Non-aureus Staphylococci Species in the Teat Canal and Milk in Four Commercial Swiss Dairy Herds
- 1Vetsuisse Faculty, Clinic for Ruminants, University of Bern, Bern, Switzerland
- 2Business Economics Group, Wageningen University, Wageningen, Netherlands
- 3Vetsuisse Faculty, Institute of Veterinary Bacteriology, University of Bern, Bern, Switzerland
Non-aureus staphylococci (NAS) are frequently found in milk samples as well as on the teat apex and in the teat canal and are known to be a cause of subclinical mastitis. The objective of this study was to investigate the relationship between NAS species colonizing the teat canal and those causing intramammary infection (IMI) in four commercial dairy herds. Teat canal swabs were obtained and thereafter milk samples were aseptically collected and evaluated for the presence of staphylococci using selective agar plates. Species identification was performed using matrix-assisted laser desorption/ionization time–of–flight mass spectrometry. The relationship between NAS species distribution and sample type (teat canal vs. milk samples) was quantified using hierarchical multivariable logistic regression models. The most prevalent NAS species in teat canal swabs were S. xylosus (35%), S. vitulinus (10%), and S. chromogenes (7%), whereas in milk samples S. chromogenes (5%), S. xylosus (5%), and S. haemolyticus (4%) were most prevalent. There were significantly higher odds for S. vitulinus (OR = 215), S. xylosus (OR = 20), S. sciuri (OR = 22), S. equorum (OR = 13), and S. succinus (OR = 10) to be present in teat canal swabs than in milk samples. Differences between herds in NAS species distribution were found and were most pronounced for S. succinus and a S. warneri-like species. This information aids in the understanding of NAS species as an etiology of IMI and should be taken into account when interpreting milk culture results.
Introduction
Non-aureus staphylococci (NAS) are frequently isolated from bovine milk samples (1–3). They are one of the main causes of subclinical mastitis in dairy herds that have successfully controlled major mastitis pathogens (1, 4) such as Staphylococcus aureus, Streptococcus uberis, Streptococcus agalactiae, Streptococcus dysgalactiae, and coliforms (5).
Non-aureus staphylococci species considered most relevant as causes of intramammary infection (IMI) are S. chromogenes, S. epidermidis, S. haemolyticus, S. simulans, and S. xylosus (6). Ecological, epidemiological and virulence behavior, as well as the antimicrobial resistance profile, vary widely between species (6–10) and even amongst strains of a given species (3, 11–14).
Multiple studies demonstrate that NAS are the most prevalent bacteria found on teat apices of lactating dairy cows (8, 15). They are also frequently isolated from teat canal samples (16–18). These findings suggest that teat apex and canal colonization may act as a reservoir for NAS species causing IMI. The importance of the teat canal as a barrier against bacterial invasion was partly supported by two recent studies which showed that, collectively, NAS were more frequently found in samples obtained by milking the quarter than in samples collected directly from the udder cistern or using the cannula technique (19, 20).
To the authors' knowledge, only one study has investigated the relationship between teat canal colonization and IMI on a NAS species level (17). Further studies to assess the extent of teat canal colonization and IMI by NAS species are needed to gain more insight into their ecology and epidemiology. Therefore, the objective of this study was to investigate the relationship of NAS species identified in milk with those colonizing the teat canal.
Materials and Methods
The study was approved by the committee for animal experimentation (KTV) of the Canton of Bern (BE58/14), Switzerland.
Selection and Characterization of Study Herds
Four commercial Swiss dairy herds (A, B, C, and D) were enrolled in the study. Herd health visits were part of the clinical service of the Clinic for Ruminants, University of Bern, and took place every 2 weeks. During these regular herd health visits, California Mastitis Tests (CMT) (21) were performed on cows with an individual composite SCC > 150,000 cells/ml, as indicated by the Swiss law (22). Quarters scoring > 1 in the CMT for the first time were sampled for bacteriological culture, species identification was subsequently performed using matrix-assisted laser desorption/ionization time–of–flight mass spectrometry (MALDI–TOF MS). Herds A, B, and C were also part of a longitudinal 1-year study to determine risk factors for NAS species IMI (23). Data collected during the herd health visits and in the aforementioned study were utilized to select herds with a known NAS status for this study. The proportion of samples NAS-positive compared to all quarter samples taken in 2013 were: A = 35.7%, B = 42.9%, C = 33.3%, D = 39.1%). Moreover, the herds had a low proportion of IMI caused by major mastitis pathogens (<20% of samples).
On farm A and B, cows were housed in free-stall barns with straw-bedded cubicles, while cows on farm C and D were housed in a tie-stall bedded on rubber mats covered with straw and sawdust, respectively. Milking was performed twice daily by one milker. For teat cleaning, all farmers used at least one disposable disinfection towel per cow. Post-milking teat disinfection with iodine-based products was performed routinely after each milking on farm A and D, sporadically (i.e., for high cell count cows) on farm B, and never on farm C.
