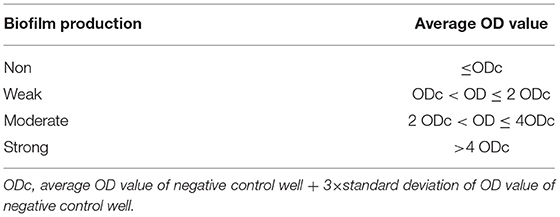Impact of Sub-MIC Eugenol on Klebsiella pneumoniae Biofilm Formation via Upregulation of rcsB
- 1College of Animal Medicine, Jilin Agricultural University, Changchun, China
- 2The Key Laboratory of New Veterinary Drug Research and Development of Jilin Province, Jilin Agricultural University, Changchun, China
- 3Animal Production Department, Faculty of Agriculture, Al-Azhar University, Nasr City, Egypt
- 4The 3nd Affiliated Clinical Hospital of Changchun University of Chinese Medicine, Changchun, China
- 5College of Life Science, Jilin Agricultural University, Changchun, China
- 6The Engineering Research Center of Bioreactor and Drug Development, Ministry of Education, Jilin Agricultural University, Changchun, China
The Rcs phosphorelay system is present in many members of the Enterobacteriaceae. The aim of this study was to illustrate the possible mechanisms of eugenol on ultimate targets of Klebsiella pneumoniae (K. pneumoniae) Rcs phosphorelay, rcsB, and impact on biofilm formation. The minimum inhibitory concentration (MIC) of eugenol against K. pneumoniae KP1 and KP1 ΔrcsB strain was determined using the 2-fold micro-dilution method. Biofilm was measured by crystal violet staining. Transcriptome sequencing was performed to investigate sub-MIC eugenol on K. pneumoniae, and gene expression at mRNA level was analyzed by RT-qPCR. In vitro biofilm formation test and molecular docking were used to evaluate the effect of eugenol and to predict potential interactions with RcsB. MicroScale Thermophoresis (MST) was conducted for further validation. MIC of eugenol against K. pneumoniae KP1 and KP1 ΔrcsB strain was both 200 μg/ml. Transcriptome sequencing and RT-qPCR results indicated that rpmg, degP, rnpA, and dapD were downregulated, while rcsB, rcsD, rcsA, yiaG, and yiaD were upregulated in the eugenol-treated group. ΔrcsB exhibited a weakened biofilm formation capacity. Additional isopropyl-β-D-thiogalactoside (IPTG) hinders biofilm formation, while sub-MIC eugenol could promote biofilm formation greatly. Docking analysis revealed that eugenol forms more hydrophobic bonds than hydrogen bonds. MST assay also showed a weak binding affinity between eugenol and RcsB. These results provide significant evidence that rcsB plays a key role in K. pneumoniae biofilm formation. Sub-MIC eugenol facilitates biofilm formation to a large extent instead of inhibiting it. Our findings reveal the potential risk of natural anti-biofilm ingredients at sub-MIC to treat drug-resistance bacteria.
Introduction
Antimicrobial resistance (AMR) has increased the challenge in today's livestock and human health globally. Klebsiella pneumoniae is one of the opportunistic pathogens that cause numerous diseases, such as bacteremia, pneumonia, septicemia, respiratory and urinary tract infections, mastitis, pyometra, and enteritis in cattle, goats, and companion animals (1). Globally, an increase in the incidence of hypervirulent and multidrug-resistant K. pneumoniae is considered a consequence of currently excessive use of antibiotics (2, 3). Although K. pneumoniae is believed to be less risky for animal production for a long time, its prevention and control have been necessitated due to the emergence of hypervirulent and multidrug-resistant strains of K. pneumoniae (4). For instance, it has also been noticed that loss of milk production as a result of mastitis and deaths of livestock is due to these multidrug-resistant strains of K. pneumoniae (5).
Biofilm resistance ability is one of the major barriers to the treatment of infectious diseases, as it prevents the antibiotics from interring the bacterial cells. The formation of bacterial biofilm is a dynamic process that depends on bacterial structure, function, and composition (6, 7). Bacterial extracellular polysaccharides (EPSs) play a major role in stabilizing the structure and function of bacterial biofilms (8).
The Regulator of Capsule Synthesis (Rcs) phosphorelay system was originally identified during a screening study on Escherichia coli for necessary genes responsible for capsular polysaccharide (CPS) synthesis (9). The Rcs phosphorelay system is composed of core proteins RcsB, RcsC, and RcsD (10). The phosphoryl group is transferred to a phosphotransfer protein RcsD (known as membrane-spanning protein) by RcsC and finally to RcsB (11). The previous study has proved that in the absence of external phosphatases, the Rcs activation signal passed to RcsB can be long-lived. Despite this, RcsB can have significant constitutive activity in the absence of RcsC or RcsD phosphorelay but through other pathways to carry out regulation. In addition, several auxiliary regulatory proteins such as RcsA, GadE, BglJ, MatA (EcpR), TviA, DctR, RflM, and RmpA act independently for phosphorylation (12, 13). The biofilm formation and regulation of CPS are majorly dependent on the Rcs phosphorelay (14, 15).
