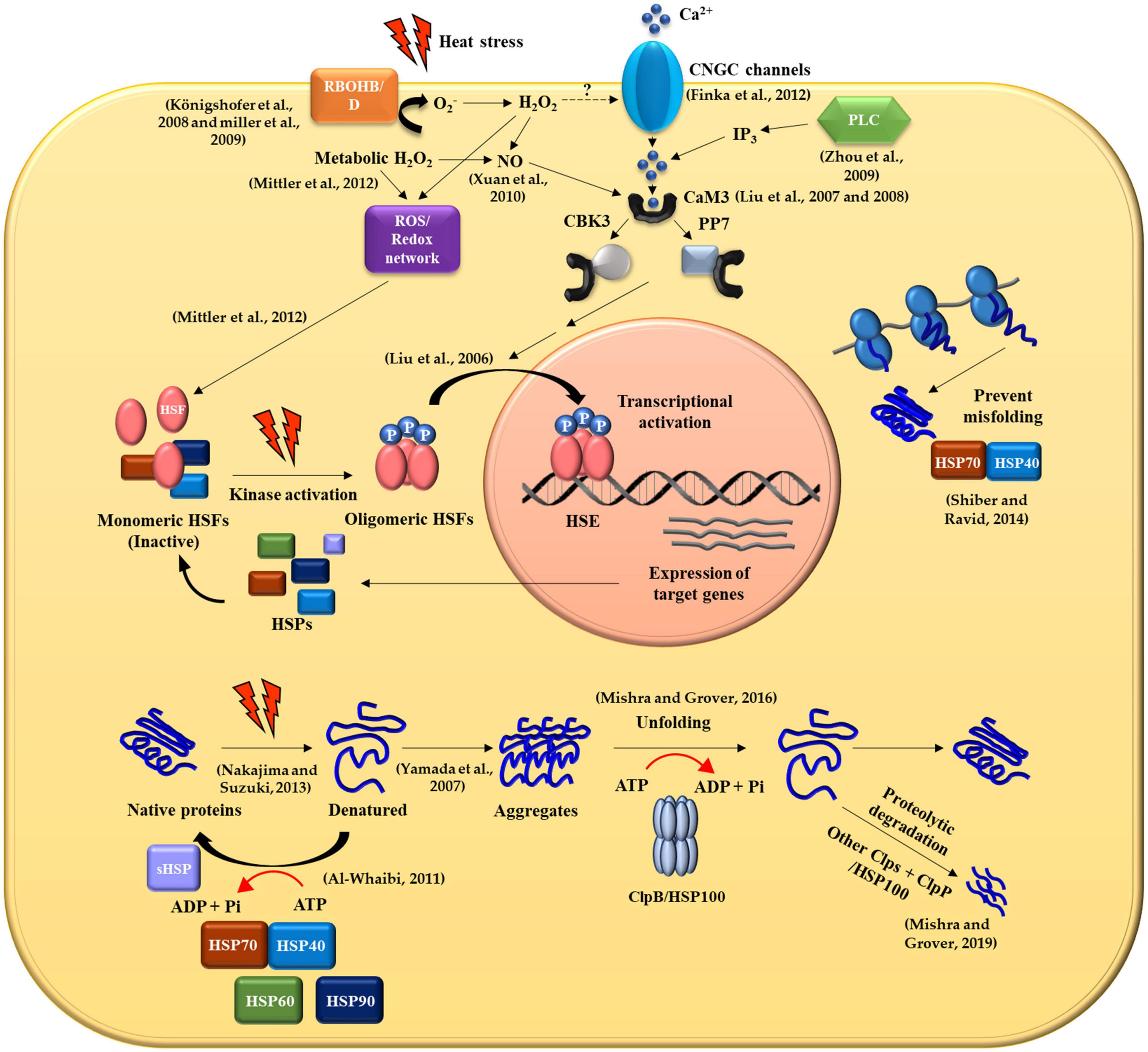
Figure 1. General molecular mechanism of heat shock protein production and transcriptional regulation in response to heat stress in plant cells.
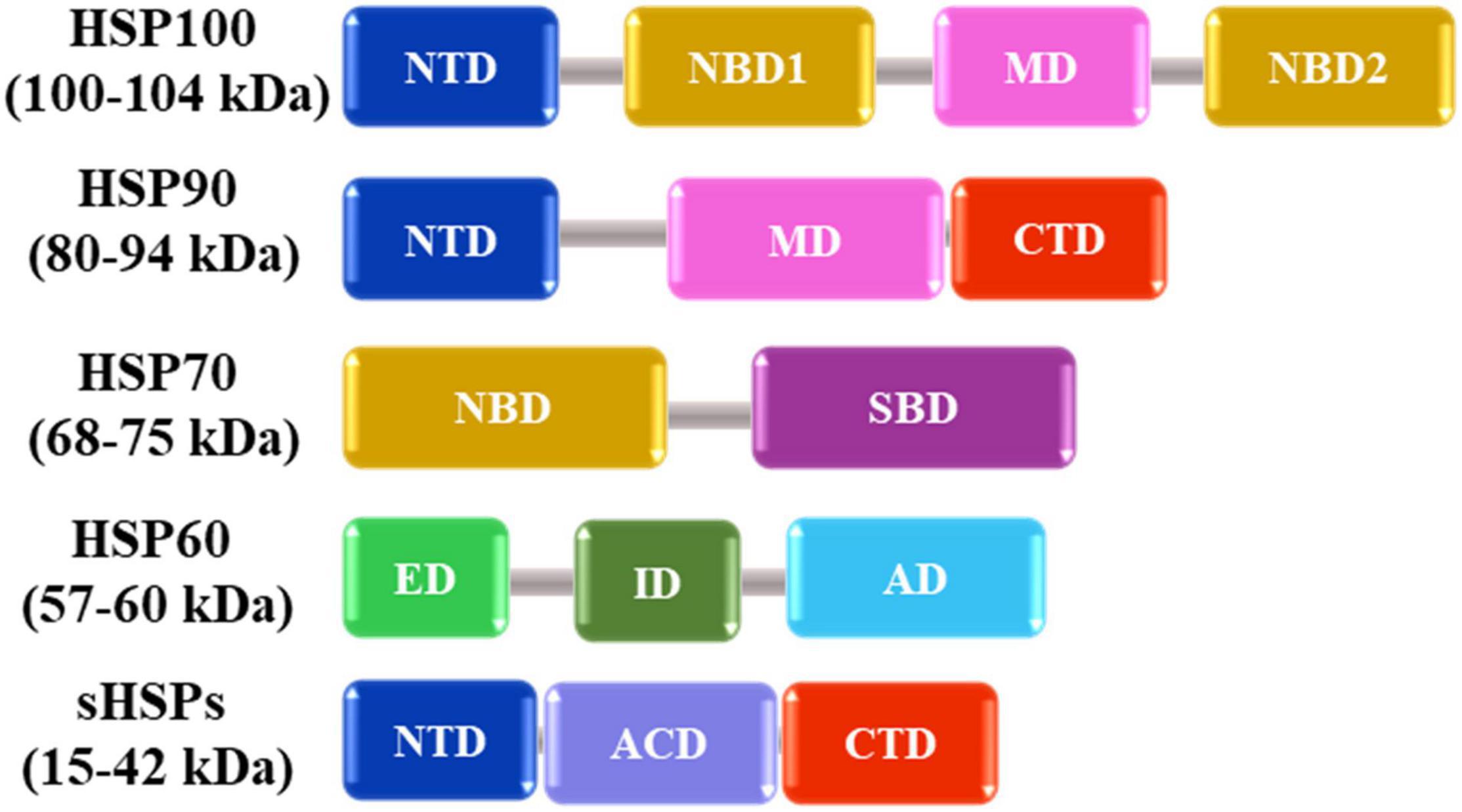
Figure 2. Schematic representation of available domains in the five major families of heat shock proteins. NTD (N-terminal domain), NBD (Nucleotide binding domain), MD (Middle domain), CTD (C-terminal domain), SBD (Substrate binding domain), ED (Equatorial domain), ID (Intermediate domain), AD (Apical domain), and ACD (Alpha-crystallin domain) are shown as boxes with different colors based on their functions. Numbers in parenthesis indicate the molecular weight distribution of each HSP family.
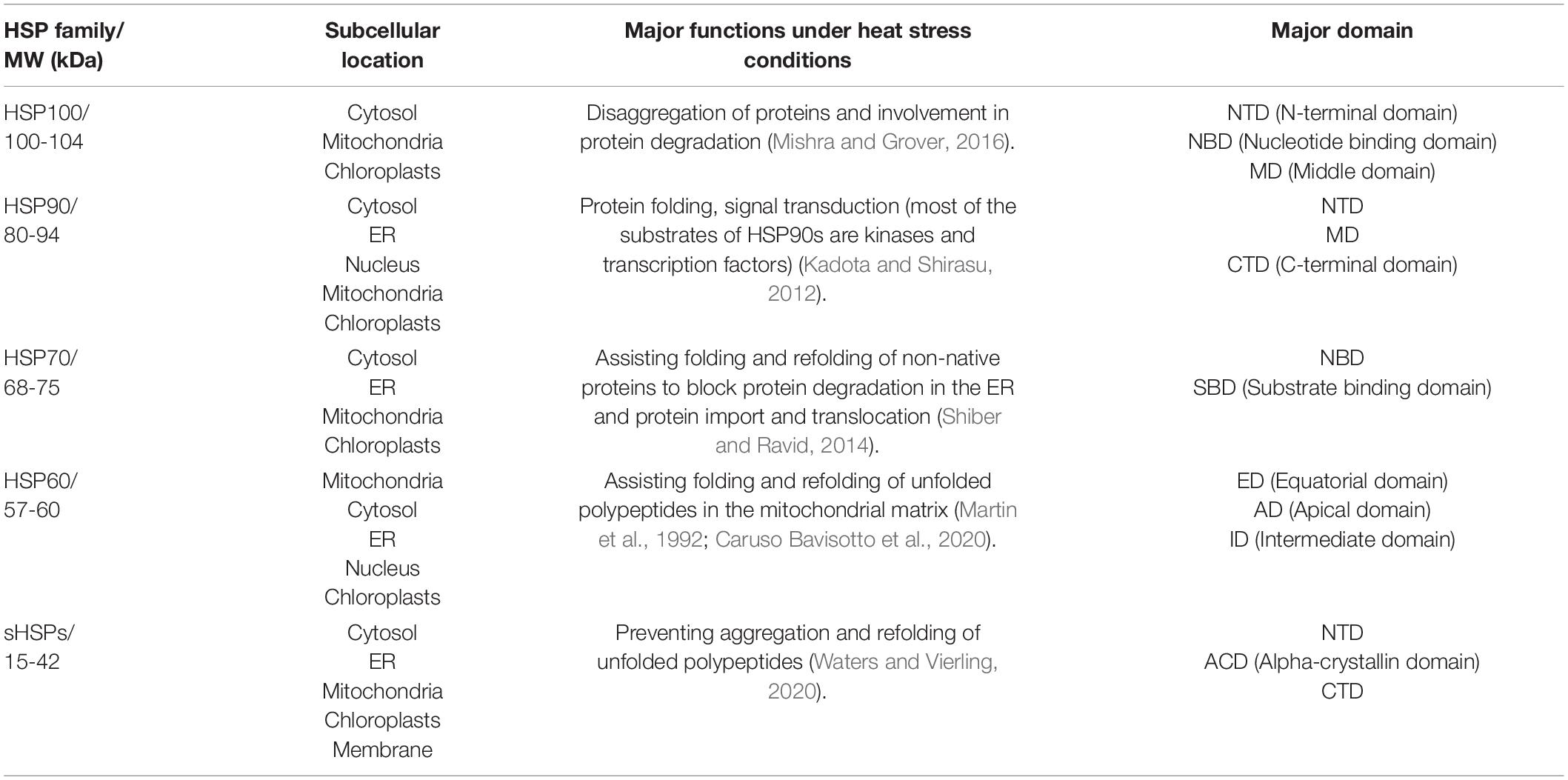
Table 1. Five major families of heat shock proteins and their major function under heat stress conditions.