Sampling Procedure
Herds were visited once in spring/summer 2015. Individual quarter milk samples and teat canal swabs were collected aseptically from all lactating cows. Teat cleaning and, if necessary, udder cleaning was performed by the milkers prior to sampling. Teats were then thoroughly disinfected with cotton swabs drenched in ethanol (70%). A sterile swab (Transwab ENT Amies Plain, MWE Medical Wire & Equipment, Corsham, Wiltshire, England) was inserted approximately 3 mm into the teat canal, rotated tree times and extracted. Subsequently, pre-milking of at least three streams of milk was performed, teat ends were disinfected again and individual quarter foremilk samples were aseptically collected. Latex gloves were worn during the whole procedure and changed after each cow. The milk samples were transported to the laboratory within 40 min at a maximum temperature of 7°C and were then frozen at −20°C until further analysis (1–3 days after sampling). Clinical mastitis cases were excluded from the study.
Laboratory Analysis
Milk samples were thawed and centrifuged at 590 × g for 10 min at 20°C. Ten microliter of milk sediment and the teat canal swabs were directly streaked on chromogenic agar plates selective for staphylococci (SaSelect, Bio-Rad, Marnes-la-Coquette, France) and incubated at 37°C for 48 h. Growth of staphylococci from both milk and teat canal swabs on the selective agar was previously validated as part of a parallel study (23). Staphylococci were differentiated from non–staphylococci species, such as Corynebacterium spp., Bacillus spp., and Aerococcus spp., based on colony shape and color. All suspected staphylococci isolates were identified to the species level by selecting one colony from each phenotypically different group on the plate and using MALDI–TOF MS (Microflex LT, Bruker Daltonics GmbH, Bremen, Germany). The extraction technique using 70% formic acid and α-Cyano-4-hydroxycinnamic acid (HCCA)-Matrix (Bruker Daltonics GmbH, Bremen, Germany) was applied. Isolates with scores ≥2.0 were identified to the species level according to manufacturer's guidelines. In a previous study, we identified two types of isolates displaying a score below 2.0 by MALDI–TOF MS (23). These isolates were found to be related to S. warneri and to S. devriesei by 16S rRNA gene-sequencing as well as by sequencing of the rpoB or dnaJ genes and were considered as S. warneri-like and S. devriesei-like, respectively (23). The reference spectra of these two species were introduced into the MALDI-TOF MS database and the updated database was used for the current study.
Teat canal swabs and milk samples with more than three different NAS species on the selective agar plates were considered as mixed staphylococcal species (23, 24). Incidentally growing non-staphylococci species were not considered for further analysis.
Definition of IMI
To maximize sensitivity, quarters were defined as having a NAS IMI if ≥100 CFU (colony forming units) per milliliter of at least one NAS species from a single sample were identified, following Dohoo et al. (25) and De Visscher et al. (26). Accordingly, teat canals were considered to be colonized if ≥1 CFU of at least one NAS species was identified.
Statistical Analyses
Raw data were entered and stored in Excel (Microsoft Excel 2010). Data management and statistical analyses were performed using the melogit command in STATA 14 (StataCorp, College Station, Texas, USA).
The bacteriological status “mixed staphylococci species” was analyzed first. The hierarchical logistic regression model for the probability of the bacteriological status “mixed staphylococci species” was as follows:
where sample type included teat canal swab vs. milk sample, herd represented the herd (A, B, C, and D) in which mixed staphylococci species were identified to allow for between-herd differences in NAS-species distribution (26–29), and u and v represented the random intercepts for quarter and cow. Covariates in the model were evaluated by the Type 3 test.
The “mixed staphylococci species” observations were subsequently excluded from the model in order to evaluate the NAS species individually. The same statistical model was applied for analyzing the NAS species but the sample type x herd interaction term was excluded because it was non-significant for all NAS species evaluated. The random quarter effect had to be removed from the model for S. chromogenes because of non-convergence. Staphylococci species with ≥5 observations in milk samples and swabs in total were analyzed individually, the remaining species were grouped and analyzed as “Others.”
The proportion of explained variance for species-specific NAS IMI at quarter and cow level was estimated based on the final NAS species-specific multilevel logistic regression model. Using the latent variable approach, by assuming that the variance at sample level was π2/3 (30), the total variance () was estimated to be:
where is the variation occurring at cow level and the variation at quarter level.
Results
Milk and teat canal samples of 97 lactating Red Holstein (including Red Factor Black Holstein) and Swiss Fleckvieh cows (herd A: n = 27, herd B: n = 15, herd C: n = 19, herd D: n = 36) were included in the analysis.
Mixed Staphylococci Species
Out of 770 quarter milk and teat canal samples, 11.3% (n = 87) contained mixed staphylococci species, consequently 88.7% (n = 683) of all samples contained zero to three staphylococci species. The final statistical model evaluating the bacterial status “mixed staphylococci species” identified a significant interaction between sample type and herd (Table 1). Mixed staphylococci species were significantly more frequently identified in teat canal swabs compared to milk samples. This was observed in all herds, except in herd C where a similar distribution was found (Figure 1).