Eugenol is a phenolic component occurring in many aromatic plants of essential oil and other sources such as cinnamon extract, clove bud oil, and many others. Many studies have demonstrated antimicrobial, antioxidant, anti-inflammatory, antifungal, antiviral, anticancer, analgesic, and antispasmodic activities for eugenol (16). The double bond located at the γ position of the methyl group and α and β positions of the side chain is responsible for eugenol-mediated antimicrobial activity (17).
The application of eugenol can prevent the growth of pathogenic biofilm formed by K. pneumoniae (18). With this background, this study aimed to investigate the impact of sub-minimum inhibitory concentration (MIC) eugenol on K. pneumoniae biofilm formation and possible interactions with RcsB.
Methods
Chemicals, Bacteria, and Culture Medium
Eugenol (purity ≥98.5%) was purchased from Shanghai Yuan Ye Bio-Technology Co., Ltd. Clinical K. pneumonia isolate KP1 and KP1ΔrcsB strain were preserved in our laboratory. Luria-Bertani (LB) medium was obtained from Qingdao Hope Bio-Technology Co., Ltd (Qingdao, China).
Determination of MIC
The MIC of eugenol against K. pneumonia wild-type KP1 and ΔrcsB strain was determined using the broth micro-dilution method. Bacterial suspension at 106 CFU/ml was used for serial dilution and inoculated to an equal volume of eugenol. The negative and positive control tubes were set as LB broth only and LB with bacteria, respectively. The value of the lowest concentration that inhibits the growth of bacteria after incubation at 37°C for 12 h was termed MIC. This assay was performed in triplicate at separate times.
Detection of Biofilm Formation
The biofilm formation of KP1 and ΔrcsB strain was determined according to a previous study with slight modifications (19). In brief, the bacteria grown in LB broth were cultured to an OD600 of 0.5 (108 CFU/ml). A 96-well microtiter plate was used for equal distribution of the suspension. The tubes containing LB medium only were set as the negative control. The plates were incubated at 37°C for 24 h to facilitate biofilm formation in a static culture. Contents were removed gently after incubation with tap water. Notably, 200 μl of phosphate buffer saline (PBS, pH 7.2) was used for wells washing four times to remove any unbound cells. To stain the remaining adherent bacteria, 50 μl of crystal violet (1%) was added to the wells followed by keeping them at room temperature for 15 min. Distilled water was used for the removal of the excess stain, and the pallet was subjected to drying. The stained biofilm was solubilized with 95% ethanol, and the total biofilm mass was quantified by measuring the OD570 using the Universal Microplate Reader (Thermo Fisher Scientific). Samples were assayed in 6 replicates and performed thrice. The previous study was adopted for interpretation of the biofilm production (Table 1) (20).
Treatment of Samples, RNA Extraction, Library Construction, and Transcriptome Sequencing
Transcriptome sequencing was described in a formerly published paper in our laboratory. In brief, K. pneumoniae was cultured to reach 0.5 at OD600. Then, eugenol was added to a final concentration of 1/4 MIC and further cultured for 1 h. In control, LB medium was added instead of eugenol. The total RNA was extracted and digested with DNase I in compliance with the manufacturer's instructions. The quality and quantity of RNA were measured using the Thermo Scientific NanoDrop 2000 UV-Vis Spectrophotometer. Complementary probe sequences were applied for the elimination of the prokaryotic rRNAs. The template for the synthesis of the first strand of cDNA was obtained by random fragmentation of the depleted rRNA into small pieces using a fragmentation buffer. Subsequently, a buffer, dNTPs, dATP, dGTP, dCUP, RNaseH, and DNA polymerase I were added for the synthesis of the second strand of cDNA. The samples were sent to Biomarker Technology for library construction and sequencing using Illumina HiSeq 2500 platform. Genome-based reads mapping reference was adopted for transcriptome analysis. High-quality reads were mapped with reference K. pneumoniae strain KP52.145 sequence (GenBank: FO834906.1) to get position and characteristic information.
RT-qPCR Analysis
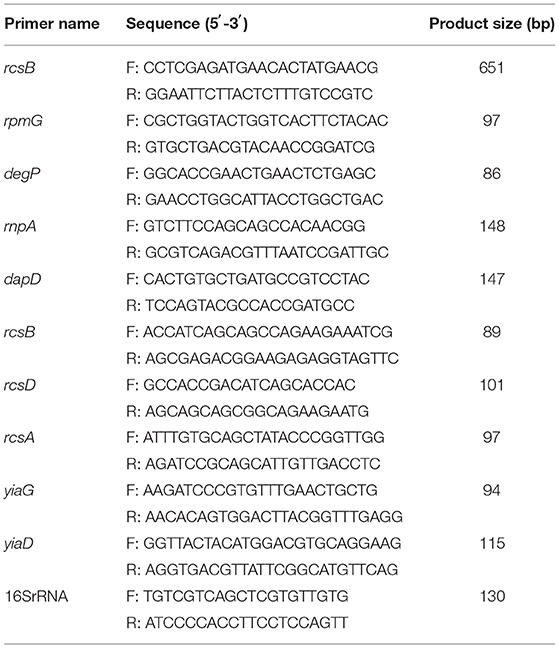
The expression of rpmG, degP, rnpA, dapD, rcsB, rcsD, rcsA, yiaG, and yiaD genes at the mRNA level was selected from transcriptome sequencing and evaluated via RT-qPCR. Bacteria were prepared as mentioned in the “Transcriptome Sequencing” section. In the control group, LB broth was added instead of eugenol. The total RNA was extracted using the method as previously described and was then reversely transcribed to cDNA. Then, cDNA was used as a template for RT-qPCR amplification in the same reaction tube using PerfectStartTM Green RT-qPCR SuperMix. RT-qPCR was carried out in an Applied Biosystems 7500 RT-qPCR System (USA). The sequences of RT-qPCR primers used are shown in Table 2. The amplifications were performed in 20 μl reaction mixtures containing 10 μl PerfectStartTM Green RT-qPCR SuperMix, 0.5 μl forward and reverse primer each, 8.0 μl nuclease-free water, and 1.0 μl template, respectively. 16SrRNA of the strain was used as a reference gene. The reaction condition was set as a two-step method as follows: one cycle at 94°C for 30 s, then 40 cycles consisting of denaturation were at 95°C for 5 s, and signal collection at 60°C for 34 s. All templates were run in triplicates.