Heat stress (HS) influences the alteration of membrane fluidity in plasma membrane (PM) in planta and activates the cyclic nucleotide-gated calcium channels (CNGCs), resulting in the movement of Ca2+ into the cytoplasm from the apoplastic space. The Ca2+ ions are associated with protein calmodulin 3 (CaM3) during HS and the Ca2+-CaM3 complex binds to either calcium/calmodulin-binding protein kinase 3 (CBK3) or phosphatase PP7 to transduce cytosol heat-stress response (HSR) signals into the nucleus by modulating phosphorylation and dephosphorylation of the heat shock transcription factors (HSFs), respectively. The elevated levels of inositol-1,4,5-triphosphate (IP3) via the phosophoinositide-signaling pathway (PLC) lead to an influx of Ca2+ into the cytoplasm from the pool of intracellular Ca2+ ions including the ER and vacuoles in response to HS and induce the same CaM3 signaling pathway. ROS are generated by respiratory burst oxidase homolog B (RbohB) and D (RbohD) during HS. RbohB/D-produced O2– is converted into H2O2, which depolarizes PM as well as inducing the ROS/Redox signaling network which is involved in the activation of HSFs. Also, H2O2 is possibly increased in plant cells due to metabolic imbalances and the production of ROS, resulting in the accumulation of nitric oxide (NO) and the activation of calcium-channels that subsequently trigger the activity of CaM3 as illustrated in the (Figure 1). Upon HS stimuli, HSP interacts with unfolded and aggregated proteins, thereby releasing HSF monomer. Heat shock factor monomers trimerize and bind to HSEs within promoter regions of heat shock genes. Heat shock factors undergo several post transcriptional modifications (PTMs) such as phosphorylation, which regulate the transactivation capacity of HSF. Under normal conditions, HSPs directly bind to HSF and provide negative feedback required to deactivate HSF. HSP70 and HSP40 together function as ATP-driven machines that prevent aggregation of misfolded polypeptides and participate in protein refolding. When denatured or misfolded proteins form aggregates, ClpB/HSP100 is crucial for protein disaggregation, refolding or degradation by protease especially during HS. Consequently, HSPs as chaperones play a pivotal role in conferring thermotolerance in plants. The dashed line indicates an unknown pathway.
Heat Shock Protein 100 Family
The caseinolytic proteinase/heat shock protein 100 (Clp/HSP100) proteins are members of the AAA+ protein group (ATPases associated with various cellular activities) that act in protein disassembly and/or protein degradation using the energy from adenosine triphosphate (ATP) hydrolysis (Sauer et al., 2004; Burton and Baker, 2005; Gul et al., 2021). In contrast to the typical molecular chaperones which function in protecting proteins from misfolding and aggregation, the Clp/Hsp100 proteins play a wide variety of functional roles in eliminating non-functional proteins and/or assisting the reassembly of denatured proteins from the aggregated protein complexes. As such, the Clp/Hsp100 proteins contribute to the maintenance of protein homeostasis in cells (Schirmer et al., 1996; Latterich and Patel, 1998; Agarwal et al., 2001; Mishra and Grover, 2019). The Clp/Hsp100 proteins consist of hexameric rings and the structural features are determined by nucleotide binding domains (NBD), spacer (linker) region, the middle domain (MD), N-terminal domain (NTD) and C-terminal domain (CTD) among diverse living organisms from prokaryotes to eukaryotes (Dougan et al., 2003; Schlieker et al., 2005; Butler et al., 2006). On the basis of the number of NBD domains, the Clp/Hsp100 family is classified into two major subclasses (class I and class II). The first class ClpA, ClpB, ClpC, and ClpD proteins that harbor two nucleotide binding domains (called ATP-binding domains) separated by spacers are clustered as large Clp proteins ranging from molecular weights of 68 to 110 kDa (Wang et al., 2004), whereas the second class including ClpM, ClpN, ClpX, and ClpY proteins that possess one NBD are grouped based on their low molecular weights ranging from 40 to 50 kDa (Wang et al., 2004; Mogk et al., 2008; Mishra and Grover, 2016). It was initially reported that the system of Clp ATPase proteins are able to hydrolyze casein in vitro (Hwang et al., 1987; Katayama-Fujimura et al., 1987). Later, further investigations on two-component protease systems revealed that the complexes of ClpA regulatory machine with an AAA+ ATPase module and a proteolytic component ClpP (Schelin et al., 2002) together with Lon protease complex serve as protein choppers for the degradation of toxic protein aggregates in cells (Wang et al., 2007). Moreover, the ClpAP complex recognizes target aggregated proteins via the guidance of the ClpS adapter that assists ClpAP to specifically bind and chop the aggregated proteins (Dougan et al., 2002). In addition to this, ClpB was initially found in bacteria and yeast, and it was later reported that plant HSPs were identified with high molecular weights of 100–110 kDa (Schirmer et al., 1994). Since plants harbor semi-autonomous organelles such as chloroplasts and mitochondria, plant ClpBs are classified into three different forms ClpB-C (cytoplasmic), ClpB-P (chloroplastic), and ClpB-M (mitochondrial) (Mishra and Grover, 2014). Although ClpB is considered to be a functional ortholog of ClpA with high similarity between the two proteins (Gottesman et al., 1990; Sanchez and Lindquist, 1990), it has been experimentally shown that ClpB could not replace the function of ClpA in protein degradation due to the lack of the LIV-GFL motif required for the interaction with ClpP (Weibezahn et al., 2004; Zolkiewski, 2006; Tessarz et al., 2008). Moreover, it was demonstrated that ClpB plays an essential role in the denaturing and/or renaturing pathway to release the native proteins from the aggregates rather than the degradation pathway as other Clps do. Of note, it has been displayed that ClpB is induced by HS in contrast to other Clps (Singh et al., 2010; Kim et al., 2012), indicating that ClpB is crucial for the protein renaturation/denaturation from aggregates especially during HS. Interestingly, the possible mechanism for assisting protein folding toward native and functional form from aggregates would be collaborated with the Hsp70 member, which is another ATP-dependent chaperone that is involved in refolding of liberated proteins by ClpB/HSP100 (Glover and Lindquist, 1998; Goloubinoff et al., 1999). However, when the aggregated proteins are interacted with other Clps and the peptidase (ClpP) system, the proteins move to the degradation pathway (Wang et al., 2004). The cellular roles of ClpB have been widely studied from prokaryotes to eukaryotes such as bacteria, yeast, and plants (Lindquist, 1986; Vierling, 1991; Wang et al., 2004). Remarkably, it has been determined that the fine-tuned expression of ClpB genes within cells is required for normal growth, development, and adaptation to environmental stresses including cold, heat, drought, and high salt (Yang et al., 2006). In particular, it has been shown that ClpB proteins are essential for rendering thermotolerance to organisms in response to HS. The loss-of-function mutant of ClpB in E. coli remarkably affected cell viability in response to abrupt HT (50 °C) with a slow growth rate at 44 °C (Squires et al., 1991). Also, ScHSP104 in Saccharomyces cerevisiae is one of the ClpB genes involved in acquiring thermotolerance: ScHSP104 deficient yeast cells grew and died at the same rate as the wild-type cells did when exposed directly to HT although the mutant cells could not acquire tolerance to heat after a mild pre-heat treatment (Sanchez and Lindquist, 1990). Plant ClpB/HSP100 proteins have been evaluated in diverse plant species including Arabidopsis (Lee et al., 2007), wheat (Campbell et al., 2001), soybean (Lee et al., 1994), maize (Nieto-Sotelo et al., 1999; Young et al., 2001), and rice (Agarwal et al., 2003). Analyses of ClpB/HSP100 proteins have been also conducted in vegetable crops such as pea, tomato, pepper, carrot, spinach, potato, banana, rapeseed, and mustard greens in response to heat and cold stresses.
Heat Shock Protein 90 Family
Heat shock protein 90 (HSP90; known as GroEL in E. coli) is one of the most abundant heat-related proteins expressed in cells accounting for 1–2% of total protein levels (Taipale et al., 2010). Heat shock protein 90 is a highly conserved molecular chaperone involved in the assembly, maturation, stabilization and activation of key signaling proteins including regulatory kinases, steroid hormone receptors and transcription factors in plant cells (Kadota and Shirasu, 2012; Chen et al., 2019). Most plants have several isoforms of HSP90 classified by their subcellular localization in the cytoplasm (HSP90.1), nucleus (HSP90.4), chloroplast (HSP90.5), mitochondria (HSP90.6), and endoplasmic reticulum (ER; HSP90.7) (Milioni and Hatzopoulos, 1997; Krishna and Gloor, 2001; Xu et al., 2012). HSP90 exists in the form of a dimer consisting of three main structural domains: NTD, which binds ATP; MD, which is important for ATP hydrolysis and client protein binding; and CTD, which mediates HSP90 dimerization and client protein binding. ATP binding to the NTD and its hydrolysis induce conformational change which is essential for chaperone activity (Krishna and Gloor, 2001; Pearl and Prodromou, 2006). HSP90 proteins play a major role in assisting the proper folding of other proteins together with HSP70s (Picard, 2002) by acting as molecular chaperones, signaling for the cellular quality control, trafficking of other HSP proteins (Pratt and Toft, 2003) and stabilizing proteins against HS (Marcu et al., 2002; Wang R. et al., 2016). Also, HSP90 proteins along with their co-chaperone HSP70s contribute to the maintenance of cellular protein homeostasis by inactivating HSF during attenuation/recovery of HSR (Hahn et al., 2011). In Arabidopsis, HSP90 and the co-chaperone SUPPRESSOR OF G2 ALLELE SKP1 (SGT1) positively regulate plant growth by stabilizing the auxin co-receptor F-box protein TIR1 under higher ambient temperature conditions (Wang R. et al., 2016), showing that HSP90 participates in plant growth control under changing thermal conditions.
Heat Shock Protein 70 Family
The heat shock protein 70 (HSP70) family (known as DnaK in E. coli), one of the most ubiquitous classes of chaperones, is highly conserved in all organisms, and also found in different cellular compartments such as the cytosol, chloroplasts, ER and mitochondria (Amir-Shapira et al., 1990; Radons, 2016; Usman et al., 2017). The HSP70 family is the central hub of the protein homeostasis network that prevents protein aggregation and uses the energy of ATP hydrolysis to solubilize, translocate and mediate the proper refolding and unfolding of proteins (Ben-Zvi et al., 2004; Imamoglu et al., 2020). Heat shock protein 70 contains two major domains: one is the N-terminal nucleotide binding domain for hydrolyzing ATP to ADP (Adenosine diphosphate) and the other is the C-terminal substrate binding domain (SBD) (Mayer, 2010). Under abiotic stress conditions such as HS, HSP70 molecular chaperones also function as ATP-driven unfolding/refolding machines that are capable of shifting substrate polypeptides between various folding states together with their co-chaperones such as HSP40 (Lee et al., 2007; Shiber and Ravid, 2014; Palakolanu et al., 2016). The significance of HSP70 regarding functional roles against HS was highlighted by transgenic plants overexpressing AtHSP70-1 and NtHSP70-1 (Sung and Guy, 2003; Cazalé et al., 2009; Cho and Choi, 2009). In addition, numerous experimental results have shown that HSP70 is involved in thermotolerance in various crops such as rice (Jung et al., 2013), tomato (Hahn et al., 2011), and pepper (Guo et al., 2014) under HS conditions.
Heat Shock Protein 60 Family
The heat shock protein 60 (HSP60) family (also known as chaperonins, Cpn, and GroEL in E. coli) typically functions inside the mitochondria together with the co-chaperone HSP10 to maintain protein homeostasis (Caruso Bavisotto et al., 2020). However, they have also been found in other subcellular compartments including the ER, cytosol, chloroplasts and nucleus, and participate in folding and aggregation of many proteins (Meng et al., 2018). Chaperonins are generally composed of two rings, stacked back to back, consisting of subunits of ∼60 kDa molecular weight (Nguyen et al., 2021). Each oligomer has three domains (1) the equatorial domain (ED), which has the ATP-biding site, (2) the apical domain (AD), which hosts client proteins and (3) the intermediate domain (ID), which transduces signals from the equatorial domain (Pipaón et al., 2021). When signals are transmitted to the ID from ATP binding and hydrolysis, conformational changes occur in the AD corresponding to the open and closed forms (Xu et al., 1997). Heat shock protein 60 proteins bind several types of proteins before folding to block their aggregation (Parsell and Lindquist, 1993) and stromal chaperones (Hsp70 and Hsp60) are involved in functional conformation of newly transferred proteins to the chloroplast (Jackson-Constan et al., 2001). Most of the HSP60 family proteins are heat inducible and also required for preventing protein aggregation, and mediating folding and refolding in mitochondria under HS conditions (Martin et al., 1992; Sharma et al., 2006).
Small Heat Shock Protein Family
Small heat shock proteins (sHSPs), which have a low molecular mass of 15-42 kDa, are very diverse in plants (Wang et al., 2004; Basha et al., 2006; Morrow and Tanguay, 2012). Small heat shock proteins have a common alpha-crystallin domain (ACD) containing 80–100 amino acid residues on the C-terminal region, and contribute to degradation of proteins with unsuitable folding (Seo et al., 2006). Small heat shock proteins are ubiquitous ATP-independent molecular chaperones that bind and stabilize misfolded or unfolding intermediates of substrate proteins in an energy-independent manner (Ferguson et al., 1990; Miernyk, 1999; Waters and Vierling, 2020).