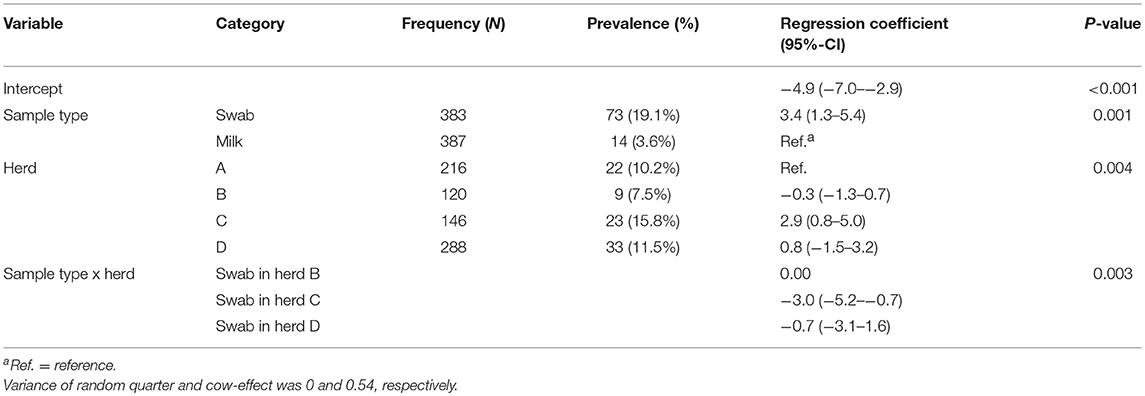
Table 1. Final hierarchical logistic regression model evaluating the presence of mixed non-aureus staphylococci species in 387 quarter milk samples and 383 quarter teat canal swabs in four commercial dairy herds in Switzerland.
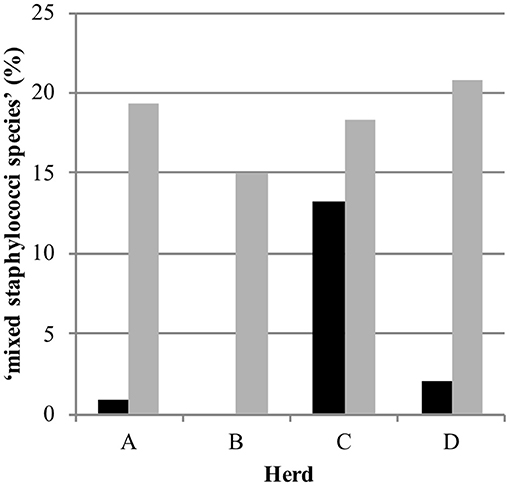
Figure 1. Distribution of the observed bacteriological status “mixed staphylococci species” in 387 quarter milk samples (black) and 383 quarter teat canal swabs (gray) in four commercial dairy herds (herds A, B, C, and D) in Switzerland.
NAS
Eighteen different NAS species were identified. The NAS species identified most often in both milk sample and teat canal of the same quarter was S. chromogenes (n = 12) (Table 2). As presented in Table 3, the most prevalent NAS species found in the teat canal was S. xylosus (34.8%, n = 108), followed by S. vitulinus (10.0%, n = 31) and S. chromogenes (7.1%, n = 22). The most prevalent NAS species found in milk was S. chromogenes (5.4%, n = 20), followed by S. xylosus (4.6%, n = 17), and S. haemolyticus (3.8%, n = 14).
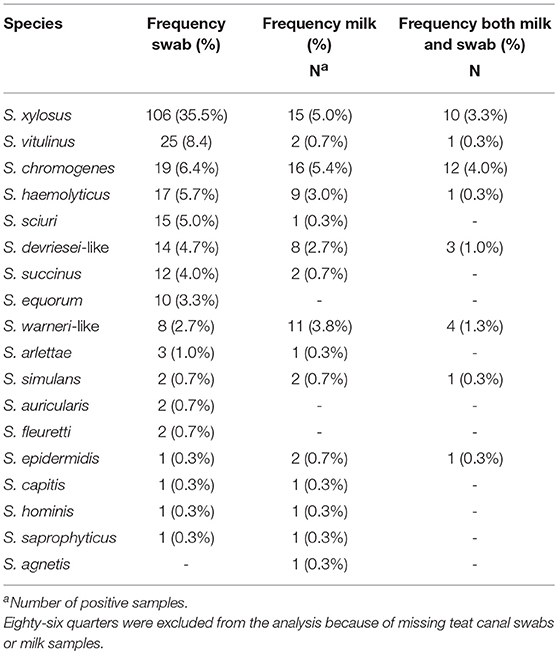
Table 2. Distribution of non-aureus staphylococci in milk samples and teat canal swabs taken from the same quarter (N = 299) in four commercial dairy herds in Switzerland.