Inhibitory Effect of Eugenol on Bacterial Biofilm Formation
The biofilm formation of different groups was as follows: plasmid pET-28a-rcsB transferred to ΔrcsB competent cell was set as ΔrcsB + RcsB group, plasmid pET-28a-rcsB transferred to KP1 competent cell was set as KP1 + RcsB group, moreover, ΔrcsB + RcsB + Eu, ΔrcsB + RcsB + isopropyl-β-D-thiogalactoside (IPTG), ΔrcsB + RcsB + IPTG + Eu, KP1 + RcsB + Eu, KP1 + RcsB + IPTG, and KP1 + RcsB + IPTG + Eu were also set, respectively. In ΔrcsB + RcsB + IPTG and KP1 + RcsB + IPTG groups, 0.1 mmol/L IPTG was first added and cultured for 4 h to contribute to the induction and then cultured for 24 h. For ΔrcsB + RcsB + IPTG + Eu and KP1 + RcsB + IPTG + Eu groups, in addition to the above treatment, eugenol was cocultured with the mixture with a final concentration of 1/4 MIC at 37°C for 24 h. The biofilm formation was determined with the same method as mentioned in the sections above. The negative and positive controls were set on the basis of tubes containing LB only and LB with ΔrcsB or KP1, respectively.
Molecular Docking
To gain further insight into the interaction between eugenol and RcsB, molecular docking was performed. ChemBioDraw Ultra 17.0 was used to draw the structure of eugenol. The three-dimensional structure of RcsB (PDB ID: 2kx7) was selected and downloaded from the Protein Data Bank. RcsB and eugenol were converted into PDBQT format using AutodockTools 1.5.6. Autodock Vina 1.1.2 was adopted for molecular docking research. To increase the accuracy of calculation, the parameter exhaustiveness was set to 20. Unless otherwise specified, all other parameters adopted default values. Finally, the conformation with the highest score was selected to analyze the results with the Discovery Studio 2019 client.
MicroScale Thermophoresis Analysis
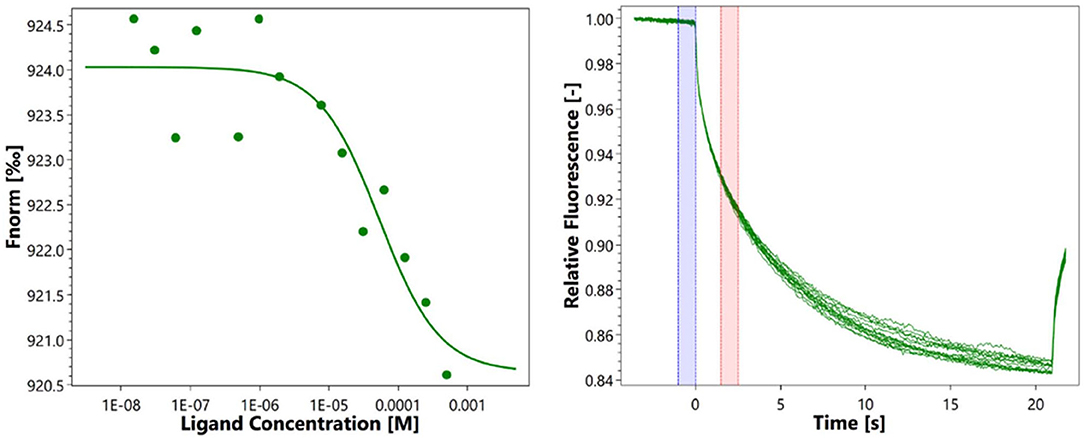
To determine the equilibration dissociation constant (KD) of eugenol to RcsB, MicroScale Thermophoresis (MST) measurements were performed by Protein Preparation and Identification Platform, Protein Research Technology Center, Tsinghua University, using a Monolith NT.115 device (201610-BR-N007, Nanotemper Technologies, Munich, Germany). RcsB, with a calculated MW of 27.5 kDa, was already expressed, purified, and stored in the laboratory. Eugenol was added to samples with a final concentration of 50 μg/ml containing increasing concentrations of RcsB (0–100 nM). Then, a 1:1 serial dilution of protein and eugenol stock solution was prepared in PBS containing 0.05% Tween-20. The samples were then transferred into specialized glass capillaries (Monolith NT.155 premium capillaries MO-K022; Nanotemper Technologies, Munich, Germany). For all experiments, standard parameters were used as recommended by the manufacturer. The data were evaluated through the thermophoresis effect using the manufacturer-supplied NT analysis software.
