- College of Life Sciences, Qingdao University, Qingdao, China
Drought, salinity, extreme temperatures, and ultraviolet (UV) radiation are major environmental factors that adversely affect plant growth and crop production. As a protective shield covering the outer epidermal cell wall of plant aerial organs, the cuticle is mainly composed of cutin matrix impregnated and sealed with cuticular waxes, and greatly contributes to the plant adaption to environmental stresses. Past decades have seen considerable progress in uncovering the molecular mechanism of plant cutin and cuticular wax biosynthesis, as well as their important roles in plant stress adaptation, which provides a new direction to drive strategies for stress-resilient crop breeding. In this review, we highlighted the recent advances in cuticle biosynthesis in plant adaptation to drought, salinity, extreme temperatures, and UV radiation stress, and discussed the current status and future directions in harnessing cuticle biosynthesis for crop improvement.
Introduction
Growing population and consumption greatly increase the global crop demand. It is expected that 70% more food is needed to feed 10 billion people by 2050 (Tilman et al., 2011). However, plant growth and global crop production are adversely affected by unfavorable environmental conditions such as water deficit (drought), salinity, extreme temperatures, and ultraviolet (UV) radiation. For instance, 10.1% loss in national cereal production was reported in drought years from 1964 to 2007 (Lesk et al., 2016). Notably, drought stress led to 20.6 and 39.3% yield reduction in wheat and maize, respectively between 1980 and 2015 (Daryanto et al., 2016; Fahad et al., 2017). As an environmental factor causing land degradation, salinity affects about 11% of the global irrigated areas and could lead to above 50% yield loss in salt-sensitive crop species including all important glycophytic crops (Zörb et al., 2019). During the past century, extreme temperatures have become more frequent and seriously affected agriculture production. It was estimated that global cereal production in extreme heat years was reduced by 9.1% on average during 1964–2007 (Lesk et al., 2016). Low-temperature stresses such as chilling (0–18°C) and freezing (<0°C) temperature significantly affect spatial distribution and production in cold-sensitive crop species like rice (Shimono et al., 2007; Wang P. et al., 2019; Zhang et al., 2019a). Similarly, increased UV radiation could induce morphological changes, alter genome stability and affect yields in sensitive crop species (Robson et al., 2019). In addition, stratospheric ozone depletion and global climate change have contributed to the increased incidence and prolonged duration of these environmental stresses during past decades. Therefore, developing crop varieties adapted to drought, salinity, extreme temperatures, and UV radiation stress is essential to secure and enhance food production.
As the true interface between plant aerial parts and surrounding environments, lipophilic cuticle synthesized by plant epidermal cells protects plant tissue from environmental stresses such as desiccation, extreme temperatures, increased UV radiation, pathogen infections, and mechanical damages, and greatly contributes to the plant adaptation to stressful terrestrial habitats (Domínguez et al., 2011; Kong et al., 2020a). Although cuticle composition varies among plant species, organs, developmental stages and environmental conditions, the cuticle is generally composed of lipids, polysaccharides and phenolic compounds (Yeats and Rose, 2013). Cuticular lipid components mainly consist of wax, cutin, and cutan polymers, which confer cuticle the hydrophobic property. As the framework of the plant cuticle, cutin polymers contain a large amount of cross-linked polyester of oxygenated long-chain (C16 or C18) fatty acids and their derivatives (Philippe et al., 2020). Cuticular wax mixtures are mainly composed of very-long-chain (VLC, >C20) fatty acids, alcohols, aldehydes, alkanes, esters, and ketones (Lee and Suh, 2015; Wang et al., 2020; Suh et al., 2022). Compared with cutin and wax, cutan polymers are less explored in structure due to their non-hydrolysable bounds (Bhanot et al., 2021; Reynoud et al., 2021). Phenolic compounds identified in cuticle fraction include hydroxycinnamic acids such as ð-coumaric, ferulic, caffeic, and ð-hydroxybenzoic acids, as well as flavonoids in some species (Fernández et al., 2016). As an adaptive innovation in land plants, lipophilic cuticle could protect plant tissues from environmental stresses, thereby gaining increasing attention in the study of plant stress tolerance (Li and Chang, 2021). Herein, we summarized the most recent progress in studies of cuticle biosynthesis in plant adaptation to drought, salinity, extreme temperatures, and UV radiation stress, and discussed the potentials, challenges and strategies in exploiting cuticle biosynthesis for crop improvement.
Cuticle biosynthesis in model and crop plants
With the contribution of cuticle biosynthetic mutants and advanced cuticle phenotyping methods, cuticle biosynthetic pathways are extensively studied in the model plant Arabidopsis thaliana (Yeats and Rose, 2013; Suh et al., 2022). As summarized in previous reviews, cuticular wax and cutin are synthesized mainly by modification of plastid-derived C16 and C18 fatty acids in the endoplasmic reticulum (ER) of plant epidermal cells (Philippe et al., 2020; Wang et al., 2020). For the cutin monomer biosynthesis, plastid C16 and C18 fatty acids were conjugated to coenzyme A (CoA) by long-chain acyl-coenzyme A synthases (LACS) and then trafficked to the ER, where the C16 and C18 acyl-CoAs could undergo aliphatic chain elongation into C20–C26 acyl-CoAs catalyzed by the fatty acid elongase (FAE) enzyme complex consisting of β-ketoacyl-CoA synthases (KCS), β-ketoacyl-CoA reductase (KCR), 3-hydroxyacyl-CoA dehydratases (HCD), and enoyl-CoA reductases (ECR) (Yeats and Rose, 2013; Fich et al., 2016; Philippe et al., 2020). These C16–C26 acyl-CoAs are oxidized at their terminal and/or midchain carbons by cytochrome P450 enzymes (CYP77 and CYP86) and the protein HOTHEAD, hydroxylated by epoxide hydrolases (EH), and finally modified into monoacylglycerol cutin monomers by glycerol-3-phosphate acyltransferases (GPAT) (Pineau et al., 2017; Philippe et al., 2020). At the same time, the phenolic compound ferulic acid converted from coumaric acid by cytochrome P450 enzyme (CYP98) is conjugated to monoacylglycerol cutin monomers under the action of the transferase enzyme DEFICIENT IN CUTIN FERULATE (DCF) (Fich et al., 2016; Philippe et al., 2020). These cutin monomers are then exported out of the cell via the plasma membrane (PM)-localized ABC type of transporters and deposited into the cuticle, where cutin synthase (CUS) proteins mediate the cutin polymerization (Fich et al., 2016; Hong et al., 2017).
For the cuticular wax biosynthesis, plastid-derived C16 and C18 acyl-CoAs were first elongated to VLC (up to C34) acyl-CoAs by the FAE complex and the ECERIFERUM2 (CER2) protein (Haslam et al., 2015). It was recently demonstrated that protein-protein interactions in the FAE complex, including extensive protein–protein interactions among Arabidopsis FAE complex proteins KCR1, PASTICCINO2 (PAS2), ECR, and PAS1, as well as specific interactions between KCS9 and PAS2 or ECR, are essential to the VLC acyl-CoAs elongation (Kim et al., 2021). These elongated VLC acyl-CoAs were then either converted into n-alkanes, n-aldehydes, secondary alcohols, and ketones via the alkane-forming pathway or modified into primary n-alcohols and esters through the alcohol-forming pathway. In the alkane-forming pathway, VLC acyl-CoAs were converted into VLC alkanes under the action of the ECERIFERUM1 (CER1)/ECERIFERUM3 (CER3)/CYTOCHROME B5 (CYTB5) complex, and then oxidized to secondary alcohols and ketones by the CYP95A family cytochrome P450 enzymes (MAH1) (Pascal et al., 2019; Tang et al., 2020). In the alcohol-forming pathway, VLC acyl-CoAs were converted into primary alcohols under actions of acyl desaturase ECERIFERUM17 (CER17) and fatty acyl-CoA reductase ECERIFERUM4 (CER4), and modified into wax esters by WAX SYNTHASE/ACYL-COA:DIACYLGLYCEROL ACYLTRANSFERASE 1 (WSD1) (Yang et al., 2017). These generated wax components such as VLC fatty acids, alcohols, aldehydes, alkanes, esters, and ketones were transported from the ER to the PM via the Golgi and trans-Golgi network (TGN)-trafficking pathways, and finally exported out of the plant cell to the cuticle via the PM-localized ATP binding cassette G (ABCG) subfamily half transporters and the lipid transfer proteins (LTPs) (Yeats and Rose, 2013; Lee and Suh, 2015; Suh et al., 2022). In addition to these straight-chain wax components, branched waxes such as iso-alkanes and iso-alcohols have been identified in Arabidopsis flowers and leaves, and are derived from the catabolism of branched-chain amino acid (BCAA) such as valine. The isobutyl-CoA synthetase ACYL-ACTIVATING ENZYME 9 (AAE9) was recently identified to connect the BCAA catabolism with branched wax biosynthesis in the plastid (Li et al., 2022).