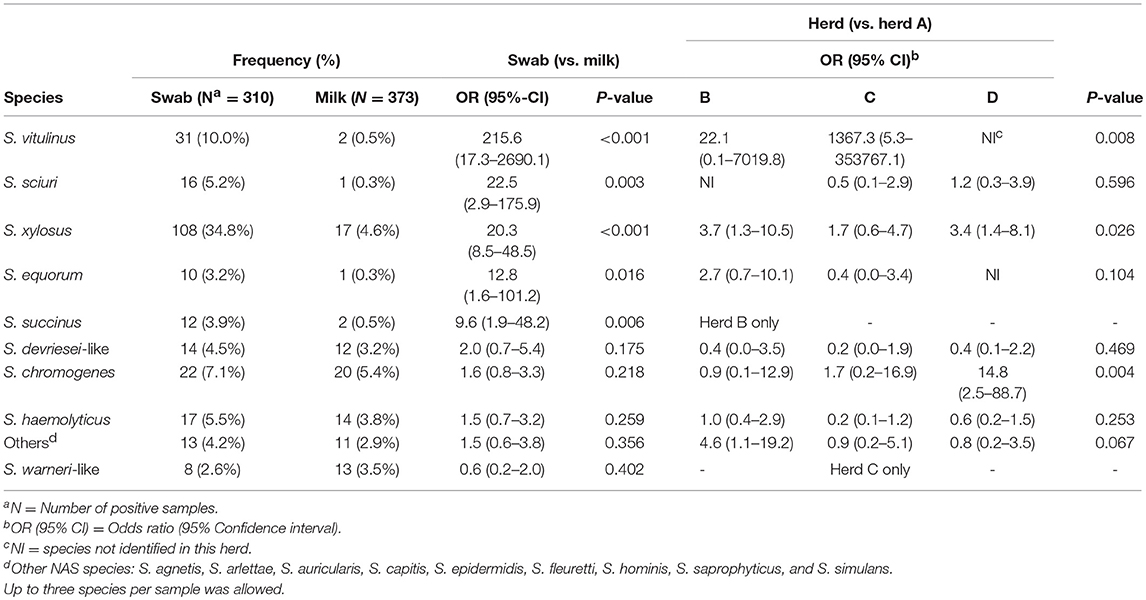
Table 3. Final multilevel multivariable logistic regression models describing the non-aureus staphylococci species distribution in teat canal swabs and milk samples (n = 683) in four commercial dairy herds in Switzerland.
The final statistical models evaluating NAS species distributions in milk samples and teat canal swabs are presented in Table 3. There were significantly higher odds for S. vitulinus (OR = 215.6), S. xylosus (OR = 20.3), S. sciuri (OR = 22.5), S. equorum (OR = 12.8), and S. succinus (OR = 9.6) to be present in teat canal swabs than in milk samples. There were no differences in occurrence of the species in question between milk samples and teat canal swabs for the other NAS species evaluated. Differences between herds concerning species distribution were observed for S. chromogenes, S. succinus, S. vitulinus, S. warneri-like, and S. xylosus.
Variance Components
Variance components of all final models are presented in Table 4 and differed between the evaluated species.
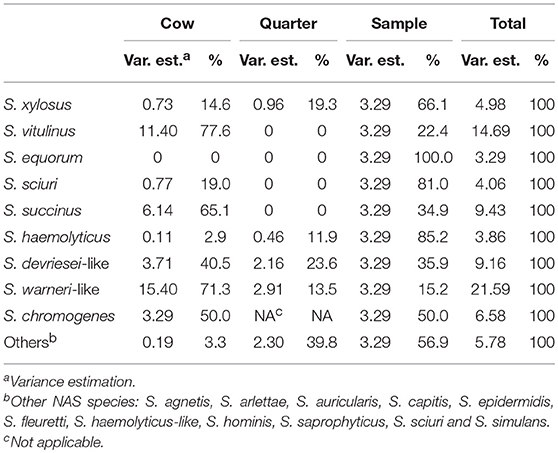
Table 4. Variance component estimation at cow, quarter and sample level (teat canal swabs vs. milk) for all 11 final multilevel logistic regression models describing the non-aureus staphylococci species distribution in four commercial dairy herds in Switzerland.
Discussion
Although the relationship between NAS species causing IMI and those colonizing the teat canal has rarely been quantified (17), a difference in distribution of NAS species between teat canal samples and milk samples was expected. Previous studies found uneven distributions of different NAS species in milk samples and teat apices, including NAS species relevant and less relevant for IMI respectively (8, 16, 27, 31). The teat canal acts as a barrier against invading microorganisms causing IMI and several studies have shown that it harbors several different bacterial species, including Staphylococcus spp., Enterobacteriaceae, Streptococcus spp., and Clostridium spp. (17, 18, 32).
For some NAS species, no significant differences between occurrence in milk samples and teat canal swabs were found in this study, which is mirrored by the distribution of the NAS species found in both sample types of the same quarter. A possible interpretation is that the NAS species without a difference between sample types are more likely to cause IMI. This was previously reported for S. chromogenes and S. haemolyticus (11, 14, 29). Molecular analysis for strain characterization is needed to determine if the isolates found in teat canals and milk samples are identical. Temporal changes in NAS species occurrence were not assessed and it therefore remains unclear if they first colonized the teat canal and caused an IMI by invading the mammary gland or vice versa. It must be noted that the lack of statistical significance for some NAS species might be due to a low statistical power. However, given the low magnitude of these point estimates, as represented by the odds ratio, the difference between teat canal swabs and milk samples will not be clinically relevant for these NAS species.
For S. chromogenes, Quirk et al. (17) showed that teat canal colonization mostly preceded IMI, whereas the opposite was true for S. xylosus. Various studies agree on the host-associated characteristics of S. chromogenes since it is mainly isolated from cows' body-sites and milk and rarely from the environment (8, 16, 28). Strain typing showed high clonality and the capacity of S. chromogenes to disseminate within herds (13). Additionally, several authors confirmed its ability to cause persistent IMI (3, 29, 33). Some S. chromogenes strains are able to adhere to and enter bovine mammary epithelial cells (12) and a difference between pathogenicity and in vivo growth capacity was observed (14), which supports the hypothesis of host adaptation.