Statistical Analysis
All data were expressed as mean ± standard deviation. Statistical analysis was tested using Graphpad Prism 8 using Student's t-test, and P < 0.05, P < 0.01, and P < 0.001 were considered statistically significant and marked as “*,” “**,” and “***,” respectively, in all comparisons.
Results
Minimum Inhibitory Concentration
As shown in Table 3, the MIC value of eugenol was 200 μg/ml against both KP1 and ΔrcsB strains.
Biofilm Formation
Crystal violet assessment confirmed that wild-type KP1 grown on LB medium at 37°C for 24 h resulted in a strong biofilm formation. However, ΔrcsB strain almost loses the biofilm formation capacity under similar conditions (Figure 1).
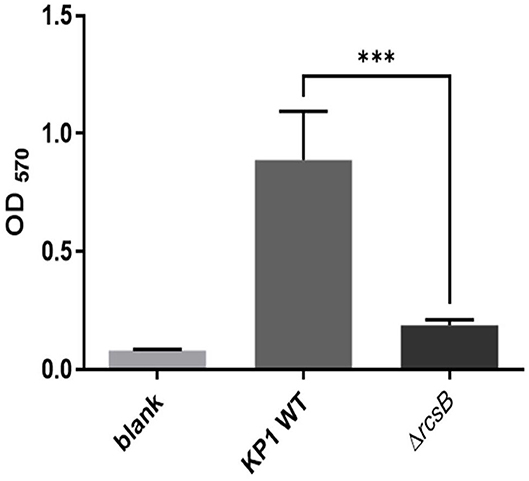
Figure 1. Biofilm formation of Klebsiella pneumonia KP1, ΔrcsB, and sterile LB broth as blank. The *** denotes P < 0.001.
Transcriptome Sequencing
The data were available in the NCBI SRA database (SRA accession: PRJNA504310). Among these, 5,779 reference genes were detected including 4,514 known genes in the control group and 4,561 genes in the eugenol treated group. Based on the false discovery rate (FDR) threshold, 890 significant DEGs were identified where 771 genes were upregulated and the rest 119 genes were downregulated between the two groups.
Expression Level of Selected Genes
The results of RT-qPCR showed that the relative expression of 50S ribosomal protein gene rpmG, serine endoprotease gene degP, and ribonuclease P protein component rnpA in the KP wild type was downregulated after eugenol treatment. No significant difference was observed compared with the nontreated group. As shown in Figure 2, the expression of 2,3,4,5-tetrahydropyridine-2,6-carboxylate N-succinyltransferase synthesis gene dapD was significantly lower than the control group. Oppositely, rcs phosphorelay system core genes rcsB, rcsD, and rcsA and transcriptional regulator gene yiaG showed a significant upregulation trend after eugenol treatment. Expression of OmpA family lipoprotein gene yiaD was also higher than control. The expression profile of all tested genes was in agreement with the FPKM dataset of transcriptome sequencing except for degP.
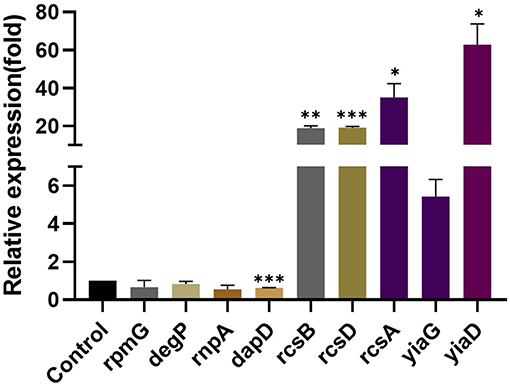
Figure 2. Relative expression of rpmG, degP, rnpA, dapD, rcsB, rcsD, rcsA, yiaG, and yiaD. The *, **, and *** denotes P < 0.05, P < 0.01, and P < 0.001 respectively.
Effect of Eugenol on Biofilm Formation
It can be seen from Figure 3 that plasmid pET-28a-rcsB transformation can hardly recover biofilm formation of ΔrcsB competent cells compared with ΔrcsB only. Besides, a decrease of biofilm in ΔrcsB + RcsB + IPTG, ΔrcsB + RcsB + Eu, ΔrcsB + RcsB + IPTG + Eu, KP1 + RcsB + Eu, and KP1 + RcsB + IPTG + Eu groups was observed after IPTG induction. However, nonsignificance was found within ΔrcsB + RcsB + IPTG, ΔrcsB + RcsB, KP1 + RcsB, and KP1 + RcsB + IPTG groups.