Transcriptional Regulation of Heat Shock Proteins in Plants Under Heat Stress
Heat-stress response is known to be controlled by complex, tight networks, including selective enhancement and repression of gene expression in various metabolic processes, production of chaperone proteins for cellular protein homeostasis and other protective molecules that prevent targets from detrimental effectors such as ROS. The regulation of this network is critical for plant cells not only to adapt to various environmental conditions linked to temperature, humidity and light, but also to protect them from proteotoxic stresses. HSFs have a central function as major regulators in HSR by regulating transcription of a wide range of genes in several signaling and metabolic pathways (von Koskull-Döring et al., 2007; Guy et al., 2008). Heat shock factors are responsible for rapid synthesis and accumulation of HSPs, molecular chaperones for preventing protein aggregation and maintaining cellular protein homeostasis (Vierling, 1991; Wang et al., 2004; Gupta et al., 2010; Schleiff and Becker, 2011). Heat shock factor activity in each cell is controlled through sophisticated and complex feedback mechanisms and protein interactions, allowing for rapid adjustment and flexibility by diverse chaperones to changing environmental conditions (Akerfelt et al., 2010).
The expression of HSPs is induced by HSFs that bind the HSEs in the promoters of heat shock responsive genes (Nover et al., 2001). Under normal conditions, monomeric HSFs are bound to HSP70 in the cytoplasm. When plants are exposed to HS, HSFs are released from HSP70-HSF complexes, and phosphorylated in the cytoplasm, and form a trimer for biding to HSEs in the nucleus (Liu et al., 2006). Overexpression of HSF genes in turn turns on almost all heat shock genes containing the HSE consensus sequence, conferring tolerance to HS. HSP70/90 plays an important role in the regulation of HSFA1 activity. HSP70/90 complex keeps HSFA1 inactive under normal conditions by repressing transactivation activity and nuclear localization of HSFA1 (Yamada et al., 2007; Hahn et al., 2011). Recently, the temperature-dependent repression (TDR) domain has been identified in the central region of HSFA1d, one of the Arabidopsis HSFA1s responsible for HS-dependent transactivation activity (Ohama et al., 2017). Overexpression of constitutively active HSFA1d, which lacks the TDR domain, induced the expression of heat shock proteins in the absence of HS, thereby conferring strong thermal stability in the overexpressing plants. Under HS conditions, HSFA1a is released from the HSFA1-HSP70/90 complex and activated. Of note, no TDR domain has been observed in mammalian HSFA1 proteins although the repression of the activities of HSFs by the HSP70/90 complex is generally conserved in both plants and animals. Activated HSFA1 directly and rapidly regulates expression levels of genes encoding important HS-responsive transcription factors (TFs) such as DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2A (DREB2A), HSFA2, HSFA7a, HSFBs, and MULTIPROTEIN BRIDGING FACTOR 1C (MBF1C) (Yoshida et al., 2011). Subsequently, DREB2A directly regulates the gene expression level of HSFA3 by creating a coactivator complex with NUCLEAR FACTOR Y, SUBUNIT A2 (NF-YA2), NF-YB3, and DNA POLYMERASE II SUBUNIT B3-1 (DPB3-1)/NF-YC10 (Chen et al., 2010; Sato et al., 2014). HSFA3 knockout or knockdown transgenic lines caused reduced expression of putative target HSP genes under HS, thus HSFA3 is regarded as an important HS-responsive TF (Schramm et al., 2008; Yoshida et al., 2008). Furthermore, HSFA2 contributes to high levels of modifications at specific histone tail residues (H3K4me2 and H3K4me3) of ascorbate peroxidase 2 (APX2), HSP22, and HSP18.2 (Sung et al., 2003; Charng et al., 2007; Lämke et al., 2016). Heat stress memory is maintained for several days, allowing plants to survive when they are exposed to the next HS conditions (Yamaguchi, 2021). Strong/rapid expression of sHSP genes including HSP21, HSP22, and HSP17.6C is observed in primed plants compared to non-primed plants (Yamaguchi et al., 2021). FORGETTER3 (FGT3)/HSFA3 is needed to retain HS memory for several days following HS exposure (Friedrich et al., 2021). A recent discovery showed that genes encoding stem cell regulators such as CLAVATA1 (CLV1), CLV3, and HSP17.6A, and the primary carbohydrate metabolism gene FRUCTOSE-BISPHOSPHATE ALDOLASE 6 (FBA6) are involved in the HS transcriptional memory in the shoot apical meristem (Olas et al., 2021). JUMONJI-C DOMAIN CONTAINING PROTEINs (JMJs) that code for H3K27me3 demethylases are regulators of heat acclimation through controlling the methylation status of HSP loci (Pan et al., 2007; Xiao et al., 2016; Yamaguchi et al., 2021; Yamaguchi and Ito, 2021).
Extreme HT causes protein misfolding and denaturation. Unfolded proteins can be degraded by the ubiquitin proteasome system or autophagy (Buchberger et al., 2010; Amm et al., 2014; Xu and Xue, 2019). It has been demonstrated that some ubiquitin E3 ligases and autophagy-related genes play a critical role in plant heat tolerance (Zhou et al., 2014; Li et al., 2015; Liu J. et al., 2016; Gil et al., 2017). Transgenic plants overexpressing ubiquitin or ubiquitin E3 ligases displayed enhanced BTT and/or ATT (Tian et al., 2014; Liu J. et al., 2016), and Zhang Y. et al. (2021) reported that silencing CARBOXYL TERMINUS OF THE HSC70-INTERACTING PROTEINS (CHIP), a chaperone-dependent ubiquitin E3 ligase caused reduced heat tolerance in tomato. CHIP plays a critical role in HSR through the misfolded proteins degradation induced by HS. Transgenic Arabidopsis seedlings overexpressing PROTEIN WITH THE RING DOMAIN AND TMEMB (PPRT1) encoding a C3HC4 zinc-finger ubiquitin E3 ligase showed enhanced BTT and ATT (Liu Y. et al., 2020). Moreover, virus-induced gene silencing (VIGS) of tomato AUTOPHAGY RELATED5 (ATG5) and ATG7 genes resulted in increased sensitivity of tomato plants to HS (Zhou et al., 2014).
Understanding the dynamic behavior involving expression levels of TFs and HSPs under HS will help understand the whole regulatory network to adapt to HT.
Expression Patterns of HSP and HSF Genes in Vegetables Under Heat Stress
Exposure to extreme temperature stresses such as heat and cold induces cellular changes in plant cells (Guy, 1999; Bita and Gerats, 2013). Plants have evolved various physiological and molecular adaptations to stresses in order to minimize damage and provide cellular homeostasis (Theocharis et al., 2012; Awasthi et al., 2015). In response to the extreme temperature stresses, plants synthesize many stress-responsive proteins including HSP and HSF by regulating gene expression (Guo et al., 2016a; Ul Haq et al., 2019). So far, many studies on gene expression patterns under heat and/or cold stresses in vegetable crops have been reported and the collected information can be seen in Table 2.
Tomato (Solanum lycopersicum L.)
Tomato is one of the most economically important vegetable crops worldwide (Campos et al., 2021). As global warming leads to extreme weather events, a number of researchers have examined the effects of heat and/or cold stresses on the expression pattern of genes such as HSPs and HSFs, which play crucial roles in thermotolerance in tomatoes (Tubiello et al., 2007).
Heat treatment has been found to induce chloroplastic SlHSP100 genes in both thermotolerant and thermosensitive tomato seedlings. The highest upregulation was observed in the genotype 17903, which showed the highest ratio of cell viability and cell membrane stability under HS, implying a crucial role for the gene in ATT (Gul et al., 2021). Besides the role of HSP100 as a chaperone, Sabehat et al. (1996) found that tomato fruits heated and then chilled showed a high level expression of both HSP70 and sHSP family genes (14–25kDa) and enhanced chilling tolerance compared to unheated fruits (Sabehat et al., 1996). Similar results were also reported by Kadyrzhanova et al. (1998) and Sabehat et al. (1998) where the expression of chloroplastic HSP21 and HSP17.6 was first decreased and re-induced when the heated fruits were transferred to low temperature. The members of SlHSP20s in tomato were also upregulated in both thermotolerant and thermosensitive lines under HS, except for SlHsp15.7 (Yu et al., 2016). Moreover, it has been reported that the expression of HSFA2, transcriptional activator of HSP expression, and HSP17-CII was highly activated in the tomato anther during its development under HS (Giorno et al., 2010).
Pepper (Capsicum annuum)
The production and consumption of pepper has steadily increased worldwide due to its nutritional benefits and spice, but it is thermosensitive (Crosby, 2008; Guo et al., 2014). As with tomato, there has been a growing body of research that explores the expression of HSP genes in pepper under temperature stress conditions. Many HSPs including CaHSP70, CaHSP60, CaHSP20, and CaHSP16.4 are upregulated in pepper under HS (Guo et al., 2015; Usman et al., 2015; Feng et al., 2019; Haq et al., 2019). HSP70 gene was significantly upregulated in the thermotolerant line compared to the thermosensitive line after 2 h of HS treatment at 42°C, indicating that the gene is quickly and sharply induced by heat shock and plays a major role in thermotolerance (Usman et al., 2015). Haq et al. (2019) observed that fifteen CaHSP60 genes were upregulated under HS and cold stress, and only CaHSP60-3 was downregulated in both thermosensitive B6 and thermotolerant R9 lines (Haq et al., 2019).
Soybean (Glycine max)
Soybeans are members of the legume family of vegetables and have been a staple of Asian cuisines for a long time. Soybean yield is severely affected by temperature stresses. Under low or high temperature stress conditions, HSPs are induced in soybean to prevent cell damage caused by the temperature stresses. Xu et al. (2013) studied the expression of GmHSP90 in relation to HS, and observed a significant upregulation of this gene in early response to HS (Xu et al., 2013). Expression patterns of soybean 61 GmHSP70 genes under HS and drought were analyzed. Among those genes, 55 GmHSP70 genes were highly upregulated during HS, and 29 GmHSP70 genes showed increased expression under both heat and drought stress conditions, indicating that most of the GmHSP70 genes play an important role in heat and drought tolerance (Zhang et al., 2015). Similarly, 47 GmHSP20 genes among 51 GmHSP20 candidates were found to be highly induced under HS and 5 genes were induced under both heat and cold conditions (Lopes-Caitar et al., 2013).
Pea (Pisum sativum)
Pea has long been important in the human diet due to its starch, protein, and fiber content and the many phytochemical substances it contains, but it is a cool season crop which is heat-sensitive (Dahl et al., 2012). Therefore, some researchers have investigated the expression of HSPs in pea during HS. DeRocher et al. (1991) observed that the HSP18.1 mRNA peaked at the beginning of the maximum temperature during 4 h gradual HS (30–42°C) period, and began to decline 6 to 8 h before the amount of HSP18.1 protein reached maximum levels, implying that sHSP levels in plants may also be self-regulated or regulated by some other heat-inducible protein.
Potato (Solanum tuberosum)
Potato is a vegetable crop that mainly grows in a temperate climate, so HS can have a negative effect on the yield by inducing physiological defects in tubers (Rykaczewska, 2017). Hence, it is important to examine the accumulation of HSPs in response to HS. Ahn et al. (2004) reported that the 18 kDa sHSP proteins were synthesized for a longer time in the heat tolerant cultivars compared to the heat sensitive cultivars under strong heat shock temperature, suggesting that sHSP plays an important role in the heat tolerance enhancement (Ahn et al., 2004). Fifteen HSPs, including three HSP70s, two HSP80s, one HSP90, one HSP100 and eight sHSPs were consistently upregulated by low temperatures at both the RNA and protein levels to reduce cellular damage and re-build cellular homeostasis in potato tubers under cold stress during postharvest storage (Lin et al., 2019).
Lettuce (Lactuca sativa)
Lettuce is an important cool season leafy vegetable with an optimal growing temperature ranging from 17 to 28°C (Holmes et al., 2019). HT can facilitate the accumulation of gibberellin (GA) which promotes lettuce bolding (Fukuda et al., 2012). Under HT, it is suggested that induced expression of genes encoding LsHSPs that interact with a calmodulin confers enhanced tolerance to heat with bolting resistance in lettuce (Liu R. et al., 2020). Recently, putative early heat responsive HSP genes were identified by transcriptome profiling in lettuce (Kang et al., 2021). Among them, sHSP and HSP70 genes were quickly and sharply induced within 1 h in response to HS, indicating that these genes could be potential candidates as the breeding targets for the development of heat-tolerant lettuce cultivars.