In the current study, S. haemolyticus was found in both milk samples and teat canal swabs, but rarely in both samples from the same quarter. The presence in both teat canal and milk samples confirms findings of other studies (8, 15, 17, 34). Presence on teat apices and in teat canals, however, seems to vary since S. haemolyticus was not found in teat canals in two separate studies by Gill et al. (18) and Taponen et al. (16). This difference might be due to the finding of Piessens et al. (28), who reported that the environment (air, slatted floor, sawdust) was the main reservoir for this species. A study from Leroy et al. (11) comparing subtypes from milk and teat apices indicated that IMI originated from strains present on the teat skin. Thus, S. haemolyticus appears to be a versatile species that can adapt to the animal (skin, teat canal, udder) as well as to the environment.
Staphylococcus xylosus was isolated more frequently in the teat canal than in milk samples and rarely in both samples from the same quarter, indicating that this species often colonizes the teat canal without necessarily causing an IMI. Together with S. chromogenes, S. xylosus is one of the most frequently identified NAS species in milk (6). The higher proportion of S. xylosus in teat canals compared to milk might be explained by a primarily environmental reservoir, as postulated by Piessens et al. (28); however, other authors have cultured the species from teat apex, udder skin, perineal tissue, and milkers' hands (16, 27, 31).
Staphylococcus warneri-like may represent a new species related to S. warneri (23) and was found in both samples from the same quarter in four of 19 cases, exclusively in study herd C. It also had a high within-herd occurrence (23) which may be a result of either a high incidence, a low cure rate, or both (3). Although not significant, this was the only NAS species found more frequently in milk samples than in teat canal swabs, which could be explained by a low cure rate and would be in accordance with Mørk et al. (3), who found that S. warneri can cause persistent IMI. Its predominant reservoir is unclear, but S. warneri has rarely been isolated from environmental samples (28) and is part of the human skin microbiota (35, 36).
In the present study, the odds to find S. vitulinus in the teat canal, rather than in milk, were the highest of all NAS species. To the authors' knowledge, S. vitulinus has not been previously found in teat canal samples but seems to be well-adapted to this particular microenvironment in the herds that we sampled. Staphylococcus vitulinus was rarely found to cause IMI (23, 26, 37) and was only recovered from a small proportion of teat apex samples (8, 27, 31). This might indicate that this species does not favor the mammary gland, but acts opportunistically.
Among the less prevalent species identified, S. sciuri, S. equorum, and S. succinus also had higher odds to be found in the teat canal vs. in milk samples. This was not the case for S. devriesei-like and “Others.” Staphylococcus equorum and S. sciuri were primarily isolated from the environment (28) and extramammary parlor-related habitats, such as clusters, liners, and milkers' gloves (8), but are also known to cause IMI (23, 28, 29). Staphylococcus equorum was among the most frequently found NAS species on teat apices (15, 27, 38) but has only been isolated once from the teat canal (17). Moreover, it was the predominant species in bulk milk in a Belgian longitudinal study (37). Quarters with dirty teat apices before calving were more likely to be infected with S. equorum and S. sciuri after parturition, supporting their environmental nature (26). Staphylococcus succinus was isolated from teat apices and rarely from the teat canal (16). Staphylococcus devriesei was frequently isolated from teat apices before parturition (38), but, to the authors' knowledge, not from the teat canal. Staphylococcus devriesei, S. devriesei-like, and S. succinus are able to cause IMI (23), but little is known about their reservoirs and epidemiology.
Variance estimates were the highest at cow level for S. chromogenes, S. devriesei-like, S. succinus, S. vitulinus, and S. warneri-like, indicating that a high proportion of the unexplained variance of IMI is determined by cow characteristics such as the efficiency of the innate non-local immune system. For S. equorum, S. haemolyticus, S. sciuri, and S. xylosus the biggest variance occurred at the sample level, which might be caused by sample location differences such as local barriers (i.e., the teat canal keratin layer) and the local immune system preventing a teat canal colonization to develop into IMI.
Some authors claim that certain NAS species might belong to the normal intramammary microbiota (39, 40) and may also play a role in the modulation of mastitis susceptibility, as recently reviewed by Derakhshani et al. (41). This concept, however, is questioned by others (42).
Mixed staphylococci species were found significantly more often in teat canal swabs samples (19.1%) than in milk samples (3.6%), except in herd C. The high presence of flies in this herd, as observed during the regular herd visits and the longitudinal study preceding the current study (23), might influence the bacteria population on teats and in teat canals since they may transmit bacteria causing IMI (43, 44). The same definition for mixed staphylococcal species was used in our above-mentioned previous study and also by Mahmmod et al. (24). In order to avoid preclusion of potentially relevant information, considering that the teat canal—similar to teat apices—harbors many different NAS species (8, 15–18) and taking into account the duality of its role—as a barrier against IMI on one side and potential reservoir for NAS species causing IMI on the other—the decision to include culture results with up to three Staphylococcus species was made.
The objective of this cross-sectional study was to assess the relationship between NAS species colonizing the teat canal and those causing IMI, thereby emphasizing the importance of S. chromogenes, S. haemolyticus, and S. xylosus as potential causative agents of IMI. In contrast, species such as S. equorum, S. vitulinus, and S. succinus seem to be more important as residents of the teat canal.