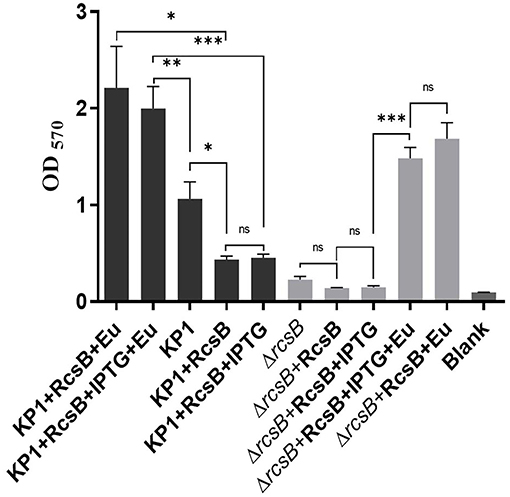
Figure 3. Effect of eugenol on biofilm formation. The *, **, and *** denotes P < 0.05, P < 0.01, and P < 0.001 respectively.
Oppositely, all groups after adding sub-MIC eugenol exhibited an obvious biofilm increase trend compared with nontreatment groups, which reflects a strong biofilm promoting effect of eugenol. Biofilm in both KP1 + RcsB + IPTG + Eu and ΔrcsB + RcsB + IPTG + Eu groups was significantly higher than that in the noneugenol group. This result was consistent with transcriptome sequencing and RT-qPCR. Moreover, a significant difference (P = 0.0075) was observed between biofilm formation of KP1 + RcsB + IPTG + Eu and KP1 regardless of IPTG inhibition. Likewise, the same trend was also shown in the KP1 + RcsB + Eu group compared with the KP1 group. Based on the above results, we found that sub-MIC eugenol contributes greatly during the biofilm formation process alone or by induction of rcsB.
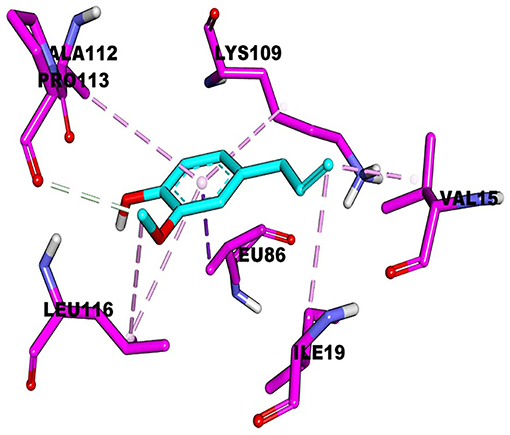
Analysis of Molecular Docking
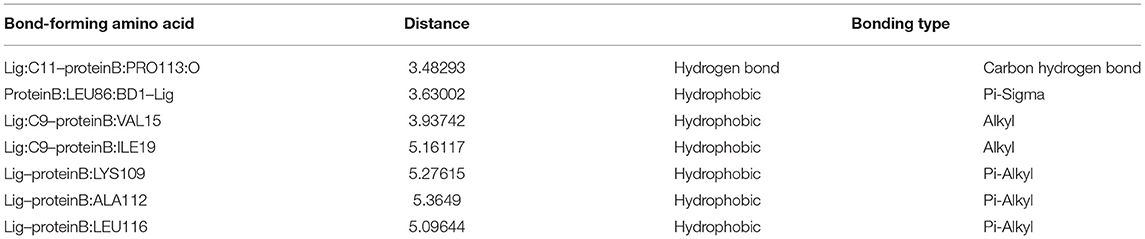
The active pockets of protein RcsB were docked by eugenol to clarify their mode of action at the molecular level. The affinity was −4.3 kcal/mol. The theoretical combination mode is shown in Figure 4. It can be seen from the surface map that small molecules and protein surfaces formed more bonds, which provides the possibility for small molecules to bind here. However, there are six hydrophobic bonds and only one hydrogen bond out of total. The interactions are shown in Table 4. All these interactions make the RcsB and eugenol pound Lig form a complex.
Binding Affinity Analysis of Eugenol to RcsB
Microscale thermophoresis measurements were used to determine the equilibration dissociation constant (KD) of eugenol to RcsB. The signal-to-noise ratio of RcsB was 7.3. The RcsB exhibits a binding affinity to eugenol with a resulting KD of 53.4 μM at a target concentration of 100 nM (Figure 5). The result of MST further proved the slight interaction of eugenol with RcsB.
Discussion
Previous investigation revealed that the production of cell surface structures was dependent on the Rcs cascade complex signal transduction system (11), and peptidoglycan synthesis was responsible for bacteria growth (21). Several virulence factors such as CPS, lipopolysaccharide (LPS), and biofilm formation-related factors responsible for pathogenicity were identified in K. pneumoniae (22). Biofilm is also considered an important factor of virulence in pathogenic bacteria. The general architecture including depth and 3D structure is determined by exopolymeric matrix component colonic acid (23). Moreover, the expression of extracellular polysaccharide colanic acid is decreased by the deactivation of the Rcs phosphorelay (9).
Biofilm is often described as a shelter that protects bacteria from hostile surroundings as well as one of the strategic ways to develop resistance against antimicrobials in which those formed biofilms are more resistant than their planktonic counterparts up to 1,000 times. It also offers a reservoir for chronic infection. However, the adjustment of biofilm to heterogeneous physicochemical conditions is necessary due to its exposure to several stresses. For instance, modifications of the LPS in Gram-negative bacteria could be caused by such adaptations. It is considered a highly dynamic structure of the bacterial outer membrane that responds to environmental changes in the surrounding (24).