Breeding for Elevated Resistance to Heat Stress
Currently, the greatest risk to crop productivity and yields associated with global climate change is being caused by extreme weather events such as extreme hot and cold weather (Reddy and Hodges, 2000). Therefore, improved tolerance to heat and cold stress might be crucial in increasing yields for most crops. Application of transgenic and genome editing technologies could help to introduce desirable abiotic stress tolerance traits into crop varieties (Sanghera et al., 2011; Lamaoui et al., 2018). In recent years, there has been an increasing effort to reveal functional roles of HSPs and HSFs using mutagenic and transgenic plants for production of crops with enhanced heat and/or cold tolerance (Table 3).
Model Plants
A number of researchers have used model plants such as Arabidopsis, tobacco and rice for functional studies (proof of concept) on genes involved in heat and cold stresses because of the ease of genetic experiments (Rensink and Buell, 2004; Koornneef and Meinke, 2010). Queitsch et al. (2000) examined transgenic Arabidopsis plants containing HSP101 antisense and/or co-suppression constructs, and found that they showed normal growth but impaired ATT and BTT, indicating HSP101 plays a pivotal role in heat tolerance in Arabidopsis. In contrast, transgenic Arabidopsis plants containing constitutively active HSF-GUS fusion proteins caused increased HSP18 expression at normal temperature by forming HSF trimers and their binding to DNA, resulting in enhanced BTT (Lee et al., 1995).
In addition, transgenic approaches with other crop genes have also been made with a fair degree of success. Genetically engineered Arabidopsis plants overexpressing HSP genes from pepper (Guo et al., 2016b; Feng et al., 2019), primula (Zhang et al., 2014), wheat (Feng et al., 2019) and David Lily (Mu et al., 2013) exhibited increased thermotolerance activity. Similar events were also observed under cold stress conditions by Wang et al. (2017) and Zhang L. et al. (2018). They introduced CsHSP17.7, CsHSP18.1, CsHSP21.8, and PfHSP17.2 from Camellia sinensis and Forrest primrose into Arabidopsis for overexpression. Transgenic plants showed increased root length and tolerance to cold stress. Furthermore, overexpression of OsHSP101 (Chang et al., 2007), ZmHSP16.9 (Sun et al., 2012), LeHSP21 (Zhang et al., 2016), BcHSP70 (Wang X. et al., 2016), AtHSP101 (Katiyar-Agarwal et al., 2003) and OsHSP18.6 (Wang et al., 2015) conferred improved HS tolerance in tobacco and rice. These results indicate that HSP genes from various crops play a key role in developing thermotolerance.
Vegetables
Vegetable crops are very susceptible to abiotic stresses such as high and low temperatures. Therefore, the development of varieties that are tolerant to heat and cold stresses is an important goal for improvement in crop productivity. Recently investigators have examined the protective roles of HSP and HSF against heat and cold stresses in transgenic vegetables. Li et al. (2003) reported that increased activity of soluble isoforms of ascorbate peroxidase (APX) and tolerance were observed in the transgenic tomato plants overexpressing AtHSFA1b-gusA fusion gene under heat and cold stress conditions. In addition, 15 heat tolerant tomato lines were isolated through screening of over 4000 ethyl methanesulfonate (EMS) Micro-Tom mutants. Among the selected heat tolerant mutants, the HT7 line displayed much higher fruit number and total pollen number with enhanced viability under HS conditions. Higher expression levels of SIHSFA1b3, which is known as a master regulator that activates HSR (Mishra et al., 2002), and HSP101 were detected in the leaves of HT7 compared to those of WT after long-term exposure to HS, suggesting that HT7 could be used as a breeding material for production of tomato with improved heat tolerance (Pham et al., 2020). Also, up and downregulated expression of HSP23.8 made it possible for each transgenic plant to display the opposite phenotype under low temperature conditions: Transgenic plants overexpressing HSP23.8 gene showed increased cold tolerance whereas decreased chilling tolerance, wilting, skin wrinkles and partial discoloration were observed in the transgenic plant with reduced expression of HSP23.8 gene (Escobar et al., 2021). Similar studies have reported that the HSP17.7 gene plays a role in the HS tolerance in potato (Ahn and Zimmerman, 2006) and carrot (Malik et al., 1999). Recently, it has been reported that HS tolerance decreases in pepper when the CaHSP60-6 gene is down-regulated by virus-induced gene silencing (VIGS) (Haq et al., 2019). In particular, CRISPR-Cas9 based gene knockout was applied to GmHSP90A2 in soybean, and the GmHSP90A2 mutant exhibited reduced heat tolerance (Huang et al., 2019). In conclusion, major HSP and HSF genes are tightly related to thermotolerance of vegetables. Thus, continuous efforts to identify detailed functions and working mechanisms of HSP and HSF genes are needed for the generation of vegetables with enhanced heat/cold tolerance traits through precise manipulation of genetic elements.
Conclusion and Future Prospects
Climate change including global warming is causing abrupt changes in weather patterns, and extreme weather events that threaten crop yields. Elevated temperatures, in particular, will have a severe influence on the productivity and yields of vegetables in agricultural fields. It is, therefore, indispensible to understand the sophisticated mechanisms vegetable crops use to adapt to changing temperature environments, from the signal perception to gene expression in reponse to HS.
As mentioned above, recent research has elucidated that an interplay of cooperative HSP, HSF, and HSR mechanisms orchestrate the expression of heat-responsive genes as the plant response to HS. Furthermore, research identifying TFs related to abiotic stresses and their molecular functions has contributed to the expansion of knowledge for the production of crops with desired traits through genetic manipulation and/or molecular breeding. Functional and cellular roles of some key TFs such as HSFA1s and DREB2A have been determined in transcriptional networks of HSR at the post-translational levels during HS. Nevertheless, the current information on the functional roles of HSP and HSF genes in vegetable crops is still insufficient for their practical application to breeding. Transcriptional regulation between HSPs and HSFs, and in-depth working mechanisms and pathways of heat-related proteins during HSR remain to be explored.
Chromatin immunoprecipitation sequencing (ChIP-seq) for protein-protein complexes and reverse ChIP for mining the upstream-gene regulatory sequences have been shown to be effective tools to investigate potential interaction networks between regulatory regions in HSE and proteins, respectively (Machanick and Bailey, 2011; Shim et al., 2021). It will be necessary to utilize these techniques to clarify the in-depth mechanism underlying the gene regulatory relationships in the HSPs and HSFs of vegetable crops during HSR. It is becoming evident that microRNAs, small RNAs, and epigenetic modulations in DNA, RNA, and protein species play a pivotal role in HS memory (Guan et al., 2013; Stief et al., 2014a,b; Lämke et al., 2016). Advances in high-throughput small RNA sequences (RNA-seq) together with methylated DNA and RNA-sequencing combined with IP will be of help in determining the functions of TFs and epigenetic regulators (Pall and Hamilton, 2008; Zhang H. et al., 2018; Shen et al., 2019; Lee et al., 2021). In addition, state-of-art next-generation sequencing (NGS) including quantitative trait loci (QTL)-sequencing, genotyping-by-sequencing (GBS), and genome-wide association studies (GWAS) have been successfully developed and adopted for deciphering comprehensive genome sequences, thus facilitating the identification of a wide variety of molecular markers corresponding to target traits in crops (Han et al., 2016; Jo et al., 2017; Lee et al., 2020; Jha et al., 2021). Candidate and/or identified genes crucial for thermotolerant-traits and HS-related pathways can be used for production of transgenic vegetable crops via genetic engineering. Furthermore, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) and dead Cas9 (dCas9) systems have been extensively introduced into crop biotechnology as powerful tools for gene/genome editing in spite of controversial GMO and non-GMO issues (Liu D. et al., 2016; Pramanik et al., 2020; Gao, 2021; Kim et al., 2021). Indeed, “Sicilian Rouge High GABA tomato” was recently developed by using the CRISPR/Cas9 gene editing technology. It contains high levels of gamma-aminobutyric acid (GABA), an amino acid believed to aid relaxation and help lower blood pressure.2 All the aforementioned technologies can be utilized for dissecting action modes and intricate networks of HSP, HSF and HSR for thormotolerance in vegetable crops.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported in part by a grant from the World Vegetable Center Korea Office (WKO #10000379) and the long-term strategic donors to the World Vegetable Center: Taiwan, United Kingdom aid from the United Kingdom government, United States Agency for International Development (USAID), Australian Centre for International Agricultural Research (ACIAR), Germany, Thailand, Philippines, South Korea and Japan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Kazuo Nakashima at JIRCAS, and Myeong-Cheoul Cho at NIHHS, RDA for indirect assistance via intellectual discussions.
Footnotes
References
Agarwal, M., Katiyar-Agarwal, S., Sahi, C., Gallie, D. R., and Grover, A. (2001). Arabidopsis thaliana Hsp100 proteins: kith and kin. Cell Stress Chaperones 6, 219–224. doi: 10.1379/1466-1268(2001)006<0219:athpka>2.0.co;2
Agarwal, M., Sahi, C., Katiyar-Agarwal, S., Agarwal, S., Young, T., Gallie, D. R., et al. (2003). Molecular characterization of rice hsp101: complementation of yeast hsp104 mutation by disaggregation of protein granules and differential expression in indica and japonica rice types. Plant Mol. Biol. 51, 543–553. doi: 10.1023/a:1022324920316
Ahn, Y.-J., Claussen, K., and Zimmerman, J. (2004). Genotypic differences in the heat-shock response and thermotolerance in four potato cultivars. Plant Sci. 166, 901–911.
Ahn, Y. J., and Zimmerman, J. L. (2006). Introduction of the carrot HSP17.7 into potato (Solanum tuberosum L.) enhances cellular membrane stability and tuberization in vitro. Plant Cell Environ. 29, 95–104. doi: 10.1111/j.1365-3040.2005.01403.x
Ahuja, I., De Vos, R. C., Bones, A. M., and Hall, R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Sci. 15, 664–674. doi: 10.1016/j.tplants.2010.08.002
Akerfelt, M., Morimoto, R. I., and Sistonen, L. (2010). Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555. doi: 10.1038/nrm2938
Al-Whaibi, M. H. (2011). Plant heat-shock proteins: a mini review. J. King Saud Univ. Sci. 23, 139–150.
Amir-Shapira, D., Leustek, T., Dalie, B., Weissbach, H., and Brot, N. (1990). Hsp70 proteins, similar to Escherichia coli DnaK, in chloroplasts and mitochondria of Euglena gracilis. Proc. Natl. Acad. Sci. U.S.A. 87, 1749–1752. doi: 10.1073/pnas.87.5.1749
Amm, I., Sommer, T., and Wolf, D. H. (2014). Protein quality control and elimination of protein waste: the role of the ubiquitin–proteasome system. Biochim. Biophys. Acta 1843, 182–196. doi: 10.1016/j.bbamcr.2013.06.031
Aranda, M. A., Escaler, M., Thomas, C. L., and Maule, A. J. (1999). A heat shock transcription factor in pea is differentially controlled by heat and virus replication. Plant J. 20, 153–161. doi: 10.1046/j.1365-313x.1999.00586.x
Awasthi, R., Bhandari, K., and Nayyar, H. (2015). Temperature stress and redox homeostasis in agricultural crops. Front. Environ. Sci. 3:11. doi: 10.3389/fenvs.2015.00011
Basha, E., Friedrich, K. L., and Vierling, E. (2006). The N-terminal Arm of Small Heat Shock Proteins Is Important for Both Chaperone Activity and Substrate Specificity. J. Biol. Chem. 281, 39943–39952.