Increasing evidence revealed that cuticle biosynthesis is tight regulated at transcriptional, post-transcriptional, and post-translational levels. For instance, Arabidopsis APETALA2-Ethylene responsive factor (AP2-EREBP)-type transcription factors (SHN1/2/3), myeloblastosis (MYB) family transcription factors (MYB16, MYB30, MYB41, MYB94, MYB96, MYB106), zinc-finger transcription factors (NFXL2), and the class IV homeodomain-leucine zipper family transcription factors (HDG1) widely get involved in the transcriptional regulation of cutin and cuticular wax biosynthesis (Yeats and Rose, 2013; Lee and Suh, 2015; Philippe et al., 2020; Suh et al., 2022). ECERIFERUM16 (CER16), RNA exosome subunit RRP45B and Ski complex components (SKI2, SKI3, SKI7, and SKI8) regulate the post-transcriptional gene silencing of CER3, a VLC fatty acid reductase gene (Lee and Suh, 2015; Yang X. et al., 2020). E3 ubiquitin ligases ECERIFERUM9 (CER9), HMG-CoA Reductase Degradation 1 (HRD1), SMALL AND GLOSSY LEAVES1 (SAGL1) and ABA-related RING-type E3 ligase (ARRE) contribute to the post-translational regulation of cuticular lipid biosynthesis (Zhao et al., 2014; Kim et al., 2019; Liu et al., 2021; Wu et al., 2021).
With the assistance of forward and reverse genetic approaches, past decades have seen great advances in functional characterization of genes essential for cuticle biosynthesis in many crop species such as Zea mays, Triticum aestivum, Hordeum vulgare, Oryza sativa, Solanum lycopersicum, Glycine max, Malus domestica, Medicago truncatula, Camellia sinensis, Sorghum bicolor, Arachis hypogaea, Cyperus esculentus, Gossypium hirsutum, Citrus sinensis, Cucumis sativus, and Brassica napus (Zhou et al., 2013; Wang et al., 2015; Zhou et al., 2015; Li et al., 2018, 2019; Lokesh et al., 2019; Cheng et al., 2020; Guo et al., 2020; Zhang et al., 2020a; Abdullah et al., 2021; Ayaz et al., 2021; Busta et al., 2021; Lu et al., 2021; Wu et al., 2022). Although cuticle composition varies among plant species, evolutionarily conserved functions were revealed in cuticle biosynthesis genes of model and crop plants. For instance, tomato cutin synthase (SlCUS1), together with its homolog in Arabidopsis and the moss Physcomitrella patens, exhibited cutin monomer polymerizing activity in vitro (Yeats et al., 2014). Expression of GmLACS2-3, soybean homolog of Arabidopsis LACS2 gene, in the Arabidopsis atlacs2 mutant could rescue its cutin-deficient phenotype (Ayaz et al., 2021). This functional conservation was also obvious for cuticular wax biosynthesis genes such as CER1, CER2, CER3, ECR, KCR, KCS1, KCS2, KCS6 in model and crop plants. For instance, silencing the SlCER1-1 gene, tomato homolog of Arabidopsis AtCER1 gene, attenuated wax alkane biosynthesis in tomato (Wu et al., 2022). Consistent with this, ectopic expression of MdKCS2 and MdCER2, apple homologs of Arabidopsis AtKCS2 and AtCER2 genes, in Arabidopsis could enhance the accumulation of cuticular wax in Arabidopsis leaves and stems (Zhong et al., 2020; Lian et al., 2021). Notably, the function of cuticular wax biosynthesis genes seems conserved among monocots and dicots. For instance, the mutation in HvKCS1 and HvKCS6, barley homologs of Arabidopsis AtKCS1 and AtKCS6, resulted in a substantial reduction in the total cuticular wax load in barley leaves (Weidenbach et al., 2014; Li et al., 2018). Similarly, reduced expression of TaECR and TaKCS6, wheat homologs of Arabidopsis AtECR and AtKCS6, was associated with significant reductions in total wax load in bread wheat (Wang X. et al., 2019; Kong et al., 2020b). In addition, the wheat nullisomic-tetrasomic substitution line lacking TaCER1-1A had significantly reduced amounts of C33 alkane (Li et al., 2019).
Recent studies revealed that regulatory genes in cuticle biosynthesis also showed functional conservation in model and crop plants. For instance, silencing MdMYB30, apple homologs of Arabidopsis AtMYB30 gene, compromised wax crystal accumulation in apple (Zhang et al., 2019c). Similarly, tomato mutant with a mutation in CUTIN DEFICIENT2 (CD2), tomato homolog of Arabidopsis ANTHOCYANINLESS2 (ANL2), exhibited cutin deficiency in tomato fruit (Nadakuduti et al., 2012). Notably, this functional conservation of cuticle biosynthesis regulatory genes is also obvious between monocots and dicots. For instance, overexpression of TaSHN1/WAX INDUCER1 (TaWIN1), wheat homologs of Arabidopsis AtSHN1, resulted in increased accumulation of wax alkanes in wheat leaves (Bi et al., 2018). Silencing wheat TaWIN1 and barley HvWIN1 could attenuate cuticular wax accumulation (Kong and Chang, 2018; McAllister et al., 2022). Similarly, overexpressing OsWR1, rice homologs of Arabidopsis AtSHN1, improved while silencing OsWR1 attenuated wax biosynthesis in rice leaves (Wang et al., 2012).
Cuticle biosynthesis in plant adaptation to drought stress
As the major transpiration barrier, the hydrophobic cuticle restricts the non-stomatal water loss and protects plant tissues from desiccation in drought conditions. Induction of cuticle biosynthesis, including activation of cuticle biosynthesis genes, by drought stress has been observed in a wide range of plant species such as A. thaliana, O. sativa, T. aestivum, Z. mays, C. sativus, S. lycopersicum, M. domestica, S. bicolor, H. vulgare, G. max, G. hirsutum, C. sinensis, and G. hirsutum (Islam et al., 2009; Weng et al., 2010; Zhou et al., 2013, 2015; Wang et al., 2015; Li et al., 2018, 2019; Lokesh et al., 2019; Guo et al., 2020; Zhang et al., 2020a; Abdullah et al., 2021; Ayaz et al., 2021; Lu et al., 2021; Wu et al., 2022). As summarized in Table 1, altered expression of cuticle biosynthesis genes such as LACS1, LACS2, CER1, CER2, CER9, KCS1, KCS2, KCS6, FAR3.1, WSD1, GPAT2, and ABCG31 could lead to the changed drought tolerance in model and crop plants. Generally, over-expression of these cuticle biosynthesis genes results in the increased accumulation of cuticular wax or cutin, which contributes to the enhanced tolerance to drought stress. For instance, over-expression of Arabidopsis wax biosynthesis gene AtCER1 and its homologs in wheat (TaCER1-A), cucumber (CsCER1) and tomato (SlCER1-1) could increase VLC alkanes accumulation and enhance drought tolerance in transgenic plants (Bourdenx et al., 2011; Wang et al., 2015; Li et al., 2018; Wu et al., 2022). Over-expression of Arabidopsis wax biosynthesis gene AtWSD1 in Arabidopsis and Camelina resulted in enhanced drought tolerance in transgenic plants (Abdullah et al., 2021). Over-expression of GmLACS2-3, soybean (Glycine max) homolog of Arabidopsis AtLACS2, in Arabidopsis could enhance the amounts of cutin and suberin but not wax, and result in the increased drought tolerance (Ayaz et al., 2021). Consistent with the results of over-expression studies, knockout or knockdown of cuticle biosynthesis genes could lead to impaired cuticle development and decreased drought tolerance in model and crop plants. For instance, silencing of SlCER1-1 in tomato reduced the amounts of n-alkanes and branched alkanes, and decreased plant drought resistance (Wu et al., 2022). Similarly, silencing of GhFAR3.1 expression in Upland cotton leaves could attenuate wax accumulation and lead to enhanced susceptibility to desiccation (Lu et al., 2021).