The small number of animals and herds sampled might limit the extrapolation of the results to the general Swiss dairy cattle population. Nevertheless,—despite not being longitudinal—this study provides new insights in the complex epidemiology and ecology of NAS. In light of the results and the previously mentioned dual role of the teat canal and the fact that NAS are frequently found on teat apices (8, 15), some relevant consequences arise. In particular, the difference in NAS distribution between sample types needs to be considered when interpreting milk culture results. This is highlighted by the fact that some NAS species were found significantly more often in teat canal swabs than in milk, which is line with other studies where isolation of NAS diminished if the teat canal was bypassed for milk sampling (19, 20). Consequently, the definition of IMI relying on culture results only is challenged, and consideration of milk SCC may be valuable in determining the causative role of NAS species (and other bacteria) isolated from conventionally collected milk samples.
Data Availability
Datasets are available on request. The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
This study was carried out in accordance with the recommendations of the committee for animal experimentation (KTV) of the Canton of Bern (BE58/14), Switzerland.
Author Contributions
BvdB and MB conceived and designed the study. JT and CD performed sample collection and part of the bacteriological analyses. AT and VP conceived and supervised the bacteriological part of the study. JT and BvdB analyzed the data and interpreted the study results together with MB. JT drafted the manuscript. BvdB and MB performed manuscript review. All authors contributed to manuscript revision and have read and approved the final manuscript.
Funding
The study was funded by internal funds of the Institute of Veterinary Bacteriology and of the Clinic for Ruminants, Vetsuisse-Faculty, University of Bern, as well as by a grant of the Department for Clinical Veterinary Medicine, Vetsuisse-Faculty, University of Bern, Switzerland.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors appreciated the collaboration of all participating farmers. Special thanks goes to Werner Lehmann from the Institute of Veterinary Bacteriology for his assistance during the study.
References
1. Piepers S, De Meulemeester L, de Kruif A, Opsomer G, Barkema HW, De Vliegher S. Prevalence and distribution of mastitis pathogens in subclinically infected dairy cows in Flanders, Belgium. J Dairy Res. (2007) 74:478–83. doi: 10.1017/S0022029907002841
2. Sampimon OC, Barkema HW, Berends IMGA, Sol J, Lam TJGM. Prevalence and herd-level risk factors for intramammary infection with coagulase-negative staphylococci in Dutch dairy herds. Vet Microbiol. (2009) 134:37–44. doi: 10.1016/j.vetmic.2008.09.010
3. Mørk T, Jørgensen HJ, Sunde M, Kvitle B, Sviland S, Waage S, et al. Persistence of staphylococcal species and genotypes in the bovine udder. Vet Microbiol. (2012) 159:171–80. doi: 10.1016/j.vetmic.2012.03.034
4. Schukken YH, González RN, Tikofsky LL, Schulte HF, Santisteban CG, Welcome FL, et al. CNS mastitis: nothing to worry about? Vet Microbiol. (2009) 134:9–14. doi: 10.1016/j.vetmic.2008.09.014
5. Djabri B, Bareille N, Beaudeau F, Seegers H. Quarter milk somatic cell count in infected dairy cows: a meta-analysis. Vet Res. (2002) 33:335–57. doi: 10.1051/vetres
6. Vanderhaeghen W, Piepers S, Leroy F, Van Coillie E, Haesebrouck F, De Vliegher S. Invited review: effect, persistence, and virulence of coagulase-negative Staphylococcus species associated with ruminant udder health. J Dairy Sci. (2014) 97:5275–93. doi: 10.3168/jds.2013-7775
7. Vanderhaeghen W, Piepers S, Leroy F, Van Coillie E, Haesebrouck F, De Vliegher S. Identification, typing, ecology and epidemiology of coagulase negative staphylococci associated with ruminants. Vet J. (2015) 203:44–51. doi: 10.1016/j.tvjl.2014.11.001
8. De Visscher A, Supré K, Haesebrouck F, Zadoks RN, Piessens V, Van Coillie E, et al. Further evidence for the existence of environmental and host-associated species of coagulase-negative staphylococci in dairy cattle. Vet Microbiol. (2014) 172:466–74. doi: 10.1016/j.vetmic.2014.06.011
9. Frey Y, Rodriguez JP, Thomann A, Schwendener S, Perreten V. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J Dairy Sci. (2013) 96:2247–57. doi: 10.3168/jds.2012-6091
10. Nobrega DB, Naushad S, Naqvi SA, Condas LAZ, Jean P, Sanders J. Prevalence and genetic basis of antimicrobial resistance in non- aureus staphylococci isolated from Canadian Dairy Herds. Front Microbiol. (2018) 9:256. doi: 10.3389/fmicb.2018.00256