The Rcs phosphorelay, presented in many Enterobacteriaceae genera, can regulate the expression of important genes in large numbers for cell division and stationary-phase sigma factor activity, maintaining the integrity of cell wall, motility, virulence, and biofilm development (11). In this study, a vital role of RcsB was noted for K. pneumoniae biofilm formation, while low production of biofilm was observed in ΔrcsB in comparison to the wild-type strain. A similar effect on the biofilm formation and growth of Edwardsiella tarda was reported by Xu et al. (25). Another report mentioned that the rcsD mutant of P. mirabilis was deficient in biofilm formation, where the contribution of Rcs phosphorelay to biofilm formation was also validated (26). Reduced biofilm production of ΔrcsB mutant is due to its fragile architecture. An easy detachment of biofilm from the surface is due to complex architecture and significant depth (25). In another study on E. coli, inactivation of the Rcs system was found that can lead to a decrement in extracellular polysaccharide colonic acid expression, which participates in overall biofilm architecture (9, 23).
In this study, lesser production of biofilm was observed. Transformation of plasmid pET-28a-rcsB did not enhance biofilm formation in either group. It is evident from the results that biofilm formation was directly affected by the expression of rcsB. However, a sharp decrease was found in KP1 competent cells transferred with plasmid pET-28a-rcsB. Although ΔrcsB had a weakened biofilm formation capacity, the biofilm of both ΔrcsB and KP1 competent cells was weakened than the original strains, indicating a competent state detrimental to bacteria. Beyond our expectation, instead of facilitating biofilm formation, IPTG induction resulted in biofilm loss during 4 h incubation. The trend was reflected by ΔrcsB + RcsB + IPTG, ΔrcsB + RcsB + IPTG + Eu, KP1 + RcsB + Eu, and KP1 + RcsB + IPTG + Eu groups. Among them, the biofilm of ΔrcsB + RcsB + IPTG was significantly lower than ΔrcsB + RcsB + IPTG + Eu. Inversely, no significant difference was found between KP1 + RcsB and KP1 + RcsB + IPTG, as well as ΔrcsB + RcsB and ΔrcsB + RcsB + IPTG groups. We speculate that induction by IPTG may create a favorable environment and let bacteria release “fake” signals to transcriptional regulatory factors to slow down the overall expression of genes essential for CPS synthesis and thus lower the biofilm level. This effect may suppress the single upregulation and a series of hereafter biofilm-related gene regulations. The differences between ΔrcsB and KP1 competent cells could be attributed to the rcsB gene existing in the KP1 chromosome, which causes the sensitivity difference between the two strains to ambient pressure. The exact mechanism needs to be further studied. Interestingly, regardless of IPTG's inhibitory effect, all groups were found to exhibit an obvious biofilm increase, especially in KP1 + RcsB + IPTG + Eu and ΔrcsB + RcsB + IPTG + Eu groups after adding sub-MIC eugenol compared with nontreatment groups, which reflect a strong biofilm promoting effect of eugenol. Through transcriptome sequencing and RT-qPCR, upregulations of mRNA expression of Rcs phosphorelay system response regulator rcsB, histidine-containing phosphotransfer domain rcsD, unstable substrate of the Lon protease rcsA, as well as a transcriptional regulator gene yiaG were analyzed and validated, which was in agreement with the in vitro biofilm formation result. Besides, molecular docking can reproduce the strength between the 3-D dimensional structure of the receptor and the ligand. An energy-binding manner between the ligand and the active site of the receptor can be evaluated. The molecular docking studies give a reasonable explanation for the interaction between RcsB and eugenol. Through analysis, eugenol could form more hydrophobic bonds that will weaken stronger bond such as hydrogen bond with RcsB. Concerning the difference, we think eugenol can target RcsB but bind normally. To better understand the binding affinity between RcsB and eugenol, MST analysis was performed. Eugenol was further proved that it can interact with RcsB.
Based on the results obtained, we found that sub-MIC eugenol contributes greatly during the biofilm formation process alone or by induction of rcsB expression, which was converse to its biological property at MIC. Many previous studies reported that eugenol significantly reduced or eradicated biofilms formed by Streptococci (27), Candida dubliniensis (28), Staphylococcus aureus (18), E. coli O157:H7 (29), Porphyromonas gingivalis (30), and Listeria monocytogenes (31).
Inhibition of biofilm formation without affecting bacterial activity at an early stage in Streptococcus mutants was reported previously (32). The absence of rcsB in Salmonella weakens biofilm formation phenotypes; besides, the absence of the phosphorelay pathway of other components such as RcsD, RcsC, and RcsA conforms to normal biofilm phenotypes (33). However, the mechanism and impact of sub-MIC eugenol against various pathogens have not yet been fully understood. Our results showed that sub-MIC natural products not only can hardly reduce or eradicate bacteria within or at the biofilm stage effectively but also will facilitate the formation at an early stage. This phenomenon offers an explanation of induced drug resistance to some extent. Therefore, more attention should be paid to the potential risk of low-dose chemical ingredient administration. The vital role of two component systems and contributions to drug resistance development and bacterial pathogenicity should not be ignored.