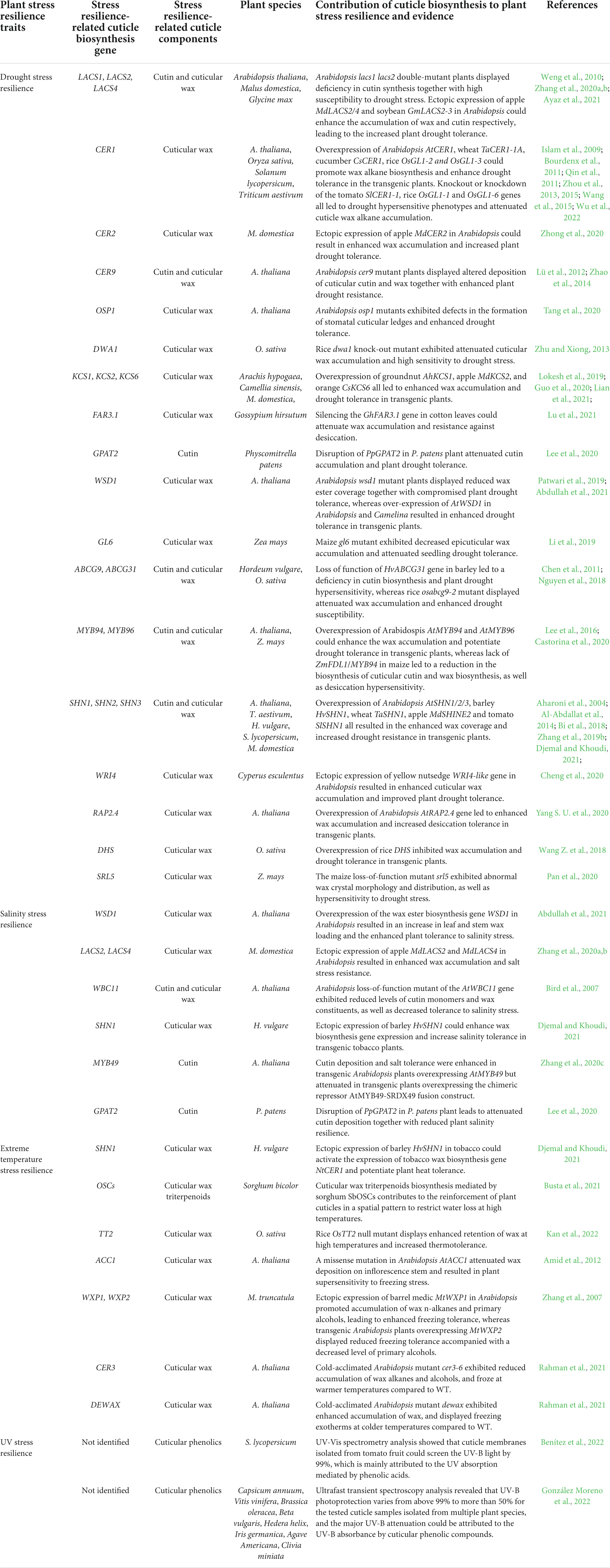
Table 1. Summary of plant resilience to drought, salinity, temperature, and ultraviolet (UV) stress contributed by cuticle biosynthesis.
Recent studies on transcription factors governing cuticle biosynthesis provide new insight into plant cuticle biosynthesis and drought stress adaptation. For instance, over-expression of Arabidopsis transcription factor genes AtSHN1, AtSHN2 and AtSHN3, as well as their homologs in barley (HvSHN1), wheat (TaSHN1), apple (MdSHINE2) and tomato (SlSHN1), could induce expression of wax biosynthesis genes and result in the increased wax accumulation and enhanced plant drought resistance (Aharoni et al., 2004; Al-Abdallat et al., 2014; Bi et al., 2018; Zhang et al., 2019b; Djemal and Khoudi, 2021). Over-expression of Arabidopsis transcription factor gene RAP2.4 could upregulate the expression of wax biosynthesis genes KCS2 and CER1, resulting in increased wax content and enhanced drought tolerance (Yang S. U. et al., 2020). In addition, the rice homeodomain leucine zipper IV (HD-ZIP IV) family of transcription factor ROC4 could positively regulate wax biosynthesis and drought tolerance by directly activating the cuticle biosynthesis gene OsBDG (Wang Z. et al., 2018). Interestingly, a RING-type E3 ligase DHS (DROUGHT HYPERSENSITIVE) negatively regulates rice wax biosynthesis and drought tolerance by targeting ROC4 for ubiquitin-mediated proteasomal degradation, suggesting the regulatory role of ubiquitin/26S proteasome (UPS) pathway in the transcriptional reprograming essential for plant cuticle biosynthesis and drought stress adaptation (Wang Z. et al., 2018).
Cuticle biosynthesis in plant adaptation to salinity stress
As a water loss barrier, the cuticle functions to reduce the transpiration rate and avoid tissue dehydration under salinity stress. Up-regulation of cuticle biosynthesis genes and elevated accumulation of wax and cutin is observed in model and crop plants in response to salinity stress. Increasing evidence revealed that plant tolerance to salinity stress could be enhanced by over-expression of cuticle biosynthesis genes and reduced by knockout or knockdown of cuticle biosynthesis genes (summarized in Table 1). For instance, over-expression of the wax ester biosynthesis gene WSD1 in Arabidopsis could increase leaf and stem wax loading, leading to enhanced plant tolerance to salinity stress (Abdullah et al., 2021). Ectopic expression of MdLACS2 and MdLACS4, apple homologs of Arabidopsis AtLACS2 and AtLACS4, could enhance the wax accumulation and salt stress resistance in transgenic Arabidopsis plants (Zhang et al., 2020a,b). Recent studies on transcription factors that regulate cuticle biosynthesis shed novel light on the involvement of plant cuticle biosynthesis in plant salinity stress adaptation. For instance, ectopic expression of HvSHN1, barley homolog of Arabidopsis transcription factor gene AtSHN1, could induce expression of wax biosynthesis gene NtCER1 and increase salinity tolerance in transgenic tobacco (Nicotiana tabacum) plants (Djemal and Khoudi, 2021). Recent transcriptome analysis revealed that the AtMYB49 could directly activate MYB41, ASFT, FACT, and CYP86B1, representative genes in the category of ‘cutin, suberin, and wax biosyntheses’ (Zhang et al., 2020c). The biochemical analysis further showed that cutin deposition and salt tolerance were higher in transgenic Arabidopsis plants overexpressing AtMYB49 but lower in transgenic plants overexpressing the chimeric repressor AtMYB49-SRDX49 fusion construct, suggesting that transcriptional activation of cutin biosynthesis by AtMYB49 contributes to the plant salt tolerance in Arabidopsis (Zhang et al., 2020c).
Cuticle biosynthesis in plant adaptation to extreme-temperature stress
Since the heat capacity of the cuticle was above the one of air, the cuticle could function as a heat sink to impair the heat transference between plant aerial organs and the surrounding environment, thereby governing the temperature increment in plant tissues at high temperatures. Differential scanning calorimetry (DSC) measurements showed that high-temperature stress could induce wax melting and glass transition, as well as increased permeability, in tomato fruit cuticle membrane (Benítez et al., 2022). As summarized in Table 1, molecular genetic studies of cuticle biosynthesis genes shed more light on plant adaptation to heat stress. A recent cuticular wax chemical analysis showed that triterpenoids enriched in adult leaf waxes of S. bicolor, a heat and drought-tolerant crop, but not its close relative Z. mays (maize) (Busta et al., 2021). Further comparative genomics and heterologous expression analyses revealed that sorghum cuticular triterpenoids are synthesized by a neofunctionalized steroid biosynthesis gene that is truncated and not expressed in maize, which leads to the reinforcement of sorghum cuticle in a spatial pattern to restrict water loss at high temperatures (Busta et al., 2021). In addition, a natural quantitative trait locus (QTL), TT2 (THERMOTOLERANCE 2), that could confer thermotolerance in rice without a yield penalty was recently found to encode a Gγ subunit that regulates the heat-triggered elevation of Ca2+, which leads to the activation of OsSCT1 (Sensing Ca2 + Transcription factor 1) protein (Kan et al., 2022). Interestingly, OsSCT1 was revealed to function as a negative regulator of cuticle biosynthesis genes such as the transcription factor gene OsWR2. Therefore, a natural loss-of-function allele TT2 fails to initiate the heat-triggered elevation of Ca2+, which results in the inactivation of SCT1 and de-repression of OsWR2 at high temperature, thereby inducing the wax biosynthesis and conferring the rice thermo-tolerance (Kan et al., 2022).
Activation of cuticle biosynthesis, such as up-regulation of cuticle biosynthesis genes, by cold stress has been observed in model and crop plants such as A. thaliana, M. truncatula, T. aestivum, B. distachyon, Thellungiella salsuginea, and Cucurbita pepo (Lee and Suh, 2015; Wang Y. et al., 2018; He et al., 2019). Interestingly, overaccumulation of cuticular wax alkanes was induced by cold stress in fruits of cold-tolerant zucchini variety “Natura,” but not the cold-sensitive variety “Sinatra,” suggesting the potential contribution of the wax alkane biosynthesis to the postharvest quality improvement of zucchini fruit during low-temperature storage (Carvajal et al., 2021). Current evidence revealed that plant tolerance to cold stress is affected by altered expression of cuticle biosynthesis genes (summarized in Table 1). For instance, Cold-acclimated Arabidopsis mutant cer3-6 exhibited reduced accumulation of wax alkanes and alcohols, and froze at warmer temperatures compared to WT. In contrast, cold-acclimated dewax mutant exhibited enhanced accumulation of wax alkanes and alcohols, and displayed freezing exotherms at colder temperatures compared to WT, indicating that cuticular wax contributes to plant tolerance to cold stress.