11. Leroy F, Van Coillie E, Braem G, Piessens V, Verbist B, De Vuyst L, et al. Short communication: subtyping of Staphylococcus haemolyticus isolates from milk and corresponding teat apices to verify the potential teat-skin origin of intramammary infections in dairy cows. J Dairy Sci. (2015) 98:7893–8. doi: 10.3168/jds.2015-9415
12. Souza FN, Piepers S, Della Libera AMMP, Heinemann MB, Cerqueira MMOP, De Vliegher S. Interaction between bovine-associated coagulase-negative staphylococci species and strains and bovine mammary epithelial cells reflects differences in ecology and epidemiological behavior. J Dairy Sci. (2016) 99:2867–74. doi: 10.3168/jds.2015-10230
13. Piessens V, De Vliegher S, Verbist B, Braem G, Van Nuffel A, De Vuyst L, et al. Intra-species diversity and epidemiology varies among coagulase-negative Staphylococcus species causing bovine intramammary infections. Vet Microbiol. (2012) 155:62–71. doi: 10.1016/j.vetmic.2011.08.005
14. Piccart K, Verbeke J, De Visscher A, Piepers S, Haesebrouck F, De Vliegher S. Local host response following an intramammary challenge with Staphylococcus fleurettii and different strains of Staphylococcus chromogenes in dairy heifers. Vet Res. (2016) 47:1–11. doi: 10.1186/s13567-016-0338-9
15. Braem G, De Vliegher S, Verbist B, Piessens V, Van Coillie E, De Vuyst L, et al. Unraveling the microbiota of teat apices of clinically healthy lactating dairy cows, with special emphasis on coagulase-negative staphylococci. J Dairy Sci. (2013) 96:1499–510. doi: 10.3168/jds.2012-5493
16. Taponen S, Björkroth J, Pyörälä S. Coagulase-negative staphylococci isolated from bovine extramammary sites and intramammary infections in a single dairy herd. J Dairy Res. (2008) 75:422–9. doi: 10.1017/S0022029908003312
17. Quirk T, Fox LK, Hancock DD, Capper J, Wenz J, Park J. Intramammary infections and teat canal colonization with coagulase-negative staphylococci after postmilking teat disinfection: species-specific responses. J Dairy Sci. (2012) 95:1906–12. doi: 10.3168/jds.2011-4898
18. Gill JJ, Sabour PM, Gong J, Yu H, Leslie K, Griffiths MW. Characterization of bacterial populations recovered from the teat canals of lactating dairy and beef cattle by 16S rRNA gene sequence analysis. FEMS Microbiol Ecol. (2006) 56:471–81. doi: 10.1111/j.1574-6941.2006.00091.x
19. Hiitiö H, Simojoki H, Kalmus P, Holopainen J, Pyörälä S, Taponen S. The effect of sampling technique on PCR-based bacteriological results of bovine milk samples. J Dairy Sci. (2016) 99:6532–6541. doi: 10.3168/jds.2015-10811
20. Friman M, Hiitiö H, Niemi M, Holopainen J, Pyörälä S, Simojoki H. The effect of a cannula milk sampling technique on the microbiological diagnosis of bovine mastitis. Vet J. (2017) 226:57–61. doi: 10.1016/j.tvjl.2017.07.003
21. Schalm O, Noorlander B. Experiments and observations leading to development of the California mastitis test. J Am Vet Med Assoc. (1957) 130:199–204.
22. Eidgenössische Departement des Innern (EDI). Verordnung des EDI über die Hygiene bei der Milchproduktion. (2017) 2005:1–14. Available online at: http://www.admin.ch/opc/de/classified-compilation/20051436/index.html
23. Dolder C, van den Borne BHP, Traversari J, Thomann A, Perreten V, Bodmer M. Quarter- and cow-level risk factors for intramammary infection with coagulase-negative staphylococci species in Swiss dairy cows. J Dairy Sci. (2017) 100:1–11. doi: 10.3168/jds.2016-11639
24. Mahmmod YS, Nonnemann B, Svennesen L, Pedersen K, Klaas IC. Typeability of MALDI-TOF assay for identification of non- aureus staphylococci associated with bovine intramammary infections and teat apex colonization. J Dairy Sci. (2018) 101:9430–8. doi: 10.3168/jds.2018-14579
25. Dohoo IR, Smith J, Andersen S, Kelton DF, Godden S. Diagnosing intramammary infections: evaluation of definitions based on a single milk sample. J Dairy Sci. (2011) 94:250–61. doi: 10.3168/jds.2010-3559
26. De Visscher A, Piepers S, Haesebrouck F, De Vliegher S. Intramammary infection with coagulase-negative staphylococci at parturition: species-specific prevalence, risk factors, and effect on udder health. J Dairy Sci. (2016) 99:6457–69. doi: 10.3168/jds.2015-10458
27. Mahmmod YS, Klaas IC, Svennesen L, Pedersen K, Ingmer H. Communications of Staphylococcus aureus and non-aureus Staphylococcus species from bovine intramammary infections and teat apex colonization. J Dairy Sci. (2018) 101:7322–33. doi: 10.3168/jds.2017-14311
28. Piessens V, Van Coillie E, Verbist B, Supré K, Braem G, Van Nuffel A, et al. Distribution of coagulase-negative Staphylococcus species from milk and environment of dairy cows differs between herds. J Dairy Sci. (2011) 94:2933–44. doi: 10.3168/jds.2010-3956
29. Supré K, Haesebrouck F, Zadoks RN, Vaneechoutte M, Piepers S, De Vliegher S. Some coagulase-negative Staphylococcus species affect udder health more than others. J Dairy Sci. (2011) 94:2329–40. doi: 10.3168/jds.2010-3741