Ben-Zvi, A., De Los Rios, P., Dietler, G., and Goloubinoff, P. (2004). Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual Hsp70 chaperones. J. Biol. Chem. 279, 37298–37303. doi: 10.1074/jbc.M405627200
Bhutia, K., Khanna, V., Meetei, T., and Bhutia, N. (2018). Effects of climate change on growth and development of chilli. Agrotechnology 7:2. doi: 10.1371/journal.pone.0257893
Bita, C., and Gerats, T. (2013). Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4:273. doi: 10.3389/fpls.2013.00273
Bourgine, B., and Guihur, A. (2021). Heat shock signaling in land plants: from plasma membrane sensing to the transcription of small heat shock proteins. Front. Plant Sci. 12:710801. doi: 10.3389/fpls.2021.710801
Buchberger, A., Bukau, B., and Sommer, T. (2010). Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell 40, 238–252. doi: 10.1016/j.molcel.2010.10.001
Burton, B. M., and Baker, T. A. (2005). Remodeling protein complexes: insights from the AAA+ unfoldase ClpX and Mu transposase. Protein Sci. 14, 1945–1954. doi: 10.1110/ps.051417505
Butler, S. M., Festa, R. A., Pearce, M. J., and Darwin, K. H. (2006). Self-compartmentalized bacterial proteases and pathogenesis. Mol. Microbiol. 60, 553–562. doi: 10.1111/j.1365-2958.2006.05128.x
Campbell, J. L., Klueva, N. Y., Zheng, H.-G., Nieto-Sotelo, J., Ho, T.-H., and Nguyen, H. T. (2001). Cloning of new members of heat shock protein HSP101 gene family in wheat (Triticum aestivum (L.) Moench) inducible by heat, dehydration, and ABA. Biochim. Biophys. Acta 1517, 270–277. doi: 10.1016/s0167-4781(00)00292-x
Campos, M. D., Félix, M. D. R., Patanita, M., Materatski, P., and Varanda, C. (2021). High throughput sequencing unravels tomato-pathogen interactions towards a sustainable plant breeding. Hortic. Res. 8:171. doi: 10.1038/s41438-021-00607-x
Caruso Bavisotto, C., Alberti, G., Vitale, A. M., Paladino, L., Campanella, C., Rappa, F., et al. (2020). Hsp60 post-translational modifications: functional and pathological consequences. Front. Mol. Biosci. 7:95. doi: 10.3389/fmolb.2020.00095
Cazalé, A.-C., Clément, M., Chiarenza, S., Roncato, M.-A., Pochon, N., Creff, A., et al. (2009). Altered expression of cytosolic/nuclear HSC70-1 molecular chaperone affects development and abiotic stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 60, 2653–2664. doi: 10.1093/jxb/erp109
Chang, C.-C., Huang, P.-S., Lin, H.-R., and Lu, C.-H. (2007). Transactivation of protein expression by rice HSP101 in planta and using Hsp101 as a selection marker for transformation. Plant Cell Physiol. 48, 1098–1107. doi: 10.1093/pcp/pcm080
Charng, Y.-Y., Liu, H.-C., Liu, N.-Y., Chi, W.-T., Wang, C.-N., Chang, S.-H., et al. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262. doi: 10.1104/pp.106.091322
Chauhan, H., Khurana, N., Nijhavan, A., Khurana, J. P., and Khurana, P. (2012). The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 35, 1912–1931. doi: 10.1111/j.1365-3040.2012.02525.x
Chen, H., Hwang, J. E., Lim, C. J., Kim, D. Y., Lee, S. Y., and Lim, C. O. (2010). Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem. Biophys. Res. Commun. 401, 238–244. doi: 10.1016/j.bbrc.2010.09.038
Cho, E. K., and Choi, Y. J. (2009). A nuclear-localized HSP70 confers thermoprotective activity and drought-stress tolerance on plants. Biotechnol. Lett. 31, 597–606. doi: 10.1007/s10529-008-9880-5
Crosby, K. M. (2008). “Pepper,” in Vegetables II: Fabaceae, Liliaceae, Solanaceae, and Umbelliferae, eds J. Prohens and F. Nuez (New York, NY: Springer), 221–248.
Dahl, W. J., Foster, L. M., and Tyler, R. T. (2012). Review of the health benefits of peas (Pisum sativum L.). Br. J. Nutr. 108(Suppl. 1), S3–S10.
DeRocher, A. E., Helm, K. W., Lauzon, L. M., and Vierling, E. (1991). Expression of a conserved family of cytoplasmic low molecular weight heat shock proteins during heat stress and recovery 1. Plant Physiol. 96, 1038–1047. doi: 10.1104/pp.96.4.1038
Dinkova, T. D., Zepeda, H., Martínez-Salas, E., Martínez, L.M., Nieto-Sotelo, J., and De Jiménez, E. S. (2005). Cap-independent translation of maize Hsp101. Plant J. 41, 722–731.
Dougan, D., Mogk, A., and Bukau, B. (2002). Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell. Mol. Life Sci. 59, 1607–1616. doi: 10.1007/pl00012487
Dougan, D. A., Weber-Ban, E., and Bukau, B. (2003). Targeted delivery of an ssrA-tagged substrate by the adaptor protein SspB to its cognate AAA+ protein ClpX. Mol. Cell 12, 373–380. doi: 10.1016/j.molcel.2003.08.012
Driedonks, N., Rieu, I., and Vriezen, W. H. (2016). Breeding for plant heat tolerance at vegetative and reproductive stages. Plant Reprod. 29, 67–79. doi: 10.1007/s00497-016-0275-9
Escobar, M. R., Ré, M. D., Sossi, M. L., Boggio, S. B., Herrfurth, C., Feussner, I., et al. (2021). Mitochondrial small heat shock protein and chilling tolerance in tomato fruit. Postharvest Biol. Technol. 175:111491.
Feder, M. E., and Hofmann, G. E. (1999). Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282.
Feng, X.-H., Zhang, H.-X., Ali, M., Gai, W.-X., Cheng, G.-X., Yu, Q.-H., et al. (2019). A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper (Capsicum annuum L.). Plant Physiol. Biochem. 142, 151–162. doi: 10.1016/j.plaphy.2019.07.001
Ferguson, D. L., Guikema, J. A., and Paulsen, G. M. (1990). Ubiquitin pool modulation and protein degradation in wheat roots during high temperature stress. Plant Physiol. 92, 740–746. doi: 10.1104/pp.92.3.740
Finka, A., Cuendet, A. F. H., Maathuis, F. J., Saidi, Y., and Goloubinoff, P. (2012). Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24, 3333–3348. doi: 10.1105/tpc.112.095844
Frank, G., Pressman, E., Ophir, R., Althan, L., Shaked, R., Freedman, M., et al. (2009). Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J. Exp. Bot. 60, 3891–3908.
Friedrich, T., Oberkofler, V., Trindade, I., Altmann, S., Brzezinka, K., Lämke, J., et al. (2021). Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat. Commun. 12:3426. doi: 10.1038/s41467-021-23786-6
Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014
Fukuda, M., Matsuo, S., Kikuchi, K., Mitsuhashi, W., Toyomasu, T., and Honda, I. (2012). Gibberellin metabolism during stem elongation stimulated by high temperature in lettuce. Acta Hortic. 932, 259–264.
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635. doi: 10.1016/j.cell.2021.01.005
Gao, F., Han, X., Wu, J., Zheng, S., Shang, Z., Sun, D., et al. (2012). A heat-activated calcium-permeable channel–Arabidopsis cyclic nucleotide-gated ion channel 6–is involved in heat shock responses. Plant J. 70, 1056–1069. doi: 10.1111/j.1365-313X.2012.04969.x
Gil, K.-E., Kim, W.-Y., Lee, H.-J., Faisal, M., Saquib, Q., Alatar, A. A., et al. (2017). ZEITLUPE contributes to a thermoresponsive protein quality control system in Arabidopsis. Plant Cell 29, 2882–2894. doi: 10.1105/tpc.17.00612
Giorno, F., Wolters-Arts, M., Grillo, S., Scharf, K.-D., Vriezen, W. H., and Mariani, C. (2010). Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers. J. Exp. Bot. 61, 453–462. doi: 10.1093/jxb/erp316
Glover, J. R., and Lindquist, S. (1998). Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82.
Goloubinoff, P., Mogk, A., Zvi, A. P. B., Tomoyasu, T., and Bukau, B. (1999). Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. U.S.A. 96, 13732–13737. doi: 10.1073/pnas.96.24.13732
Gottesman, S., Clark, W., and Maurizi, M. (1990). The ATP-dependent Clp protease of Escherichia coli. Sequence of clpA and identification of a Clp-specific substrate. J. Biol. Chem. 265, 7886–7893.
Guan, Q., Lu, X., Zeng, H., Zhang, Y., and Zhu, J. (2013). Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 74, 840–851. doi: 10.1111/tpj.12169
Gul, S., Shah, K. N., Rana, R. M., Khan, M. A., El-Shehawi, A. M., and Elseehy, M. M. (2021). Phylogenetic and expression dynamics of tomato ClpB/Hsp100 gene under heat stress. PLoS One 16:e0255847. doi: 10.1371/journal.pone.0255847
Guo, M., Liu, J.-H., Lu, J.-P., Zhai, Y.-F., Wang, H., Gong, Z.-H., et al. (2015). Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 6:806. doi: 10.3389/fpls.2015.00806
Guo, M., Liu, J.-H., Ma, X., Luo, D.-X., Gong, Z.-H., and Lu, M.-H. (2016a). The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7:114. doi: 10.3389/fpls.2016.00114
Guo, M., Liu, J.-H., Ma, X., Zhai, Y.-F., Gong, Z.-H., and Lu, M.-H. (2016b). Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 252, 246–256. doi: 10.1016/j.plantsci.2016.07.001
Guo, M., Zhai, Y.-F., Lu, J.-P., Chai, L., Chai, W.-G., Gong, Z.-H., et al. (2014). Characterization of CaHsp70-1, a pepper heat-shock protein gene in response to heat stress and some regulation exogenous substances in Capsicum annuum L. Int. J. Mol. Sci. 15, 19741–19759. doi: 10.3390/ijms151119741
Guo, S.-J., Zhou, H.-Y., Zhang, X.-S., Li, X.-G., and Meng, Q.-W. (2007). Overexpression of CaHSP26 in transgenic tobacco alleviates photoinhibition of PSII and PSI during chilling stress under low irradiance. J. Plant Physiol. 164, 126–136. doi: 10.1016/j.jplph.2006.01.004
Gupta, S. C., Sharma, A., Mishra, M., Mishra, R. K., and Chowdhuri, D. K. (2010). Heat shock proteins in toxicology: how close and how far? Life Sci. 86, 377–384. doi: 10.1016/j.lfs.2009.12.015
Guy, C. (1999). Molecular responses of plants to cold shock and cold acclimation. J. Mol. Microbiol. Biotechnol. 1, 231–242.
Guy, C., Kaplan, F., Kopka, J., Selbig, J., and Hincha, D. K. (2008). Metabolomics of temperature stress. Physiol. Plant. 132, 220–235. doi: 10.1111/j.1399-3054.2007.00999.x
Hahn, A., Bublak, D., Schleiff, E., and Scharf, K.-D. (2011). Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23, 741–755. doi: 10.1105/tpc.110.076018
Han, K., Jeong, H.-J., Yang, H.-B., Kang, S.-M., Kwon, J.-K., Kim, S., et al. (2016). An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 23, 81–91. doi: 10.1093/dnares/dsv038
Hansen, J., Sato, M., Hearty, P., Ruedy, R., Kelley, M., Masson-Delmotte, V., et al. (2016). Ice melt, sea level rise and superstorms: evidence from paleoclimate data, climate modeling, and modern observations that 2 C global warming could be dangerous. Atmos. Chem. Phys. 16, 3761–3812.
Haq, S. U., Khan, A., Ali, M., Gai, W.-X., Zhang, H.-X., Yu, Q.-H., et al. (2019). Knockdown of CaHSP60-6 confers enhanced sensitivity to heat stress in pepper (Capsicum annuum L.). Planta 250, 2127–2145. doi: 10.1007/s00425-019-03290-4
He, M., He, C.-Q., and Ding, N.-Z. (2018). Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 9:1771. doi: 10.3389/fpls.2018.01771
Holmes, S. C., Wells, D. E., Pickens, J. M., and Kemble, J. M. (2019). Selection of heat tolerant lettuce (Lactuca sativa L.) cultivars grown in deep water culture and their marketability. Horticulturae 5:50.
Huang, Y., Xuan, H., Yang, C., Guo, N., Wang, H., Zhao, J., et al. (2019). GmHsp90A2 is involved in soybean heat stress as a positive regulator. Plant Sci. 285, 26–33. doi: 10.1016/j.plantsci.2019.04.016
Hwang, B. J., Park, W. J., Chung, C. H., and Goldberg, A. L. (1987). Escherichia coli contains a soluble ATP-dependent protease (Ti) distinct from protease La. Proc. Natl. Acad. Sci. U.S.A. 84, 5550–5554. doi: 10.1073/pnas.84.16.5550
Imamoglu, R., Balchin, D., Hayer-Hartl, M., and Hartl, F. U. (2020). Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein. Nat. Commun. 11:365. doi: 10.1038/s41467-019-14245-4
Jackson-Constan, D., Akita, M., and Keegstra, K. (2001). Molecular chaperones involved in chloroplast protein import. Biochim. Biophys. Acta 1541, 102–113. doi: 10.1016/s0167-4889(01)00148-3
Jacob, P., Hirt, H., and Bendahmane, A. (2017). The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 15, 405–414. doi: 10.1111/pbi.12659
Jha, U. C., Nayyar, H., Palakurthi, R., Jha, R., Valluri, V., Bajaj, P., et al. (2021). Major QTLs and potential candidate genes for heat stress tolerance identified in Chickpea (Cicer arietinum L.). Front. Plant Sci. 12:655103. doi: 10.3389/fpls.2021.655103
Jo, J., Purushotham, P. M., Han, K., Lee, H.-R., Nah, G., and Kang, B.-C. (2017). Development of a genetic map for onion (Allium cepa L.) using reference-free genotyping-by-sequencing and SNP assays. Front. Plant Sci. 8:1606. doi: 10.3389/fpls.2017.01606
Jung, K.-H., Gho, H.-J., Nguyen, M. X., Kim, S.-R., and An, G. (2013). Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress. Funct. Integr. Genomics 13, 391–402. doi: 10.1007/s10142-013-0331-6
Kadota, Y., and Shirasu, K. (2012). The HSP90 complex of plants. Biochim. Biophys. Acta 1823, 689–697.