Cuticle biosynthesis in plant adaptation to ultraviolet radiation stress
Due to stratospheric ozone depletion during past decades, solar UV radiation that reached the earth surface has increased, especially in the Arctic and Northern biosphere. Based on the wavelength region, UV radiation could be divided into UV-A (315–400 nm), UV-B (280–315 nm), and UV-C (200–280 nm) (Robson et al., 2019). Through enhancing the generation of reactive oxygen species (ROS) to damage macromolecules such as DNA, proteins and lipids, energetic UV-B is harmful to land plants (Robson et al., 2019). Upon exposure to the UV-B radiation, increases in cuticle thickness and wax deposition have been reported in multiple plant species such as Cucumis sativus, P. vulgaris, H. vulgare, Coffea arabica, and C. canephora. As major UV-screening compounds in land plants, phenolic acids, and flavonoids have been identified in the cuticular wax fraction, cutin matrix and even cell walls. Current evidence showed that flavonoids could function as UV-A attenuators, whereas phenolic acids mainly contribute to the UV-B and UV-C photoprotection. Indeed, UV-Vis spectra analysis revealed that cuticle membranes isolated from tomato fruit could screen the UV-B light by 99%, which is mainly attributed to the UV absorption mediated by phenolic acids but not cuticular waxes (Benítez et al., 2022). Ultrafast transient spectroscopy analysis showed that the UV-B photoprotection varies from above 99% to more than 50% for the tested cuticle samples isolated from multiple plant species, and the major UV-B attenuation could be attributed to the UV-B absorbance by cuticular phenolic compounds (González Moreno et al., 2022). Quantum chemical computational analyses further revealed that UV-B energy absorbed by cuticular phenolic compounds can be either released as blue fluorescence via the radiative mechanism or dissipated via the conformational isomerization of phenolic compounds (González Moreno et al., 2022).
Harnessing cuticle biosynthesis for crop adaptation to environmental stress
Environmental stresses such as drought, salinity, extreme temperatures, and UV radiation adversely affect plant growth and crop production, which has become increasingly serious under stratospheric ozone depletion and global climate change. As an adaptive innovation in land plants, hydrophobic cuticle shields plant tissues from environmental stresses associated with terrestrialization and represents a valuable resource for crop improvement. Cuticle-associated traits have been employed in traditional crop breeding (Petit et al., 2017). For instance, glaucousness determined by epicuticular wax deposition has been selected as a target trait for crop drought tolerance in traditional breeding. Indeed, a recent study revealed that increased leaf wax n-alkane concentration has been selected for enhanced productivity in five wheat cultivars developed over the past half-century (Liu et al., 2019). In addition, advanced breeding strategies and approaches established in recent years have paved the new path for harnessing cuticle biosynthesis to improve crop adaptation to stressful environments (Bailey-Serres et al., 2019; Varshney et al., 2020; Gao, 2021).
Cuticle-associated genetic variants have been isolated from natural populations in multiple crop species such as Z. mays, O. sativa, S. lycopersicum, C. sativus, and B. napus through visual screen for altered reflectance (glaucousness, glossiness, or bloom) of plant aerial organs. As summarized in prior reviews, a set of phenotyping techniques such as toluidine blue soaking, gas chromatography coupled with mass spectrometry (GC-MS), gas chromatography equipped with flame ionization detector (GC-FID), Fourier transform infrared (FTIR) imaging, and spectroscopy, matrix-assisted laser desorption/ionization (MALDI) imaging and leaf radiometric measurements have been developed to analyze cuticle properties, chemical composition and physical structure, which greatly contribute to the characterization and high-throughput phenotyping of cuticle-related genetic variants in crop plants (Petit et al., 2017; Camarillo-Castillo et al., 2021). In addition to natural genetic diversity, cuticle-associated genetic variants have been obtained from the induced mutant population of crop plants Z. mays, O. sativa, S. lycopersicum, and H. vulgare. Chemical, irradiation and insertion mutagenesis methods are available for generating the induced mutant populations. EMS (ethyl methanesulfonate) chemical mutagenesis and irradiation mutagenesis with x/y-rays or fast neutrons are widely employed to generate induced mutation in commercial crop breeding. At the same time, T-DNA insertion mutant populations have been constructed in model and crop plants, including A. thaliana, Brachypodium distachyon, Z. mays, S. lycopersicum, and O. sativa, from which cuticle-associated genetic mutants were identified (Choi et al., 2022).
Map-based cloning of cuticle-associated genetic mutation leads to the identification of cuticle genes in model and crop plants, which has been extensively discussed in previous reviews. Although many cuticle genes identified in the model plant Arabidopsis control cuticle-associated traits in a Mendelian manner, cuticle-associated traits in natural crop populations are usually controlled by quantitative trait loci (QTLs). Conventional QTL mapping and genome-wide association studies (GWAS) were employed for identifying these cuticle-related QTLs in crop species. For instance, the natural QTL TT2 (THERMOTOLERANCE 2) conferring rice thermotolerance was identified by QTL mapping as a G-protein γ subunit gene controlling cuticular wax biosynthesis in rice plants (Kan et al., 2022). Similarly, genome-wide association study of natural variation for maize leaf cuticular conductance identifies cuticle genes associated with enhanced crop productivity under drought stress. Through employing the combined chemical analyses, heterologous expression, and comparative genomics, Busta et al. (2021) demonstrated that a neofunctionalized steroid biosynthesis gene that is truncated and not expressed in maize is essential to the biosynthesis of cuticular triterpenoids and thermotolerance in sorghum plants, suggesting that combined metabolomic, transcriptomic, and genomic analyses contribute to the identification of cuticle genes. These cuticle-related QTLs can be used as makers in crop genomic breeding (GB) to facilitate the marker-assisted selection (MAS) and marker-assisted backcrossing (MABC) (Varshney et al., 2020). Through combining chemical or irradiation mutagenesis with genome-wide screening, targeting induced local lesions in genomes (TILLING) could effectively induce mutation in cuticle-related QTLs and genes, and represents a promising non-transgenic method for improving cuticle-associated traits in crop species. For instance, a drought-insensitive rice mutant (ditl1) harboring a mutation in the LOC_Os05g48260 gene, a putative rice ortholog of WSD1, was selected by drought stress screening in the rice TILLING population (Choi et al., 2022).
As summarized in Table 1, genetic engineering of cuticle genes was widely employed for improving crop tolerance to drought, salinity and extreme temperature stress. For instance, overexpression of groundnut wax biosynthesis gene AhKCS1 led to enhanced leaf epicuticular wax accumulation and increased drought tolerance in transgenic groundnut plants (Lokesh et al., 2019). Overexpression of wheat transcription factor gene TaSHN1 and tomato SlSHN1 resulted in the enhanced wax coverage and increased drought resistance in transgenic wheat and tomato plants, respectively (Al-Abdallat et al., 2014; Bi et al., 2018). In addition, ectopic expression of cuticle genes identified from model and crop plants was conducted to improve crop stress tolerance. For instance, ectopic expression of Arabidopsis AtMYB96 and AtWSD1 in Camelina plants could potentiate plant drought tolerance (Lee et al., 2014; Abdullah et al., 2021). Similarly, ectopic expression of wheat cuticle gene TaCER1-1A in rice plants could promote rice wax alkane biosynthesis and enhance rice leaf tolerance to desiccation stress (Li et al., 2019). For the biosafety of transgenic crops, many marker-free transgenic approaches have been established. Cao et al. (2020) reported the development of marker-free and transgene insertion site-defined (MFTID) transgenic wheat lines with improved grain storability and fatty acid content through suppressing lipoxygenase (LOX) gene expression in wheat grains, which provides a new avenue for the genetic engineering of cuticle genes in crop species.
Through using sequence-specific nucleases (SSNs), plant genome editing (GE) could introduce precise DNA mutations such as insertion, deletion and base substitution into target genome regions (Manghwar et al., 2019; Gao, 2021). As the most recently developed GE technique, CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 (CRISPR associated nuclease 9) system relies on the programmable guide RNA (gRNA) to guide the Cas9 nuclease to the DNA targets, where DNA double-strand breaks (DSBs) are generated and precise GE is achieved via endogenous DNA repair pathways (Manghwar et al., 2019; Gao, 2021). CRISPR-Cas9 system has been successfully applied in generating genome-edited crop plants with altered cuticle traits. For instance, precise editing of the OsPYL9 gene, one of the ABA receptor genes in rice, by CRISPR-Cas9 system resulted in the overaccumulation of cuticular wax and enhanced drought tolerance in genome-edited rice plants (Usman et al., 2020). Similarly, multiplex knockout of transcription factor gene MYB186 and its paralogs MYB138 and MYB38 in hybrid poplar by CRISPR-Cas9 system with a single gRNA led to a glabrous phenotype accompanied with the absence of wax triterpenes in trichomeless leaves (Bewg et al., 2022). These studies paved a new path for generating genome-edited crop plants with improved cuticle traits and enhanced stress tolerance in future research.