30. Dohoo I, Martin W, Stryhn H. Veterinary Epidemiologic Research. 2nd ed. Charlottetown, PE: VER Inc. (2009).
31. Adkins PRF, Dufour S, Spain JN, Calcutt MJ, Reilly TJ, Stewart GC, et al. Molecular characterization of non- aureus Staphylococcus spp. from heifer intramammary infections and body sites. J Dairy Sci. (2018) 101:5388–403. doi: 10.3168/jds.2017-13910
32. Paduch JH, Mohr E, Krömker V. The association between teat end hyperkeratosis and teat canal microbial load in lactating dairy cattle. Vet Microbiol. (2012) 158:353–9. doi: 10.1016/j.vetmic.2012.02.032
33. Fry PR, Middleton JR, Dufour S, Perry J, Scholl D, Dohoo I. Association of coagulase-negative staphylococcal species, mammary quarter milk somatic cell count, and persistence of intramammary infection in dairy cattle. J Dairy Sci. (2014) 97:4876–85. doi: 10.3168/jds.2013-7657
34. De Visscher A, Piepers S, Supré K, Haesebrouck F, De Vliegher S. Short communication: species group-specific predictors at the cow and quarter level for intramammary infection with coagulase-negative staphylococci in dairy cattle throughout lactation. J Dairy Sci. (2015) 98:5448–53. doi: 10.3168/jds.2014-9088
35. Dekio I, Hayashi H, Sakamoto M, Kitahara M, Nishikawa T, Suematsu M, et al. Detection of potentially novel bacterial components of the human skin microbiota using culture-independent molecular profiling. J Med Microbiol. (2005) 54:1231–8. doi: 10.1099/jmm.0.46075-0
36. Rasmussen TT, Kirkeby LP, Poulsen K, Reinholdt J, Kilian M. Resident aerobic microbiota of the adult human nasal cavity. Apmis. (2000) 108:663–75. doi: 10.1034/j.1600-0463.2000.d01-13.x
37. De Visscher A, Piepers S, Haesebrouck F, Supré K, De Vliegher S. Coagulase-negative Staphylococcus species in bulk milk: prevalence, distribution, and associated subgroup- and species-specific risk factors. J Dairy Sci. (2017) 100:1–14. doi: 10.3168/jds.2016-11476
38. De Visscher A, Piepers S, Haesebrouck F, De Vliegher S. Teat apex colonization with coagulase-negative Staphylococcus species before parturition: distribution and species-specific risk factors. J Dairy Sci. (2016) 99:1427–39. doi: 10.3168/jds.2015-10326
39. Kuehn JS, Gorden PJ, Munro D, Rong R, Dong Q, Plummer PJ, et al. Bacterial community profiling of milk samples as a means to understand culture-negative bovine clinical mastitis. PLoS ONE. (2013) 8:0061959. doi: 10.1371/journal.pone.0061959
40. Oikonomou G, Machado VS, Santisteban C, Schukken YH, Bicalho RC. Microbial diversity of bovine mastitic milk as described by pyrosequencing of metagenomic 16s rDNA. PLoS ONE. (2012) 7:0047671. doi: 10.1371/journal.pone.0047671
41. Derakhshani H, Fehr KB, Sepehri S, Francoz D, De Buck J, Barkema HW, et al. Invited review: microbiota of the bovine udder: contributing factors and potential implications for udder health and mastitis susceptibility. J Dairy Sci. (2018) 101:10605–25. doi: 10.3168/jds.2018-14860
42. Rainard P. Mammary microbiota of dairy ruminants: fact or fiction? Vet Res. (2017) 48:1–10. doi: 10.1186/s13567-017-0429-2
43. Anderson KL, Lyman R, Moury K, Ray D, Watson DW, Correa MT. Molecular epidemiology of Staphylococcus aureus mastitis in dairy heifers. J Dairy Sci. (2012) 95:4921–30. doi: 10.3168/jds.2011-4913
Keywords: intramammary infection, teat canal, non-aureus staphylococci, species distribution, mastitis
Citation: Traversari J, van den Borne BHP, Dolder C, Thomann A, Perreten V and Bodmer M (2019) Non-aureus Staphylococci Species in the Teat Canal and Milk in Four Commercial Swiss Dairy Herds. Front. Vet. Sci. 6:186. doi: 10.3389/fvets.2019.00186
Received: 27 February 2019; Accepted: 24 May 2019;
Published: 12 June 2019.
Edited by:
Francisco Ruiz-Fons, Spanish National Research Council (CSIC), SpainReviewed by:
John R. Middleton, University of Missouri, United StatesJean-Philippe Roy, Université de Montréal, Canada
Copyright © 2019 Traversari, van den Borne, Dolder, Thomann, Perreten and Bodmer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia Traversari, jtraversari@vetclinics.uzh.ch
†Present Address: Julia Traversari, Vetsuisse Faculty, Clinic for Ruminants, University of Zürich, Zurich, Switzerland
 Julia Traversari
Julia Traversari Bart H. P. van den Borne
Bart H. P. van den Borne Claudio Dolder1
Claudio Dolder1  Vincent Perreten
Vincent Perreten Michèle Bodmer
Michèle Bodmer