Kadyrzhanova, D. K., Vlachonasios, K. E., Ververidis, P., and Dilley, D. R. (1998). Molecular cloning of a novel heat induced/chilling tolerance related cDNA 1 in tomato fruit by use of mRNA differential display 2. Plant Mol. Biol. 36, 885–895. doi: 10.1023/a:1005954909011
Kang, Y., Jang, S.-W., Lee, H. J., Barchenger, D. W., and Jang, S. (2021). Expression profiling of heat shock protein genes as putative early heat-responsive members in lettuce. Horticulturae 7:312.
Katayama-Fujimura, Y., Gottesman, S., and Maurizi, M. (1987). A multiple-component, ATP-dependent protease from Escherichia coli. J. Biol. Chem. 262, 4477–4485.
Katiyar-Agarwal, S., Agarwal, M., and Grover, A. (2003). Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol. Biol. 51, 677–686. doi: 10.1023/a:1022561926676
Kim, M., Lee, U., Small, I., Des Francs-Small, C. C., and Vierling, E. (2012). Mutations in an Arabidopsis mitochondrial transcription termination factor–related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. Plant Cell 24, 3349–3365. doi: 10.1105/tpc.112.101006
Kim, Y.-C., Kang, Y., Yang, E.-Y., Cho, M.-C., Schafleitner, R., Lee, J. H., et al. (2021). Applications and major achievements of genome editing in vegetable crops: a review. Front. Plant Sci. 12:688980. doi: 10.3389/fpls.2021.688980
Königshofer, H., Tromballa, H. W., and LØppert, H. G. (2008). Early events in signalling high-temperature stress in tobacco BY2 cells involve alterations in membrane fluidity and enhanced hydrogen peroxide production. Plant Cell Environ. 31, 1771–1780. doi: 10.1111/j.1365-3040.2008.01880.x
Koornneef, M., and Meinke, D. (2010). The development of Arabidopsis as a model plant. Plant J. 61, 909–921.
Kotak, S., Larkindale, J., Lee, U., Von Koskull-Döring, P., Vierling, E., and Scharf, K.-D. (2007). Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10, 310–316. doi: 10.1016/j.pbi.2007.04.011
Krishna, P., and Gloor, G. (2001). The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6, 238–246.
Kumar, S. V., and Wigge, P. A. (2010). H2A. Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147. doi: 10.1016/j.cell.2009.11.006
Lamaoui, M., Jemo, M., Datla, R., and Bekkaoui, F. (2018). Heat and drought stresses in crops and approaches for their mitigation. Front. Chem. 6:26. doi: 10.3389/fchem.2018.00026
Lämke, J., Brzezinka, K., Altmann, S., and Bäurle, I. (2016). A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 35, 162–175. doi: 10.15252/embj.201592593
Latterich, M., and Patel, S. (1998). The AAA team: related ATPases with diverse functions. Trends Cell Biol. 8, 65–71.
Lee, H.-J., and Seo, P. J. (2021). Ca2+ talyzing initial responses to environmental stresses. Trends Plant Sci. 26, 849–870. doi: 10.1016/j.tplants.2021.02.007
Lee, H. Y., Jang, S., Yu, C. R., Kang, B. C., Chin, J. H., and Song, K. (2020). Population structure and genetic diversity of Cucurbita moschata based on genome-wide high-quality SNPs. Plants 10:56. doi: 10.3390/plants10010056
Lee, J. H., Hübel, A., and Schöffl, F. (1995). Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermotolerance in transgenic Arabidopsis. Plant J. 8, 603–612. doi: 10.1046/j.1365-313x.1995.8040603.x
Lee, J. R., Lee, S. S., Jang, H. H., Lee, Y. M., Park, J. H., Park, S.-C., et al. (2009). Heat-shock dependent oligomeric status alters the function of a plant-specific thioredoxin-like protein, AtTDX. Proc. Natl. Acad. Sci. U.S.A. 106, 5978–5983. doi: 10.1073/pnas.0811231106
Lee, K., Leister, D., and Kleine, T. (2021). Arabidopsis mitochondrial transcription termination factor mTERF2 promotes splicing of group IIB introns. Cells 10:315. doi: 10.3390/cells10020315
Lee, U., Rioflorido, I., Hong, S. W., Larkindale, J., Waters, E. R., and Vierling, E. (2007). The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. Plant J. 49, 115–127. doi: 10.1111/j.1365-313X.2006.02940.x
Lee, Y., Nagao, R. T., and Key, J. L. (1994). A soybean 101-kD heat shock protein complements a yeast HSP104 deletion mutant in acquiring thermotolerance. Plant Cell 6, 1889–1897. doi: 10.1105/tpc.6.12.1889
Li, H. Y., Chang, C. S., Lu, L. S., Liu, C. A., Chan, M.-T., and Charng, Y.-Y. (2003). Over-expression of Arabidopsis thaliana heat shock factor gene (AtHsfA1b) enhances chilling tolerance in transgenic tomato (vol 44, pg 129, 2003). Bot. Bull. Acad. Sin. 44, 129–140.
Li, M., Ji, L., Jia, Z., Yang, X., Meng, Q., and Guo, S. (2018). Constitutive expression of CaHSP22.5 enhances chilling tolerance in transgenic tobacco by promoting the activity of antioxidative enzymes. Funct. Plant Biol. 45, 575–585. doi: 10.1071/FP17226
Li, X.-M., Chao, D.-Y., Wu, Y., Huang, X., Chen, K., Cui, L.-G., et al. (2015). Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 47, 827–833. doi: 10.1038/ng.3305
Lin, Q., Xie, Y., Guan, W., Duan, Y., Wang, Z., and Sun, C. (2019). Combined transcriptomic and proteomic analysis of cold stress induced sugar accumulation and heat shock proteins expression during postharvest potato tuber storage. Food Chem. 297:124991. doi: 10.1016/j.foodchem.2019.124991
Lindquist, S. (1986). The heat-shock response. Annu. Rev. Biochem. 55, 1151–1191. doi: 10.1098/rstb.2016.0525
Liu, D., Hu, R., Palla, K. J., Tuskan, G. A., and Yang, X. (2016). Advances and perspectives on the use of CRISPR/Cas9 systems in plant genomics research. Curr. Opin. Plant Biol. 30, 70–77. doi: 10.1016/j.pbi.2016.01.007
Liu, J., Zhang, C., Wei, C., Liu, X., Wang, M., Yu, F., et al. (2016). The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice. Plant Physiol. 170, 429–443. doi: 10.1104/pp.15.00879
Liu, H. T., Gao, F., Li, G. L., Han, J. L., Liu, D. L., Sun, D. Y., et al. (2008). The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J. 55, 760–773. doi: 10.1111/j.1365-313X.2008.03544.x
Liu, H. T., Li, G. L., Chang, H., Sun, D. Y., Zhou, R. G., and Li, B. (2007). Calmodulin-binding protein phosphatase PP7 is involved in thermotolerance in Arabidopsis. Plant Cell Environ. 30, 156–164. doi: 10.1111/j.1365-3040.2006.01613.x
Liu, R., Su, Z., Zhou, H., Huang, Q., Fan, S., Liu, C., et al. (2020). LsHSP70 is induced by high temperature to interact with calmodulin, leading to higher bolting resistance in lettuce. Sci. Rep. 10:15155. doi: 10.1038/s41598-020-72443-3
Liu, Y., Xiao, S., Sun, H., Pei, L., Liu, Y., Peng, L., et al. (2020). AtPPRT1, an E3 ubiquitin ligase, enhances the thermotolerance in Arabidopsis. Plants 9:1074. doi: 10.3390/plants9091074
Liu, Y., Gampert, L., Nething, K., and Steinacker, J. M. (2006). Response and function of skeletal muscle heat shock protein 70. Front. Biosci. 11, 2802–2827. doi: 10.2741/2011
Lopes-Caitar, V. S., De Carvalho, M. C. C. G., Darben, L. M., Kuwahara, M. K., Nepomuceno, A. L., Dias, W. P., et al. (2013). Genome-wide analysis of the Hsp 20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genomics 14:577. doi: 10.1186/1471-2164-14-577
Machanick, P., and Bailey, T. L. (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697.
Mahesh, U., Mamidala, P., Rapolu, S., Aragao, F. J. L., Souza, M. T., Rao, P. J. M., et al. (2013). Constitutive overexpression of small HSP24.4 gene in transgenic tomato conferring tolerance to high-temperature stress. Mol. Breed. 32, 687–697.
Malerba, M., Crosti, P., and Cerana, R. (2010). Effect of heat stress on actin cytoskeleton and endoplasmic reticulum of tobacco BY-2 cultured cells and its inhibition by Co 2+. Protoplasma 239, 23–30. doi: 10.1007/s00709-009-0078-z
Malik, M. K., Slovin, J. P., Hwang, C. H., and Zimmerman, J. L. (1999). Modified expression of a carrot small heat shock protein gene, Hsp17.7, results in increased or decreased thermotolerance ‡. Plant J. 20, 89–99.
Marcu, M. G., Doyle, M., Bertolotti, A., Ron, D., Hendershot, L., and Neckers, L. (2002). Heat shock protein 90 modulates the unfolded protein response by stabilizing IRE1alpha. Mol. Cell Biol. 22, 8506–8513. doi: 10.1128/MCB.22.24.8506-8513.2002
Martin, J., Horwich, A. L., and Hartl, F. U. (1992). Prevention of protein denaturation under heat stress by the chaperonin Hsp60. Science 258, 995–998. doi: 10.1126/science.1359644
Matsuura, H., Ishibashi, Y., Shinmyo, A., Kanaya, S., and Kato, K. (2010). Genome-wide analyses of early translational responses to elevated temperature and high salinity in Arabidopsis thaliana. Plant Cell Physiol. 51, 448–462. doi: 10.1093/pcp/pcq010
Mayer, M. P. (2010). Gymnastics of molecular chaperones. Mol. Cell 39, 321–331. doi: 10.1016/j.molcel.2010.07.012
McClung, C. R., and Davis, S. J. (2010). Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Curr. Biol. 20, R1086–R1092. doi: 10.1016/j.cub.2010.10.035
Meng, Q., Li, B. X., and Xiao, X. (2018). Toward developing chemical modulators of Hsp60 as potential therapeutics. Front. Mol. Biosci. 5:35. doi: 10.3389/fmolb.2018.00035
Miernyk, J. A. (1999). Protein folding in the plant cell. Plant Physiol. 121, 695–703. doi: 10.1104/pp.121.3.695
Milioni, D., and Hatzopoulos, P. (1997). Genomic organization of hsp90 gene family in Arabidopsis. Plant Mol. Biol. 35, 955–961. doi: 10.1023/a:1005874521528
Miller, G., Schlauch, K., Tam, R., Cortes, D., Torres, M. A., Shulaev, V., et al. (2009). The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2:ra45. doi: 10.1126/scisignal.2000448
Miller, G., Suzuki, N., Rizhsky, L., Hegie, A., Koussevitzky, S., and Mittler, R. (2007). Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 144, 1777–1785. doi: 10.1104/pp.107.101436
Mishra, R. C., and Grover, A. (2014). Intergenic sequence between Arabidopsis caseinolytic protease B-cytoplasmic/heat shock protein100 and choline kinase genes functions as a heat-inducible bidirectional promoter. Plant Physiol. 166, 1646–1658. doi: 10.1104/pp.114.250787
Mishra, R. C., and Grover, A. (2016). ClpB/Hsp100 proteins and heat stress tolerance in plants. Crit. Rev. Biotechnol. 36, 862–874. doi: 10.3109/07388551.2015.1051942
Mishra, R. W. E., and Grover, A. (2019). Voyaging around ClpB/Hsp100 proteins and plant heat tolerance. Proc. Indian Natl. Sci. Acad. 85, 791–802.