Concluding remarks and perspectives
In this review, we highlighted recent advances in cuticle biosynthesis and its roles in plant adaptation to drought, salinity, extreme temperatures, and UV radiation stress, and discussed the current strategies and future directions in harnessing cuticle biosynthesis for crop improvement. As shown in Figure 1, cuticle-related genetic diversity could be either identified from natural populations or artificially induced, which would facilitate crop breeding for cuticle-associated traits through breeding approaches MAS and MABC. In addition, crop plants with improved cuticle-related traits and stress tolerance could be generated by non-transgenic TILLING, genetic engineering and genome editing of cuticle biosynthesis genes. Although the past decade has seen great progress in the molecular biology of plant cuticle biosynthesis, we still have a long way to go toward fully understanding the mechanism of cuticle biosynthesis in plant adaptation to drought, salinity, extreme temperatures, and UV radiation stress, as well as its application in crop improvement. For instance, most of our knowledge about plant cuticle biosynthesis comes from the study of model plants Arabidopsis, mechanisms of cuticle biosynthesis in important crops remain to be explored. Furthermore, the expression of some cuticle biosynthesis genes could be induced by environmental stress, but the underlying mechanisms of cuticle biosynthesis responding to environmental stress remains to be disclosed. Moreover, cuticle generally shields plant tissues from environmental stress, but the exact roles and mechanisms of cuticle structure and components in plant adaptation to specific environmental stress are poorly understood. In addition, overexpression of cuticle biosynthesis gene usually enhances crop stress resilience with a yield penalty due to the altered metabolic flux allocation, breeding crop variety with a ‘smart cuticle’ that confer crop plants improved stress tolerance at a low cost of energy and metabolic flux might be essential to balancing crop yield and resilience. With the advance in the knowledge of plant cuticle biosynthesis in plant adaptation to drought, salinity, extreme temperatures and UV radiation stress, generating this ‘smart cuticle’ with improved structure and optimized composition would certainly provide new avenues for crop improvement under adverse environments.
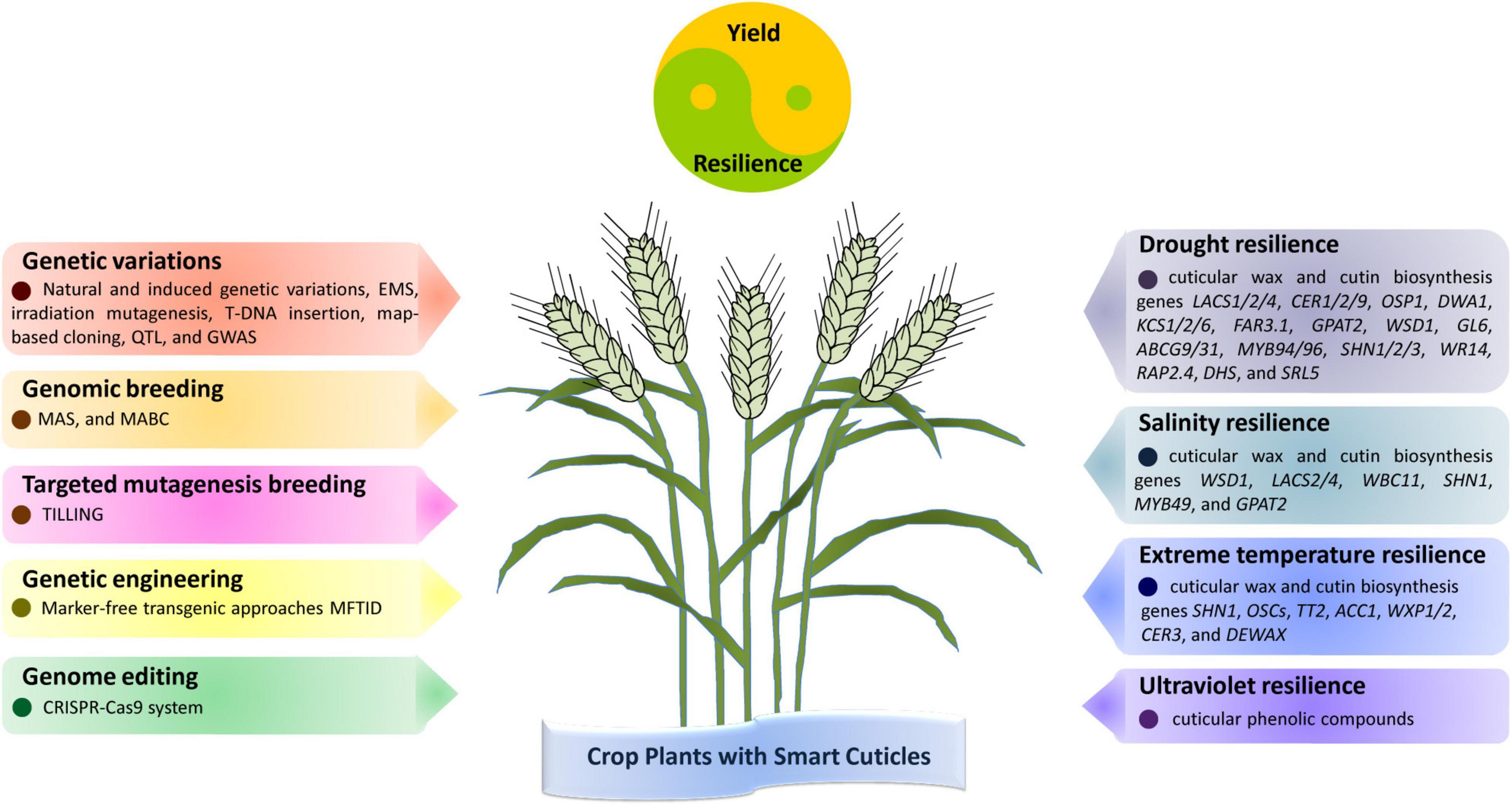
Figure 1. Strategies and targets in harnessing cuticle biosynthesis for crop resilience to drought, salinity, temperature, and ultraviolet (UV) stress. Cuticle-related genetic variations could be either identified from natural populations or artificially induced, which would facilitate crop breeding for cuticle-associated traits through genomic breeding. In addition, crop plants with improved resilience to drought, salinity, temperature, and UV stress could be generated by targeted mutagenesis, genetic engineering, and genome editing of cuticle biosynthesis genes. These crop plants with smart cuticles would display improved performance in yield and resilience under drought, salinity, temperature, and UV stress.
Author contributions
CC, LL, and XW wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (31701412), the Natural Science Foundation of Shandong Province (ZR2017BC109), Qingdao Science and Technology Bureau Fund (17-1-1-50-jch), and Qingdao University Fund (DC1900005385).