Conclusion
rcsB is critical and plays a positive role in K. pneumonia biofilm formation. Sub-MIC eugenol exerted a biofilm-promoting property via upregulating Rcs-related genes. Our study provides a new perspective and evidence for natural antibacterial ingredient research and the potential risk of sub-MIC natural ingredients to treat drug-resistance bacteria.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
EE and Z-nT performed the experiment. EE, YW, and Y-mW contributed to drafting and revising the manuscript. QW, X-yJ, and Y-mW participated in the design of the study and statistical analysis. H-xM provided constructive advice and checked the final manuscript. All authors have read and approved the final manuscript.
Funding
This study was supported by the Science and Technology Project of Jilin Provincial Department of Education 13th Five-Year Plan (Grant number: JJKH20200359KJ), Introduction of Excellent Doctor Protocol of Jilin Agricultural University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the Jilin Provincial Key Laboratory of New Veterinary Drug Innovation Research and Development for support and other members who made contributions to this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.945491/full#supplementary-material
Abbreviations
AMR, antimicrobial resistance; K. pneumoniae, Klebsiella pneumoniae; MIC, minimum inhibitory concentration; MST, MicroScale Thermophoresis; Rcs, regulator of capsule synthesis; EPS, extracellular polysaccharides; IPTG, isopropyl-β-D-thiogalactoside; CPS, capsular polysaccharide; LB, Luria-Bertani; FDR, false discovery rates; DEG, differential expression gene; LPS, lipopolysaccharide.
References
1. Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, et al. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis. (2002) 8:160–6. doi: 10.3201/eid0802.010025
2. Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. (2016) 7:895. doi: 10.3389/fmicb.2016.00895
3. Zhang L, LI Y, Shen W, Wang SM, Wang G, Zhou Y. Whole-genome sequence of a carbapenem-resistant hypermucoviscous Klebsiella pneumoniae isolate SWU01 with capsular serotype K47 belonging to ST11 from a patient in China. J Glob Antimicrob Resist. (2017) 11:87–9. doi: 10.1016/j.jgar.2017.09.001
4. Wang YM, Dong WL, Odah KA, Kong LC, Ma HX. Transcriptome analysis reveals ai-2 relevant genes of multi-drug resistant Klebsiella pneumoniae in response to eugenol at sub-MIC. Front Microbiol. (2019) 10:1159. doi: 10.3389/fmicb.2019.01159
5. Hisaeda K, Arima H, Sonobe T, Nasu M, Hagiwara K, Kirisawa R, et al. Changes in acute-phase proteins and cytokines in serum and milk whey from dairy cows with naturally occurring peracute mastitis caused by Klebsiella pneumoniae and the relationship to clinical outcome. J Vet Med Sci. (2011) 73:1399–404. doi: 10.1292/jvms.10-0403
6. Lianou A, Koutsoumanis KP. Strain variability of the biofilm-forming ability of Salmonella enterica under various environmental conditions. Int J Food Microbiol. (2012) 160:171–8. doi: 10.1016/j.ijfoodmicro.2012.10.002
7. Mizan MF, Jahid IK, Ha SD. Microbial biofilms in seafood: a food-hygiene challenge. Food Microbiol. (2015) 49:41–55. doi: 10.1016/j.fm.2015.01.009
8. Xie T, Liao Z, Lei H, Fang X, Wang J, Zhong Q. Antibacterial activity of food-grade chitosan against Vibrio parahaemolyticus biofilms. Microb Pathog. (2017) 110:291–7. doi: 10.1016/j.micpath.2017.07.011
9. Gottesman S, Stout V. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol Microbiol. (1991) 5:1599–606. doi: 10.1111/j.1365-2958.1991.tb01906.x
10. Wall E, Majdalani N, Gottesman S. The complex Rcs regulatory cascade. Annu Rev Microbiol. (2018) 72:111–39. doi: 10.1146/annurev-micro-090817-062640
11. Clarke DJ. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol. (2010) 5:1173–84. doi: 10.2217/fmb.10.83
12. Venkatesh GR, Kembou Koungni FC, Paukner A, Stratmann T, Blissenbach B, Schnetz K. BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS. J Bacteriol. (2010) 192:6456–64. doi: 10.1128/JB.00807-10
13. Pannen D, Fabisch M, Gausling L, Schnetz K. Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli. J Biol Chem. (2016) 291:2357–70. doi: 10.1074/jbc.M115.696815
14. Fang N, Yang H, Fang H, Liu L, Zhang Y, Wang L, et al. RcsAB is a major repressor of Yersinia biofilm development through directly acting on hmsCDE, hmsT, and hmsHFRS. Sci Rep. (2015) 5:9566. doi: 10.1038/srep09566
15. Walker KA, Miner TA, Palacios M, Trzilova D, Frederick DR, Broberg CA, et al. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio. (2019) 10. doi: 10.1128/mBio.00089-19