Mishra, S. K., Tripp, J., Winkelhaus, S., Tschiersch, B., Theres, K., Nover, L., et al. (2002). In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 16, 1555–1567. doi: 10.1101/gad.228802
Mittler, R., Finka, A., and Goloubinoff, P. (2012). How do plants feel the heat? Trends Biochem. Sci. 37, 118–125.
Mogk, A., Haslberger, T., Tessarz, P., and Bukau, B. (2008). Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem. Soc. Trans. 36, 120–125. doi: 10.1042/BST0360120
Morimoto, R. I. (1998). Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12, 3788–3796. doi: 10.1101/gad.12.24.3788
Morrow, G., and Tanguay, R. M. (2012). Small heat shock protein expression and functions during development. Int. J. Biochem. Cell Biol. 44, 1613–1621. doi: 10.1016/j.biocel.2012.03.009
Mu, C., Zhang, S., Yu, G., Chen, N., Li, X., and Liu, H. (2013). Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS One 8:e82264. doi: 10.1371/journal.pone.0082264
Müller, J., Menzel, D., and Šamaj, J. (2007). Cell-type-specific disruption and recovery of the cytoskeleton in Arabidopsis thaliana epidermal root cells upon heat shock stress. Protoplasma 230, 231–242. doi: 10.1007/s00709-006-0239-2
Nakajima, Y., and Suzuki, S. (2013). Environmental stresses induce misfolded protein aggregation in plant cells in a microtubule-dependent manner. Int. J. Mol. Sci. 14, 7771–7783. doi: 10.3390/ijms14047771
Narberhaus, F., Waldminghaus, T., and Chowdhury, S. (2006). RNA thermometers. FEMS Microbiol. Rev. 30, 3–16.
Nguyen, B., Ma, R., Tang, W. K., Shi, D., and Tolia, N. H. (2021). Crystal structure of P. falciparum Cpn60 bound to ATP reveals an open dynamic conformation before substrate binding. Sci. Rep. 11:5930. doi: 10.1038/s41598-021-85197-3
Nieto-Sotelo, J., Kannan, K., Martınez, L., and Segal, C. (1999). Characterization of a maize heat-shock protein 101 gene, HSP101, encoding a ClpB/Hsp100 protein homologue. Gene 230, 187–195. doi: 10.1016/s0378-1119(99)00060-8
Nover, L., Bharti, K., Döring, P., Mishra, S. K., Ganguli, A., and Scharf, K.-D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2
Ohama, N., Sato, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65.
Olas, J. J., Apelt, F., Annunziata, M. G., John, S., Richard, S. I., Gupta, S., et al. (2021). Primary carbohydrate metabolism genes participate in heat-stress memory at the shoot apical meristem of Arabidopsis thaliana. Mol. Plant 14, 1508–1524. doi: 10.1016/j.molp.2021.05.024
Ortiz, R., Braun, H.-J., Crossa, J., Crouch, J. H., Davenport, G., Dixon, J., et al. (2008). Wheat genetic resources enhancement by the International Maize and Wheat Improvement Center (CIMMYT). Genet. Resour. Crop Evol. 55, 1095–1140.
Osakabe, Y., Arinaga, N., Umezawa, T., Katsura, S., Nagamachi, K., Tanaka, H., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25, 609–624. doi: 10.1105/tpc.112.105700
Palakolanu, S. R., Chakradhar, T., Reddy, R. A., Nitnavare, R. B., Mahanty, S., and Reddy, M. K. (2016). “Role of heat shock proteins in improving heat stress tolerance in crop plants,” in Heat Shock Proteins and Plants. Heat Shock Proteins, Vol. 10, eds A. Asea, P. Kaur, and S. Calderwood (Cham: Springer), 283–307.
Pall, G. S., and Hamilton, A. J. (2008). Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3, 1077–1084. doi: 10.1038/nprot.2008.67
Pan, G., Tian, S., Nie, J., Yang, C., Ruotti, V., Wei, H., et al. (2007). Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1, 299–312. doi: 10.1016/j.stem.2007.08.003
Park, S. K., Jung, Y. J., Lee, J. R., Lee, Y. M., Jang, H. H., Lee, S. S., et al. (2009). Heat-shock and redox-dependent functional switching of an h-type Arabidopsis thioredoxin from a disulfide reductase to a molecular chaperone. Plant Physiol. 150, 552–561. doi: 10.1104/pp.109.135426
Parsell, D., and Lindquist, S. (1993). The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Ann. Rev. Genet. 27, 437–496. doi: 10.1146/annurev.ge.27.120193.002253
Pearl, L. H., and Prodromou, C. (2006). Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75, 271–294. doi: 10.1146/annurev.biochem.75.103004.142738
Pham, D., Hoshikawa, K., Fujita, S., Fukumoto, S., Hirai, T., Shinozaki, Y., et al. (2020). A tomato heat-tolerant mutant shows improved pollen fertility and fruit-setting under long-term ambient high temperature. Environ. Exp. Bot. 178:104150.
Picard, D. (2002). Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. 59, 1640–1648. doi: 10.1007/pl00012491
Pipaón, S., Gragera, M., Bueno-Carrasco, M. T., García-Bernalt Diego, J., Cantero, M., Cuéllar, J., et al. (2021). Chaperonins: nanocarriers with biotechnological applications. Nanomaterials 11:503. doi: 10.3390/nano11020503
Pramanik, D., Shelake, R. M., Kim, M. J., and Kim, J.-Y. (2020). CRISPR-mediated engineering across the central dogma in plant biology for basic research and crop improvement. Mol. Plant 14, 127–150. doi: 10.1016/j.molp.2020.11.002
Pratt, W. B., and Toft, D. O. (2003). Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228, 111–133. doi: 10.1177/153537020322800201
Queitsch, C., Hong, S.-W., Vierling, E., and Lindquist, S. (2000). Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12, 479–492. doi: 10.1105/tpc.12.4.479
Radons, J. (2016). The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones 21, 379–404. doi: 10.1007/s12192-016-0676-6
Rampino, P., Mita, G., Pataleo, S., De Pascali, M., Di Fonzo, N., and Perrotta, C. (2009). Acquisition of thermotolerance and HSP gene expression in durum wheat (Triticum durum Desf.) cultivars. Environ. Exp. Bot. 66, 257–264.
Reddy, K., and Hodges, H. (2000). Climate Change and Global Crop Productivity, eds K. R. Reddy and H. F. Hodges (Wallingford: CABI Publishing).
Rensink, W. A., and Buell, C. R. (2004). Arabidopsis to rice. Applying knowledge from a weed to enhance our understanding of a crop species. Plant Physiol. 135, 622–629. doi: 10.1104/pp.104.040170
Rodríguez, V. M., Soengas, P., Alonso-Villaverde, V., Sotelo, T., Cartea, M. E., and Velasco, P. (2015). Effect of temperature stress on the early vegetative development of Brassica oleracea L. BMC Plant Biol. 15:145. doi: 10.1186/s12870-015-0535-0
Rucińiska-Sobkowiak, R. (2010). [Oxidative stress in plants exposed to heavy metals]. Postepy Biochem. 56, 191–200.
Ruelland, E., and Zachowski, A. (2010). How plants sense temperature. Environ. Exp. Bot. 69, 225–232.
Rykaczewska, K. (2017). Impact of heat and drought stresses on size and quality of the potato yield. Plant Soil Environ. 63, 40–46.
Sabehat, A., Lurie, S., and Weiss, D. (1998). Expression of small heat-shock proteins at low temperatures: a possible role in protecting against chilling injuries. Plant Physiol. 117, 651–658. doi: 10.1104/pp.117.2.651
Sabehat, A., Weiss, D., and Lurie, S. (1996). The correlation between heat-shock protein accumulation and persistence and chilling tolerance in tomato fruit. Plant Physiol. 110, 531–537. doi: 10.1104/pp.110.2.531
Saidi, Y., Finka, A., Muriset, M., Bromberg, Z., Weiss, Y. G., Maathuis, F. J., et al. (2009). The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 21, 2829–2843. doi: 10.1105/tpc.108.065318
Sanchez, Y., and Lindquist, S. L. (1990). HSP104 required for induced thermotolerance. Science 248, 1112–1115. doi: 10.1126/science.2188365
Sanghera, G. S., Wani, S. H., Hussain, W., and Singh, N. B. (2011). Engineering cold stress tolerance in crop plants. Curr. Genomics 12, 30–43. doi: 10.2174/138920211794520178
Sato, H., Mizoi, J., Tanaka, H., Maruyama, K., Qin, F., Osakabe, Y., et al. (2014). Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 26, 4954–4973. doi: 10.1105/tpc.114.132928
Sauer, R. T., Bolon, D. N., Burton, B. M., Burton, R. E., Flynn, J. M., Grant, R. A., et al. (2004). Sculpting the proteome with AAA+ proteases and disassembly machines. Cell 119, 9–18. doi: 10.1016/j.cell.2004.09.020
Scharf, K.-D., Berberich, T., Ebersberger, I., and Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta 1819, 104–119.
Schelin, J., Lindmark, F., and Clarke, A. K. (2002). The clpP multigene family for the ATP-dependent Clp protease in the cyanobacterium Synechococcus c cThe GenBank accession numbers for the sequences of clpPII-clpX and clpR-clpPIII reported in this paper are U92039 and AJ132005, respectively. Microbiology 148, 2255–2265.
Schirmer, E. C., Glover, J. R., Singer, M. A., and Lindquist, S. (1996). HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21, 289–296.
Schirmer, E. C., Lindquist, S., and Vierling, E. (1994). An Arabidopsis heat shock protein complements a thermotolerance defect in yeast. Plant Cell 6, 1899–1909. doi: 10.1105/tpc.6.12.1899
Schleiff, E., and Becker, T. (2011). Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 12, 48–59. doi: 10.1038/nrm3027
Schlieker, C., Zentgraf, H., Dersch, P., and Mogk, A. (2005). ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol. Chem. 386, 1115–1127. doi: 10.1515/BC.2005.128
Schramm, F., Larkindale, J., Kiehlmann, E., Ganguli, A., Englich, G., Vierling, E., et al. (2008). A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 53, 264–274. doi: 10.1111/j.1365-313X.2007.03334.x
Seo, J. S., Lee, Y.-M., Park, H. G., and Lee, J.-S. (2006). The intertidal copepod Tigriopus japonicus small heat shock protein 20 gene (Hsp20) enhances thermotolerance of transformed Escherichia coli. Biochem. Biophys. Res. Commun. 340, 901–908. doi: 10.1016/j.bbrc.2005.12.086
Sharma, S., Reddy, P. V. J., Rohilla, M. S., and Tiwari, P. K. (2006). Expression of HSP60 homologue in sheep blowfly Lucilia cuprina during development and heat stress. J. Therm. Biol. 31, 546–555.
Shen, L., Liang, Z., Wong, C. E., and Yu, H. (2019). Messenger RNA modifications in plants. Trends Plant Sci. 24, 328–341. doi: 10.1016/j.tplants.2019.01.005
Shiber, A., and Ravid, T. (2014). Chaperoning proteins for destruction: diverse roles of Hsp70 chaperones and their co-chaperones in targeting misfolded proteins to the proteasome. Biomolecules 4, 704–724. doi: 10.3390/biom4030704
Shim, S., Park, C.-M., and Seo, P. J. (2021). iRegNet: an integrative regulatory network analysis tool for Arabidopsis thaliana. Plant Physiol. 187, 1292–1299. doi: 10.1093/plphys/kiab389
Singh, A., Singh, U., Mittal, D., and Grover, A. (2010). Genome-wide analysis of rice ClpB/HSP100, ClpC and ClpD genes. BMC Genomics 11:95. doi: 10.1186/1471-2164-11-95
Smertenko, A., DrÃber, P., Viklický, V., and Opatrný, Z. (1997). Heat stress affects the organization of microtubules and cell division in Nicotiana tabacum cells. Plant Cell Environ. 20, 1534–1542.