Acknowledgments
We thank Engin Yol for the kind invitation to write this review. We are also grateful to two reviewers for their very helpful comments and suggestions on this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdullah, H. M., Rodriguez, J., Salacup, J. M., Castañeda, I. S., Schnell, D. J., Pareek, A., et al. (2021). Increased cuticle waxes by overexpression of WSD1 improves osmotic stress tolerance in Arabidopsis thaliana and Camelina sativa. Int. J. Mol. Sci. 22:5173. doi: 10.3390/ijms22105173
Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16, 2463–2480. doi: 10.1105/tpc.104.022897
Al-Abdallat, A. M., Al-Debei, H. S., Ayad, J. Y., and Hasan, S. (2014). Over-expression of SlSHN1 gene improves drought tolerance by increasing cuticular wax accumulation in tomato. Int. J. Mol. Sci. 15, 19499–19515. doi: 10.3390/ijms151119499
Amid, A., Lytovchenko, A., Fernie, A. R., Warren, G., and Thorlby, G. J. (2012). The sensitive to freezing 3 mutation of Arabidopsis thaliana is a cold-sensitive allele of homomeric acetyl-CoA carboxylase that results in cold-induced cuticle deficiencies. J. Exp. Bot. 63, 5289–5299. doi: 10.1093/jxb/ers191
Ayaz, A., Huang, H., Zheng, M., Zaman, W., Li, D., Saqib, S., et al. (2021). Molecular cloning and functional analysis of GmLACS2-3 reveals its involvement in cutin and suberin biosynthesis along with abiotic stress tolerance. Int. J. Mol. Sci. 22:9175. doi: 10.3390/ijms22179175
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G., and Schroeder, J. I. (2019). Genetic strategies for improving crop yields. Nature 575, 109–118. doi: 10.1038/s41586-019-1679-0
Benítez, J. J., González Moreno, A., Guzmán-Puyol, S., Heredia-Guerrero, J. A., Heredia, A., and Domínguez, E. (2022). The response of tomato fruit cuticle membranes against heat and light. Front. Plant. Sci. 12:807723. doi: 10.3389/fpls.2021.807723
Bewg, W. P., Harding, S. A., Engle, N. L., Vaidya, B. N., Zhou, R., Reeves, J., et al. (2022). Multiplex knockout of trichome-regulating MYB duplicates in hybrid poplar using a single gRNA. Plant Physiol. 189, 516–526. doi: 10.1093/plphys/kiac128
Bhanot, V., Fadanavis, S. V., and Panwar, J. (2021). Revisiting the architecture, biosynthesis and functional aspects of the plant cuticle: there is more scope. Environ. Exp. Bot. 183:104364. doi: 10.1016/j.envexpbot.2020.104364
Bi, H., Shi, J., Kovalchuk, N., Luang, S., Bazanova, N., Chirkova, L., et al. (2018). Overexpression of the TaSHN1 transcription factor in bread wheat leads to leaf surface modifications, improved drought tolerance, and no yield penalty under controlled growth conditions. Plant Cell Environ. 41, 2549–2566. doi: 10.1111/pce.13339
Bird, D., Beisson, F., Brigham, A., Shin, J., Greer, S., Jetter, R., et al. (2007). Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 52, 485–498. doi: 10.1111/j.1365-313X.2007.03252.x
Bourdenx, B., Bernard, A., Domergue, F., Pascal, S., Léger, A., Roby, D., et al. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156, 29–45. doi: 10.1104/pp.111.172320
Busta, L., Schmitz, E., Kosma, D. K., Schnable, J. C., and Cahoon, E. B. (2021). A co-opted steroid synthesis gene, maintained in sorghum but not maize, is associated with a divergence in leaf wax chemistry. Proc. Natl. Acad. Sci. U. S. A. 118:e2022982118. doi: 10.1073/pnas.2022982118
Camarillo-Castillo, F., Huggins, T. D., Mondal, S., Reynolds, M. P., Tilley, M., and Hays, D. B. (2021). High-resolution spectral information enables phenotyping of leaf epicuticular wax in wheat. Plant Methods 17:58. doi: 10.1186/s13007-021-00759-w
Cao, X., Dong, Z., Tian, D., Dong, L., Qian, W., Liu, J., et al. (2020). Development and characterization of marker-free and transgene insertion site-defined transgenic wheat with improved grain storability and fatty acid content. Plant Biotechnol. J. 18, 129–140. doi: 10.1111/pbi.13178
Carvajal, F., Castro-Cegrí, A., Jiménez-Muñoz, R., Jamilena, M., Garrido, D., and Palma, F. (2021). Changes in morphology, metabolism and composition of cuticular wax in Zucchini fruit during postharvest cold storage. Front. Plant Sci. 12:778745. doi: 10.3389/fpls.2021.778745
Castorina, G., Domergue, F., Chiara, M., Zilio, M., Persico, M., Ricciardi, V., et al. (2020). Drought-responsive ZmFDL1/MYB94 regulates cuticle biosynthesis and cuticle-dependent leaf permeability. Plant Physiol. 184, 266–282. doi: 10.1104/pp.20.00322
Chen, G., Komatsuda, T., Ma, J. F., Li, C., Yamaji, N., and Nevo, E. (2011). A functional cutin matrix is required for plant protection against water loss. Plant Signal. Behav. 6, 1297–1299. doi: 10.4161/psb.6.9.17507
Cheng, C., Hu, S., Han, Y., Xia, D., Huang, B. L., Wu, W., et al. (2020). Yellow nutsedge WRI4-like gene improves drought tolerance in Arabidopsis thaliana by promoting cuticular wax biosynthesis. BMC Plant Biol. 20:498. doi: 10.1186/s12870-020-02707-7
Choi, S. Y., Lee, Y. J., Seo, H. U., Kim, J. H., and Jang, C. S. (2022). Physio-biochemical and molecular characterization of a rice drought-insensitive TILLING line 1 (ditl1) mutant. Physiol. Plant 174:e13718. doi: 10.1111/ppl.13718
Daryanto, S., Wang, L., and Jacinthe, P. A. (2016). Global synthesis of drought effects on maize and wheat production. PLoS One 11:e0156362. doi: 10.1371/journal.pone.0156362
Djemal, R., and Khoudi, H. (2021). The barley SHN1-type transcription factor HvSHN1 imparts heat, drought and salt tolerances in transgenic tobacco. Plant Physiol. Biochem. 164, 44–53. doi: 10.1016/j.plaphy.2021.04.018
Domínguez, E., Heredia-Guerrero, J. A., and Heredia, A. (2011). The biophysical design of plant cuticles: an overview. New Phytol. 189, 938–949. doi: 10.1111/j.1469-8137.2010.03553.x
Fahad, S., Bajwa, A. A., Nazir, U., Anjum, S. A., Farooq, A., Zohaib, A., et al. (2017). Crop production under drought and heat stress: plant responses and management options. Front. Plant Sci. 8:1147. doi: 10.3389/fpls.2017.01147
Fernández, V., Guzmán-Delgado, P., Graça, J., Santos, S., and Gil, L. (2016). Cuticle structure in relation to chemical composition: re-assessing the prevailing model. Front. Plant Sci. 7:427. doi: 10.3389/fpls.2016.00427
Fich, E. A., Segerson, N. A., and Rose, J. K. (2016). The plant polyester cutin: biosynthesis, structure, and biological roles. Annu. Rev. Plant Biol. 67, 207–233. doi: 10.1146/annurev-arplant-043015-111929
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635. doi: 10.1016/j.cell.2021.01.005
González Moreno, A., de Cózar, A., Prieto, P., Domínguez, E., and Heredia, A. (2022). Radiationless mechanism of UV deactivation by cuticle phenolics in plants. Nat. Commun. 13:1786. doi: 10.1038/s41467-022-29460-9
Guo, W., Wu, Q., Yang, L., Hu, W., Liu, D., and Liu, Y. (2020). Ectopic expression of CsKCS6 from navel orange promotes the production of very-long-chain fatty acids (VLCFAs) and increases the abiotic stress tolerance of Arabidopsis thaliana. Front. Plant Sci. 11:564656. doi: 10.3389/fpls.2020.564656
Haslam, T. M., Haslam, R., Thoraval, D., Pascal, S., Delude, C., Domergue, F., et al. (2015). ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol. 167, 682–692. doi: 10.1104/pp.114.253195
He, J., Tang, S., Yang, D., Chen, Y., Ling, L., Zou, Y., et al. (2019). Chemical and transcriptomic analysis of cuticle lipids under cold stress in Thellungiella salsuginea. Int. J. Mol. Sci. 20:4519. doi: 10.3390/ijms20184519
Hong, L., Brown, J., Segerson, N. A., Rose, J. K., and Roeder, A. H. (2017). CUTIN SYNTHASE 2 maintains progressively developing cuticular ridges in Arabidopsis sepals. Mol. Plant. 10, 560–574. doi: 10.1016/j.molp.2017.01.002
Islam, M. A., Du, H., Ning, J., Ye, H., and Xiong, L. (2009). Characterization of Glossy 1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol. Biol. 70, 443–456. doi: 10.1007/s11103-009-9483-0
Kan, Y., Mu, X. R., Zhang, H., Gao, J., Shan, J. X., Ye, W. W., et al. (2022). TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nat. Plants 8, 53–67. doi: 10.1038/s41477-021-01039-0
Kim, H., Yu, S. I., Jung, S. H., Lee, B. H., and Suh, M. C. (2019). The F-Box protein SAGL1 and ECERIFERUM3 regulate cuticular wax biosynthesis in response to changes in humidity in Arabidopsis. Plant Cell 31, 2223–2240. doi: 10.1105/tpc.19.00152
Kim, J., Kim, R. J., Lee, S. B., and Chung Suh, M. (2021). Protein-protein interactions in fatty acid elongase complexes are important for very-long-chain fatty acid synthesis. J. Exp. Bot. [online ahead of print]. doi: 10.1093/jxb/erab543.