16. Guimaraes AC, Meireles LM, Lemos MF, Guimaraes MCC, Endringer DC, Fronza M, et al. Antibacterial activity of terpenes and terpenoids present in essential oils. Molecules. (2019) 24:2471. doi: 10.3390/molecules24132471
17. Nazzaro F, Fratianni F, De Martino L, Coppola R, De Feo V. Effect of essential oils on pathogenic bacteria. Pharmaceuticals. (2013) 6:1451–74. doi: 10.3390/ph6121451
18. Yadav MK, Chae SW, Im GJ, Chung JW, Song JJ. Eugenol: a phyto-compound effective against methicillin-resistant and methicillin-sensitive Staphylococcus aureus clinical strain biofilms. PLoS ONE. (2015) 10:e0119564. doi: 10.1371/journal.pone.0119564
19. Cree RG, Phillips I, Noble WC. Adherence characteristics of coagulase-negative staphylococci isolated from patients with infective endocarditis. J Med Microbiol. (1995) 43:161–8. doi: 10.1099/00222615-43-3-161
20. Stepanovic S, Vukovic D, Hola V, Di Bonaventura G, Djukic S, Cirkovic I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. APMIS. (2007) 115:891–9. doi: 10.1111/j.1600-0463.2007.apm_630.x
21. Joseleau-Petit D, Liebart JC, Ayala JA, D'Ari R. Unstable Escherichia coli L forms revisited: growth requires peptidoglycan synthesis. J Bacteriol. (2007) 189:6512–20. doi: 10.1128/JB.00273-07
22. Zou Q, Li Y. Hypervirulent Klebsiella pneumoniae. N Engl J Med. (2021) 385:833. doi: 10.1056/NEJMicm2101602
23. Danese PN, Pratt LA, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol. (2000) 182:3593–6. doi: 10.1128/JB.182.12.3593-3596.2000
24. Szczesny M, Beloin C, Ghigo JM. Increased osmolarity in biofilm triggers RcsB-dependent lipid A palmitoylation in Escherichia coli. mBio. (2018) 9. doi: 10.1128/mBio.01415-18
25. Xu Y, Xu T, Wang B, Dong X, Sheng A, Zhang XH, et al. mutation in rcsB, a gene encoding the core component of the Rcs cascade, enhances the virulence of Edwardsiella tarda. Res Microbiol. (2014) 165:226–32. doi: 10.1016/j.resmic.2014.02.006
26. Liaw SJ, Lai HC, Wang WB. Modulation of swarming and virulence by fatty acids through the RsbA protein in Proteus mirabilis. Infect Immun. (2004) 72:6836–45. doi: 10.1128/IAI.72.12.6836-6845.2004
27. Yadav MK, Park SW, Chae SW, Song JJ, Kim HC. Antimicrobial activities of Eugenia caryophyllata extract and its major chemical constituent eugenol against Streptococcus pneumoniae. APMIS. (2013) 121:1198–206. doi: 10.1111/apm.12067
28. De Paula SB, Bartelli TF, Dl Raimo V, Santos JP, Morey AT, Bosini MA, et al. Effect of Eugenol on cell surface hydrophobicity, adhesion, and biofilm of Candida tropicalis and Candida dubliniensis isolated from oral cavity of HIV-infected patients. Evid Based Complement Alternat Med. (2014) 2014:505204. doi: 10.1155/2014/505204
29. Kim YG, Lee JH, Gwon G, Kim SI, Park JG, Lee J. Essential oils and eugenols inhibit biofilm formation and the virulence of Escherichia coli O157:H7. Sci Rep. (2016) 6:36377. doi: 10.1038/srep36377
30. Zhang Y, Wang Y, Zhu X, Cao P, Wei S, Lu Y. Antibacterial and antibiofilm activities of eugenol from essential oil of Syzygium aromaticum (L.) Merr. and L. M. Perry (clove) leaf against periodontal pathogen Porphyromonas gingivalis. Microb Pathog. (2017) 113:396–402. doi: 10.1016/j.micpath.2017.10.054
31. Upadhyay A, Upadhyaya I, Kollanoor-Johny A, Venkitanarayanan K. Antibiofilm effect of plant derived antimicrobials on Listeria monocytogenes. Food Microbiol. (2013) 36:79–89. doi: 10.1016/j.fm.2013.04.010
32. Adil M, Singh K, Verma PK, Khan AU. Eugenol-induced suppression of biofilm-forming genes in Streptococcus mutans: an approach to inhibit biofilms. J Glob Antimicrob Resist. (2014) 2:286–92. doi: 10.1016/j.jgar.2014.05.006
Keywords: eugenol, sub-MIC, biofilm formation, Klebsiella pneumoniae, rcsB
Citation: Elken EM, Tan Z-n, Wang Q, Jiang X-y, Wang Y, Wang Y-m and Ma H-x (2022) Impact of Sub-MIC Eugenol on Klebsiella pneumoniae Biofilm Formation via Upregulation of rcsB. Front. Vet. Sci. 9:945491. doi: 10.3389/fvets.2022.945491
Received: 16 May 2022; Accepted: 08 June 2022;
Published: 12 July 2022.
Edited by:
Yi Wu, Nanjing Agricultural University, ChinaReviewed by:
Yun peng Fan, Northwest A&F University, ChinaHongbin Si, Guangxi University, China
Liwei Guo, Yangtze University, China
Copyright © 2022 Elken, Tan, Wang, Jiang, Wang, Wang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-xia Ma, hongxia0731001@163.com; Yi-ming Wang, wym27149@126.com
†These authors have contributed equally to this work
 Emad Mohammed Elken
Emad Mohammed Elken Zi-ning Tan1,2†
Zi-ning Tan1,2†  Xiu-yun Jiang
Xiu-yun Jiang Hong-xia Ma
Hong-xia Ma