Squires, C., Pedersen, S., Ross, B., and Squires, C. (1991). ClpB is the Escherichia coli heat shock protein F84. 1. J. Bacteriol. 173, 4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W.-R., and Bäurle, I. (2014a). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807. doi: 10.1105/tpc.114.123851
Stief, A., Brzezinka, K., Lämke, J., and Bäurle, I. (2014b). Epigenetic responses to heat stress at different time scales and the involvement of small RNAs. Plant Signal. Behav. 9:e970430. doi: 10.4161/15592316.2014.970430
Sun, L., Liu, Y., Kong, X., Zhang, D., Pan, J., Zhou, Y., et al. (2012). ZmHSP16.9, a cytosolic class I small heat shock protein in maize (Zea mays), confers heat tolerance in transgenic tobacco. Plant Cell Rep. 31, 1473–1484. doi: 10.1007/s00299-012-1262-8
Sung, D. Y., and Guy, C. L. (2003). Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiol. 132, 979–987. doi: 10.1104/pp.102.019398
Sung, D.-Y., Kaplan, F., Lee, K.-J., and Guy, C. L. (2003). Acquired tolerance to temperature extremes. Trends Plant Sci. 8, 179–187. doi: 10.1016/S1360-1385(03)00047-5
Suri, S. S., and Dhindsa, R. S. (2008). A heat-activated MAP kinase (HAMK) as a mediator of heat shock response in tobacco cells. Plant Cell Environ. 31, 218–226. doi: 10.1111/j.1365-3040.2007.01754.x
Suzuki, N., Koussevitzky, S., Mittler, R., and Miller, G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. doi: 10.1111/j.1365-3040.2011.02336.x
Taipale, M., Jarosz, D. F., and Lindquist, S. (2010). HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528. doi: 10.1038/nrm2918
Tessarz, P., Mogk, A., and Bukau, B. (2008). Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 68, 87–97. doi: 10.1111/j.1365-2958.2008.06135.x
Theocharis, A., Clément, C., and Ait Barka, E. (2012). Physiological and molecular changes in plants at low temperatures. Planta 235, 1091–1105.
Tian, F., Gong, J., Zhang, J., Feng, Y., Wang, G., Guo, Q., et al. (2014). Overexpression of monoubiquitin improves photosynthesis in transgenic tobacco plants following high temperature stress. Plant Sci. 226, 92–100. doi: 10.1016/j.plantsci.2014.03.006
Tian, F., Hu, X.-L., Yao, T., Yang, X., Chen, J.-G., Lu, M.-Z., et al. (2021). Recent advances in the roles of HSFs and HSPs in heat stress response in woody plants. Front. Plant Sci. 12:704905. doi: 10.3389/fpls.2021.704905
Tubiello, F. N., Soussana, J. F., and Howden, S. M. (2007). Crop and pasture response to climate change. Proc. Natl. Acad. Sci. U.S.A. 104, 19686–19690. doi: 10.1073/pnas.0701728104
Ul Haq, S., Khan, A., Ali, M., Khattak, A. M., Gai, W. X., Zhang, H. X., et al. (2019). Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 20:5321. doi: 10.3390/ijms20215321
Usman, M. G., Rafii, M., Mohammad Yusoff, M., Yusuff, O., Ismail, M., and Miah, G. (2017). Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnol. Genet. Eng. Rev. 33, 26–39. doi: 10.1080/02648725.2017.1340546
Usman, M. G., Rafii, M. Y., Ismail, M. R., Malek, M. A., and Latif, M. A. (2015). Expression of target gene Hsp70 and membrane stability determine heat tolerance in chili pepper. J. Am. Soc. Hortic. Sci. 140, 144–150.
van Oort, P. A. J., and Zwart, S. J. (2018). Impacts of climate change on rice production in Africa and causes of simulated yield changes. Glob. Change Biol. 24, 1029–1045. doi: 10.1111/gcb.13967
Vierling, E. (1991). The roles of heat shock proteins in plants. Annu. Rev. Plant Biol. 42, 579–620.
von Koskull-Döring, P., Scharf, K.-D., and Nover, L. (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. doi: 10.1016/j.tplants.2007.08.014
Wahid, A., Gelani, S., Ashraf, M., and Foolad, M. R. (2007). Heat tolerance in plants: an overview. Environ. Exp. Bot. 61, 199–223.
Wang, A., Yu, X., Mao, Y., Liu, Y., Liu, G., Liu, Y., et al. (2015). Overexpression of a small heat-shock-protein gene enhances tolerance to abiotic stresses in rice. Plant Breed. 134, 384–393.
Wang, K. H., Sauer, R. T., and Baker, T. A. (2007). ClpS modulates but is not essential for bacterial N-end rule degradation. Genes Dev. 21, 403–408.
Wang, L., Guo, Y., Jia, L., Chu, H., Zhou, S., Chen, K., et al. (2014). Hydrogen peroxide acts upstream of nitric oxide in the heat shock pathway in Arabidopsis seedlings. Plant Physiol. 164, 2184–2196. doi: 10.1104/pp.113.229369
Wang, L., Zhao, C. M., Wang, Y. J., and Liu, J. (2005). Overexpression of chloroplast-localized small molecular heat-shock protein enhances chilling tolerance in tomato plant. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 31, 167–174.
Wang, M., Zou, Z., Li, Q., Xin, H., Zhu, X., Chen, X., et al. (2017). Heterologous expression of three Camellia sinensis small heat shock protein genes confers temperature stress tolerance in yeast and Arabidopsis thaliana. Plant Cell Rep. 36, 1125–1135. doi: 10.1007/s00299-017-2143-y
Wang, R., Zhang, Y., Kieffer, M., Yu, H., Kepinski, S., and Estelle, M. (2016). HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun. 7:10269. doi: 10.1038/ncomms10269
Wang, X., Yan, B., Shi, M., Zhou, W., Zekria, D., Wang, H., et al. (2016). Overexpression of a Brassica campestris HSP70 in tobacco confers enhanced tolerance to heat stress. Protoplasma 253, 637–645. doi: 10.1007/s00709-015-0867-5
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Waters, E. R., and Vierling, E. (2020). Plant small heat shock proteins – evolutionary and functional diversity. New Phytol. 227, 24–37. doi: 10.1111/nph.16536
Weibezahn, J., Tessarz, P., Schlieker, C., Zahn, R., Maglica, Z., Lee, S., et al. (2004). Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665. doi: 10.1016/j.cell.2004.11.027
Xiao, J., Lee, U.-S., and Wagner, D. (2016). Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 34, 41–53. doi: 10.1016/j.pbi.2016.08.002
Xu, F. Q., and Xue, H. W. (2019). The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 42, 2931–2944. doi: 10.1111/pce.13633
Xu, J., Xue, C., Xue, D., Zhao, J., Gai, J., Guo, N., et al. (2013). Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS One 8:e69810. doi: 10.1371/journal.pone.0069810
Xu, Z., Horwich, A. L., and Sigler, P. B. (1997). The crystal structure of the asymmetric GroEL–GroES–(ADP)7 chaperonin complex. Nature 388, 741–750. doi: 10.1038/41944
Xu, Z.-S., Li, Z.-Y., Chen, Y., Chen, M., Li, L.-C., and Ma, Y.-Z. (2012). Heat shock protein 90 in plants: molecular mechanisms and roles in stress responses. Int. J. Mol. Sci. 13, 15706–15723. doi: 10.3390/ijms131215706
Xuan, Y., Zhou, S., Wang, L., Cheng, Y., and Zhao, L. (2010). Nitric oxide functions as a signal and acts upstream of AtCaM3 in thermotolerance in Arabidopsis seedlings. Plant Physiol. 153, 1895–1906. doi: 10.1104/pp.110.160424
Yamada, K., Fukao, Y., Hayashi, M., Fukazawa, M., Suzuki, I., and Nishimura, M. (2007). Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J. Biol. Chem. 282, 37794–37804. doi: 10.1074/jbc.M707168200
Yamaguchi, N. (2021). Heat memory in plants: histone modifications, nucleosome positioning and miRNA accumulation alter heat memory gene expression. Genes Genet. Syst. 96, 229–235. doi: 10.1266/ggs.21-00040
Yamaguchi, N., and Ito, T. (2021). JMJ histone demethylases balance H3K27me3 and H3K4me3 levels at the HSP21 locus during heat acclimation in Arabidopsis. Biomolecules 11:852. doi: 10.3390/biom11060852
Yamaguchi, N., Matsubara, S., Yoshimizu, K., Seki, M., Hamada, K., Kamitani, M., et al. (2021). H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat. Commun. 12:3480. doi: 10.1038/s41467-021-23766-w
Yang, J.-Y., Sun, Y., Sun, A.-Q., Yi, S.-Y., Qin, J., Li, M.-H., et al. (2006). The involvement of chloroplast HSP100/ClpB in the acquired thermotolerance in tomato. Plant Mol. Biol. 62, 385–395. doi: 10.1007/s11103-006-9027-9
Yoshida, T., Ohama, N., Nakajima, J., Kidokoro, S., Mizoi, J., Nakashima, K., et al. (2011). Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics 286, 321–332. doi: 10.1007/s00438-011-0647-7
Yoshida, T., Sakuma, Y., Todaka, D., Maruyama, K., Qin, F., Mizoi, J., et al. (2008). Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 368, 515–521. doi: 10.1016/j.bbrc.2008.01.134
Young, T. E., Ling, J., Geisler-Lee, C. J., Tanguay, R. L., Caldwell, C., and Gallie, D. R. (2001). Developmental and thermal regulation of the maize heat shock protein, HSP101. Plant Physiol. 127, 777–791.
Yu, J., Cheng, Y., Feng, K., Ruan, M., Ye, Q., Wang, R., et al. (2016). Genome-wide identification and expression profiling of tomato Hsp20 gene family in response to biotic and abiotic stresses. Front. Plant Sci. 7:1215. doi: 10.3389/fpls.2016.01215
Zandalinas, S. I., Fritschi, F. B., and Mittler, R. (2021). Global warming, climate change, and environmental pollution: recipe for a multifactorial stress combination disaster. Trends Plant Sci. 26, 588–599. doi: 10.1016/j.tplants.2021.02.011
Zhang, H., Lang, Z., and Zhu, J.-K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhang, L., Hu, W., Gao, Y., Pan, H., and Zhang, Q. (2018). A cytosolic class II small heat shock protein, PfHSP17.2, confers resistance to heat, cold, and salt stresses in transgenic Arabidopsis. Genet. Mol. Biol. 41, 649–660. doi: 10.1590/1678-4685-GMB-2017-0206
Zhang, H., Zhu, J., Gong, Z., and Zhu, J.-K. (2021). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119.
Zhang, Y., Lai, X., Yang, S., Ren, H., Yuan, J., Jin, H., et al. (2021). Functional analysis of tomato CHIP ubiquitin E3 ligase in heat tolerance. Sci. Rep. 11:1713. doi: 10.1038/s41598-021-81372-8
Zhang, J., Chen, H., Wang, H., Li, B., Yi, Y., Kong, F., et al. (2016). Constitutive expression of a tomato small heat shock protein gene LeHSP21 improves tolerance to high-temperature stress by enhancing antioxidation capacity in tobacco. Plant Mol. Biol. Report. 34, 399–409.
Zhang, L., Zhang, Q., Gao, Y., Pan, H., Shi, S., and Wang, Y. (2014). Overexpression of heat shock protein gene PfHSP21.4 in Arabidopsis thaliana enhances heat tolerance. Acta Physiol. Plant. 36, 1555–1564.
Zhang, L., Zhao, H.-K., Dong, Q.-L., Zhang, Y.-Y., Wang, Y.-M., Li, H.-Y., et al. (2015). Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Front. Plant Sci. 6:773. doi: 10.3389/fpls.2015.00773
Zhang, N., Zhao, H., Shi, J., Wu, Y., and Jiang, J. (2020). Functional characterization of class I SlHSP17.7 gene responsible for tomato cold-stress tolerance. Plant Sci. 298:110568. doi: 10.1016/j.plantsci.2020.110568
Zhang, W., Zhou, R.-G., Gao, Y.-J., Zheng, S.-Z., Xu, P., Zhang, S.-Q., et al. (2009). Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 149, 1773–1784. doi: 10.1104/pp.108.133744
Zhou, J., Wang, J., Yu, J.-Q., and Chen, Z. (2014). Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 5:174.
Zhou, R., Li, B., Liu, H., and Sun, D. (2009). Progress in the participation of Ca2+–calmodulin in heat shock signal transduction. Prog. Nat. Sci. 19, 1201–1208.
Keywords: global warming, heat shock factor, heat shock protein, heat stress, thermotolerance, vegetables
Citation: Kang Y, Lee K, Hoshikawa K, Kang M and Jang S (2022) Molecular Bases of Heat Stress Responses in Vegetable Crops With Focusing on Heat Shock Factors and Heat Shock Proteins. Front. Plant Sci. 13:837152. doi: 10.3389/fpls.2022.837152
Received: 16 December 2021; Accepted: 09 March 2022;
Published: 11 April 2022.
Edited by:
Mostafa Abdelwahed Abdelrahman, Aswan University, EgyptReviewed by:
Shabir Hussain Wani, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaVivek Ambastha, Migal – Galilee Research Institute, Israel
Copyright © 2022 Kang, Lee, Hoshikawa, Kang and Jang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seonghoe Jang, seonghoe.jang@worldveg.org, orcid.org/0000-0001-5018-3480
‡These authors have contributed equally to this work
 Yeeun Kang1‡
Yeeun Kang1‡ Kwanuk Lee
Kwanuk Lee Seonghoe Jang
Seonghoe Jang