Kong, L., and Chang, C. (2018). Suppression of wheat TaCDK8/TaWIN1 interaction negatively affects germination of Blumeria gramini sf. sp. tritici by interfering with very-long-chain aldehyde biosynthesis. Plant Mol. Biol. 96, 165–178. doi: 10.1007/s11103-017-0687-4
Kong, L., Liu, Y., Zhi, P., Wang, X., Xu, B., Gong, Z., et al. (2020a). Origins and evolution of cuticle biosynthetic machinery in land plants. Plant Physiol. 184, 1998–2010. doi: 10.1104/pp.20.00913
Kong, L., Zhi, P., Liu, J., Li, H., Zhang, X., Xu, J., et al. (2020b). Epigenetic activation of Enoyl-CoA Reductase by an acetyltransferase complex triggers wheat wax biosynthesis. Plant Physiol. 183, 1250–1267. doi: 10.1104/pp.20.00603
Lee, S. B., Kim, H., Kim, R. J., and Suh, M. C. (2014). Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep. 33, 1535–1546. doi: 10.1007/s00299-014-1636-1
Lee, S. B., Kim, H. U., and Suh, M. C. (2016). MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis. Plant Cell Physiol. 57, 2300–2311. doi: 10.1093/pcp/pcw147
Lee, S. B., and Suh, M. C. (2015). Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 34, 557–572. doi: 10.1007/s00299-015-1772-2
Lee, S. B., Yang, S. U., Pandey, G., Kim, M. S., Hyoung, S., Choi, D., et al. (2020). Occurrence of land-plant-specific glycerol-3-phosphate acyltransferases is essential for cuticle formation and gametophore development in Physcomitrella patens. New Phytol. 225, 2468–2483. doi: 10.1111/nph.16311
Lesk, C., Rowhani, P., and Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Li, C., Haslam, T. M., Krüger, A., Schneider, L. M., Mishina, K., Samuels, L., et al. (2018). The β-Ketoacyl-CoA synthase HvKCS1, encoded by Cer-zh, plays a key role in synthesis of barley leaf wax and germination of barley powdery mildew. Plant Cell Physiol. 59, 806–822. doi: 10.1093/pcp/pcy020
Li, H., and Chang, C. (2021). Evolutionary insight of plant cuticle biosynthesis in bryophytes. Plant Signal. Behav. 16:1943921. doi: 10.1080/15592324.2021.1943921
Li, L., Du, Y., He, C., Dietrich, C. R., Li, J., Ma, X., et al. (2019). Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J. Exp. Bot. 70, 3089–3099. doi: 10.1093/jxb/erz131
Li, S., Yang, X., Huang, H., Qiao, R., Jenks, M. A., Zhao, H., et al. (2022). Arabidopsis ACYL-ACTIVATING ENZYME 9 (AAE9) encoding an isobutyl-CoA synthetase is a key factor connecting branched-chain amino acid catabolism with iso-branched wax biosynthesis. New Phytol. 233, 2458–2470. doi: 10.1111/nph.17941
Lian, X. Y., Gao, H. N., Jiang, H., Liu, C., and Li, Y. Y. (2021). MdKCS2 increased plant drought resistance by regulating wax biosynthesis. Plant Cell Rep. 40, 2357–2368. doi: 10.1007/s00299-021-02776-4
Liu, S., Tong, M., Zhao, L., Li, X., and Kunst, L. (2021). The ARRE RING-Type E3 ubiquitin ligase negatively regulates cuticular wax biosynthesis in Arabidopsis thaliana by controlling ECERIFERUM1 and ECERIFERUM3 protein levels. Front. Plant Sci. 12:752309. doi: 10.3389/fpls.2021.752309
Liu, X., Feakins, S. J., Ma, X. F., Anderson, J. D., Vidal, E., and Blancaflor, E. B. (2019). Crop breeding has increased the productivity and leaf wax n-alkane concentration in a series of five winter wheat cultivars developed over the last 60 years. J. Plant Physiol. 243:153056. doi: 10.1016/j.jplph.2019.153056
Lokesh, U., Venkatesh, B., Kiranmai, K., Nareshkumar, A., Amarnathareddy, V., Rao, G. L., et al. (2019). Overexpression of β-Ketoacyl Co-A Synthase 1 gene improves tolerance of drought susceptible groundnut (Arachis hypogaea L.) cultivar K-6 by increased leaf epicuticular wax accumulation. Front. Plant Sci. 9:1869. doi: 10.3389/fpls.2018.01869
Lü, S., Zhao, H., Des Marais, D. L., Parsons, E. P., Wen, X., Xu, X., et al. (2012). Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 159, 930–944. doi: 10.1104/pp.112.198697
Lu, Y., Cheng, X., Jia, M., Zhang, X., Xue, F., Li, Y., et al. (2021). Silencing GhFAR3.1 reduces wax accumulation in cotton leaves and leads to increased susceptibility to drought stress. Plant Direct 5:e00313. doi: 10.1002/pld3.313
Manghwar, H., Lindsey, K., Zhang, X., and Jin, S. (2019). CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24, 1102–1125. doi: 10.1016/j.tplants.2019.09.006
McAllister, T., Campoli, C., Eskan, M., Liu, L., and McKim, S. M. (2022). A gene encoding a SHINE1/WAX INDUCER1 transcription factor controls cuticular wax in barley. Agronomy 12:1088. doi: 10.3390/agronomy12051088
Nadakuduti, S. S., Pollard, M., Kosma, D. K., Allen, C., Ohlrogge, J. B., and Barry, C. S. (2012). Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of CUTIN DEFICIENT2 in epidermal cell function in tomato. Plant Physiol. 159, 945–960. doi: 10.1104/pp.112.198374
Nguyen, V., Lee, S. B., Suh, M. C., An, G., and Jung, K. H. (2018). OsABCG9 is an important ABC transporter of cuticular wax deposition in rice. Front. Plant Sci. 9:960. doi: 10.3389/fpls.2018.00960
Pan, Z., Liu, M., Zhao, H., Tan, Z., Liang, K., Sun, Q., et al. (2020). ZmSRL5 is involved in drought tolerance by maintaining cuticular wax structure in maize. J. Integr. Plant Biol. 62, 1895–1909. doi: 10.1111/jipb.12982
Pascal, S., Bernard, A., Deslous, P., Gronnier, J., Fournier-Goss, A., Domergue, F., et al. (2019). Arabidopsis CER1-LIKE1 functions in a cuticular very-long-chain alkane-forming complex. Plant Physiol. 179, 415–432. doi: 10.1104/pp.18.01075
Patwari, P., Salewski, V., Gutbrod, K., Kreszies, T., Dresen-Scholz, B., Peisker, H., et al. (2019). Surface wax esters contribute to drought tolerance in Arabidopsis. Plant J. 98, 727–744. doi: 10.1111/tpj.14269
Petit, J., Bres, C., Mauxion, J. P., Bakan, B., and Rothan, C. (2017). Breeding for cuticle- associated traits in crop species: traits, targets, and strategies. J. Exp. Bot. 68, 5369–5387. doi: 10.1093/jxb/erx341
Philippe, G., Sørensen, I., Jiao, C., Sun, X., Fei, Z., Domozych, D. S., et al. (2020). Cutin and suberin: assembly and origins of specialized lipidic cell wall scaffolds. Curr. Opin. Plant Biol. 55, 11–20. doi: 10.1016/j.pbi.2020.01.008
Pineau, E., Xu, L., Renault, H., Trolet, A., Navrot, N., Ullmann, P., et al. (2017). Arabidopsis thaliana EPOXIDE HYDROLASE1 (AtEH1) is a cytosolic epoxide hydrolase involved in the synthesis of poly-hydroxylated cutin monomers. New Phytol. 215, 173–186. doi: 10.1111/nph.14590
Qin, B. X., Tang, D., Huang, J., Li, M., Wu, X. R., Lu, L. L., et al. (2011). Rice OsGL1-1 is involved in leaf cuticular wax and cuticle membrane. Mol. Plant 4, 985–995. doi: 10.1093/mp/ssr028
Rahman, T., Shao, M., Pahari, S., Venglat, P., Soolanayakanahally, R., Qiu, X., et al. (2021). Dissecting the roles of cuticular wax in plant resistance to shoot dehydration and low-temperature stress in Arabidopsis. Int. J. Mol. Sci 22:1554. doi: 10.3390/ijms22041554
Reynoud, N., Petit, J., Bres, C., Lahaye, M., Rothan, C., Marion, D., et al. (2021). The complex architecture of plant cuticles and its relation to multiple biological functions. Front. Plant Sci. 12:782773. doi: 10.3389/fpls.2021.782773
Robson, T. M., Aphalo, P. J., Banaś, A. K., Barnes, P. W., Brelsford, C. C., Jenkins, G. I., et al. (2019). A perspective on ecologically relevant plant-UV research and its practical application. Photochem. Photobiol. Sci. 18, 970–988. doi: 10.1039/c8pp00526e
Shimono, H., Hasegawa, T., and Iwama, K. (2007). Modeling the effects of water temperature on rice growth and yield under a cool climate: I. model development. Agron. J. 99, 1327–1337. doi: 10.2134/agronj2006.0337
Suh, M. C., Uk Kim, H., and Nakamura, Y. (2022). Plant lipids: trends and beyond. J. Exp. Bot. 73, 2715–2720. doi: 10.1093/jxb/erac125
Tang, J., Yang, X., Xiao, C., Li, J., Chen, Y., Li, R., et al. (2020). GDSL lipase occluded stomatal pore 1 is required for wax biosynthesis and stomatal cuticular ledge formation. New Phytol. 228, 1880–1896. doi: 10.1111/nph.16741
Tilman, D., Balzer, C., Hill, J., and Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U. S. A. 108, 20260–20264. doi: 10.1073/pnas.1116437108
Usman, B., Nawaz, G., Zhao, N., Liao, S., Liu, Y., and Li, R. (2020). Precise editing of the OsPYL9 gene by RNA-guided Cas9 nuclease confers enhanced drought tolerance and grain yield in rice (Oryza sativa L.) by regulating circadian rhythm and abiotic stress responsive proteins. Int. J. Mol. Sci. 21:7854. doi: 10.3390/ijms21217854
Varshney, R. K., Sinha, P., Singh, V. K., Kumar, A., Zhang, Q., and Bennetzen, J. L. (2020). 5Gs for crop genetic improvement. Curr. Opin. Plant Biol. 56, 190–196. doi: 10.1016/j.pbi.2019.12.004
Wang, P., Hu, T., Kong, F., Xu, J., and Zhang, D. (2019). Rice exposure to cold stress in china: how has its spatial pattern changed under climate change? Eur. J. Agron. 103, 73–79. doi: 10.1016/j.eja.2018.11.004
Wang, W., Zhang, Y., Xu, C., Ren, J., Liu, X., Black, K., et al. (2015). Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol. Biol. 87, 219–233. doi: 10.1007/s11103-014-0271-0
Wang, X., Kong, L., Zhi, P., and Chang, C. (2020). Update on cuticular wax biosynthesis and its roles in plant disease resistance. Int. J. Mol. Sci. 21:5514. doi: 10.3390/ijms21155514
Wang, X., Zhi, P., Fan, Q., Zhang, M., and Chang, C. (2019). Wheat CHD3 protein TaCHR729 regulates the cuticular wax biosynthesis required for stimulating germination of Blumeria graminis f.sp. tritici. J. Exp. Bot. 70, 701–713. doi: 10.1093/jxb/ery377
Wang, Y., Sun, Y., You, Q., Luo, W., Wang, C., Zhao, S., et al. (2018). Three fatty Acyl-Coenzyme A reductases, BdFAR1, BdFAR2 and BdFAR3, are Involved in cuticular wax primary alcohol biosynthesis in Brachypodium distachyon. Plant Cell Physiol. 59, 527–543. doi: 10.1093/pcp/pcx211
Wang, Y., Wan, L., Zhang, L., Zhang, Z., Zhang, H., Quan, R., et al. (2012). An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 78, 275–288. doi: 10.1007/s11103-011-9861-2
Wang, Z., Tian, X., Zhao, Q., Liu, Z., Li, X., Ren, Y., et al. (2018). The E3 Ligase DROUGHT HYPERSENSITIVE negatively regulates cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in rice. Plant Cell 30, 228–244. doi: 10.1105/tpc.17.00823
Weidenbach, D., Jansen, M., Franke, R. B., Hensel, G., Weissgerber, W., Ulferts, S., et al. (2014). Evolutionary conserved function of barley and Arabidopsis 3-KETOACYL-CoA SYNTHASES in providing wax signals for germination of powdery mildew fungi. Plant Physiol. 166, 1621–1633. doi: 10.1104/pp.114.246348
Weng, H., Molina, I., Shockey, J., and Browse, J. (2010). Organ fusion and defective cuticle function in a lacs1 lacs2 double mutant of Arabidopsis. Planta 231, 1089–1100. doi: 10.1007/s00425-010-1110-4
Wu, H., Liu, L., Chen, Y., Liu, T., Jiang, Q., Wei, Z., et al. (2022). Tomato SlCER1-1 catalyzes the synthesis of wax alkanes which increases the drought tolerance and fruit storability. Hortic. Res. 9:uhac004. doi: 10.1093/hr/uhac004
Wu, P., Gao, H., Liu, J., Kosma, D. K., Lü, S., and Zhao, H. (2021). Insight into the roles of the ER-associated degradation E3 ubiquitin ligase HRD1 in plant cuticular lipid biosynthesis. Plant Physiol. Biochem. 167, 358–365. doi: 10.1016/j.plaphy.2021.08.021
Yang, S. U., Kim, H., Kim, R. J., Kim, J., and Suh, M. C. (2020). AP2/DREB transcription factor RAP2.4 activates cuticular wax biosynthesis in Arabidopsis leaves under drought. Front. Plant Sci. 11:895. doi: 10.3389/fpls.2020.00895
Yang, X., Feng, T., Li, S., Zhao, H., Zhao, S., Ma, C., et al. (2020). CER16 inhibits post-transcriptional gene silencing of CER3 to regulate alkane biosynthesis. Plant Physiol. 182, 1211–1221. doi: 10.1104/pp.19.01002
Yang, X., Zhao, H., Kosma, D. K., Tomasi, P., Dyer, J. M., Li, R., et al. (2017). The acyl desaturase CER17 is involved in producing wax unsaturated primary alcohols and cutin monomers. Plant Physiol. 173, 1109–1124. doi: 10.1104/pp.16.01956
Yeats, T. H., Huang, W., Chatterjee, S., Viart, H. M., Clausen, M. H., Stark, R. E., et al. (2014). Tomato Cutin Deficient 1 (CD1) and putative orthologs comprise an ancient family of cutin synthase-like (CUS) proteins that are conserved among land plants. Plant J. 77, 667–675. doi: 10.1111/tpj.12422
Yeats, T. H., and Rose, J. K. (2013). The formation and function of plant cuticles. Plant Physiol. 163, 5–20. doi: 10.1104/pp.113.222737
Zhang, C. L., Hu, X., Zhang, Y. L., Liu, Y., Wang, G. L., You, C. X., et al. (2020a). An apple long-chain acyl-CoA synthetase 2 gene enhances plant resistance to abiotic stress by regulating the accumulation of cuticular wax. Tree Physiol. 40, 1450–1465. doi: 10.1093/treephys/tpaa079
Zhang, C. L., Zhang, Y. L., Hu, X., Xiao, X., Wang, G. L., You, C. X., et al. (2020b). An apple long-chain acyl-CoA synthetase, MdLACS4, induces early flowering and enhances abiotic stress resistance in Arabidopsis. Plant Sci. 297:110529. doi: 10.1016/j.plantsci.2020.110529
Zhang, J., Li, X. M., Lin, H. X., and Chong, K. (2019a). Crop improvement through temperature resilience. Annu. Rev. Plant Biol. 70, 753–780. doi: 10.1146/annurev-arplant-050718-100016
Zhang, J. Y., Broeckling, C. D., Sumner, L. W., and Wang, Z. Y. (2007). Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 64, 265–278. doi: 10.1007/s11103-007-9150-2
Zhang, P., Wang, R., Yang, X., Ju, Q., Li, W., Lü, S., et al. (2020c). The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 43, 1925–1943. doi: 10.1111/pce.13784
Zhang, Y. L., Zhang, C. L., Wang, G. L., Wang, Y. X., Qi, C. H., Zhao, Q., et al. (2019c). The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 19:362. doi: 10.1186/s12870-019-1918-4
Zhang, Y. L., Zhang, C. L., Wang, G. L., Wang, Y. X., Qi, C. H., You, C. X., et al. (2019b). Apple AP2/EREBP transcription factor MdSHINE2 confers drought resistance by regulating wax biosynthesis. Planta 249, 1627–1643. doi: 10.1007/s00425-019-03115-4
Zhao, H., Zhang, H., Cui, P., Ding, F., Wang, G., Li, R., et al. (2014). The putative E3 ubiquitin ligase ECERIFERUM9 regulates abscisic acid biosynthesis and response during seed germination and postgermination growth in Arabidopsis. Plant Physiol. 165, 1255–1268. doi: 10.1104/pp.114.239699
Zhong, M. S., Jiang, H., Cao, Y., Wang, Y. X., You, C. X., Li, Y. Y., et al. (2020). MdCER2 conferred to wax accumulation and increased drought tolerance in plants. Plant Physiol. Biochem. 149, 277–285. doi: 10.1016/j.plaphy.2020.02.013
Zhou, L., Ni, E., Yang, J., Zhou, H., Liang, H., Li, J., et al. (2013). Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS One 8:e65139. doi: 10.1371/journal.pone.0065139
Zhou, X., Li, L., Xiang, J., Gao, G., Xu, F., Liu, A., et al. (2015). OsGL1-3 is involved in cuticular wax biosynthesis and tolerance to water deficit in rice. PLoS One 10:e116676. doi: 10.1371/journal.pone.0116676
Zhu, X., and Xiong, L. (2013). Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc. Natl. Acad. Sci. U. S. A. 110, 17790–17795. doi: 10.1073/pnas.1316412110
Keywords: cuticle, drought, salinity, extreme temperatures, ultraviolet radiation, crop improvement
Citation: Liu L, Wang X and Chang C (2022) Toward a smart skin: Harnessing cuticle biosynthesis for crop adaptation to drought, salinity, temperature, and ultraviolet stress. Front. Plant Sci. 13:961829. doi: 10.3389/fpls.2022.961829
Received: 05 June 2022; Accepted: 01 July 2022;
Published: 25 July 2022.
Edited by:
Engin Yol, Akdeniz University, TurkeyReviewed by:
Muhammad Azhar Nadeem, Sivas University of Science and Technology, TurkeyHatice Sari, Akdeniz University, Turkey
Copyright © 2022 Liu, Wang and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng Chang, cc@qdu.edu.cn
 Lang Liu
Lang Liu Cheng Chang
Cheng Chang