Identification of Compounds With Antiviral Activity Against SARS-CoV-2 in the MMV Pathogen Box Using a Phenotypic High-Throughput Screening Assay
- 1Brazilian Biosciences National Laboratory (LNBio), Brazilian Center for Research in Energy and Materials (CNPEM), Campinas, Brazil
- 2Department of Genetics, Microbiology and Immunology, Institute of Biology, State University of Campinas (UNICAMP), Campinas, Brazil
- 3Department of Cellular and Structural Biology, Institute of Biology, State University of Campinas (UNICAMP), Campinas, Brazil
- 4Laboratory of Emerging Viruses, Department of Genetics, Microbiology and Immunology, Institute of Biology, University of Campinas, Campinas, Brazil
- 5Biodiversity Research Center, Federal University of Roraima (UFRR), Roraima, Brazil
- 6Medicines for Malaria Venture (MMV), Geneva, Switzerland
- 7Ilum School of Science, Campinas, Brazil
Until December 2021, the COVID-19 pandemic has caused more than 5.5 million deaths. Vaccines are being deployed worldwide to mitigate severe disease and death, but continued transmission and the emergence of SARS-CoV-2 variants indicate that specific treatments against COVID-19 are still necessary. We screened 400 compounds from the Medicines for Malaria Venture (MMV) Pathogen Box seeking for molecules with antiviral activity against SARS-CoV-2 by using a high-throughput screening (HTS) infection assay in Vero CCL81 cells. On resupply of 15 selected hit compounds, we confirmed that 7 of them presented a dose-dependent cytoprotective activity against SARS-CoV-2-induced cytopathic effect (CPE) in the micromolar range. They were validated in low-throughput infection assays using four different cell lines, including the human lung Calu-3 cell line. MMV000063, MMV024937, MMV688279, and MMV688991 reduced viral load in cell culture, assessed by RT-qPCR and viral plaque assay, while MMV688279 and MMV688991 (also known as nitazoxanide) were the most promising, reducing SARS-CoV-2 load by at least 100-fold at 20 µM in almost all cell types tested. Our results indicate that active anti-SARS-CoV-2 molecules exist within the repertoire of antiviral, antiparasitic and antimicrobial compounds available to date. Although the mode of action by which MMV688279 and MMV688991 reduce SARS-CoV-2 replication is yet unknown, the fact that they were active in different cell types holds promise not only for the discovery of new therapeutic targets, but also for the development of novel antiviral medicines against COVID-19.
1 Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) was first identified in the city of Wuhan, China end of 2019 (1, 2) as the causative agent of COVID-19 (3). SARS-CoV-2 is a Betacoronavirus member of Coronaviridae family like other respiratory viral pathogens such as SARS-CoV and MERS-CoV. The COVID-19 pandemic completed its first year in March 2021 with more than 108 million confirmed cases and approximately 2.4 million deaths (“4). COVID-19 patients experience a variety of symptoms such as fever, dry cough and tiredness, or more severe symptoms such as difficulty breathing, chest pain, and loss of smell and taste (5). Among COVID-19 clinical manifestations, a decrease in oxygen saturation is the main cause of hospitalization (6). A combination of high transmissibility and a greater chance of developing severe disease in risk groups (i.e. elderly, obese or diabetic people) led to the eventual collapse of public health care systems and a high mortality rate in many countries (7).
Treatment of COVID-19 patients is largely supportive. Several therapeutic strategies have been tested in both animal models and patients, including anti-inflammatory compounds such dexamethasone (8), convalescent plasma (9), monoclonal therapy (10), and antiviral compounds such as remdesivir, hydroxychloroquine, lopinavir, interferon beta-1a (11), and nitazoxanide (12). Unfortunately, most treatments have shown limited efficacy in reducing symptoms or accelerating patient recovery, especially after patients evolve to more severe forms of COVID-19. Difficulties associated with mass vaccination during the COVID-19 pandemic (13) and the growing evidences indicating that emerging SARS-CoV-2 variants may be more transmissible and capable of escaping previous established immune responses (14, 15) support the continued effort to discover active compounds against SARS-CoV-2. This includes antiviral molecules that may be developed into a treatment or preventive strategy against SARS-CoV-2 infection. HTS has been used extensively for the screening and repurposing of FDA-approved drugs as candidate treatments against SARS-CoV-2 infection (16), resulting in many clinical trials and candidate compounds testing (17, 18). Here, we used a cell culture-based HTS assay to screen 400 compounds from the Pathogen Box, provided by Medicines for Malaria Venture (MMV), against SARS-CoV-2 infection. The Pathogen Box contains anti-parasitic and antimicrobial compounds that were not covered in reported HTS campaigns against SARS-CoV-2 or its viral protein targets (19–22). We identified 7 hit compounds with cytoprotective properties that were validated in four different cell types, thus confirming their antiviral activity against SARS-CoV-2. The dihydroquinazoline (MMV688279) and nitaxozanide (MMV688991) were the most potent and promising antiviral compounds present in the Pathogen Box.
2 Materials and Methods
2.1 Virus and Cell lines
Low-passage strain HIAE-02 SARS-CoV-2/SP02/human/2020/BRA (GenBank accession number MT126808.1) was isolated in Brazil and provided by Prof. Edison Luiz Durigon (USP-SP, Brazil). We produced SARS-CoV-2 stocks in Vero CCL81 cells (BCRJ, #0245) cultured to a confluent monolayer in 75 cm2 culture flask and infected with approximately 2.5x105 PFU of SARS-CoV-2. HTS assays were performed in Vero CCL81 cells (BCRJ, # 0245). Human cell lineages HuH 7.5 (provided by Dr. Claudia N. Duarte dos Santos, Fiocruz-PR, Brazil), Caco-2 (BCRJ, # 0059), Calu-3 (provided by Patrícia R. M. Rocco - Federal University of Rio de Janeiro, Brazil) were used to validate results obtained in primary assays. Vero CCL81 and HuH 7.5 were maintained with Dulbecco’s Modified Essential Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine, and 1% penicillin/streptomycin. Caco-2 and Calu-3 were cultured with DMEM/F-12 1:1 supplemented with 20% FBS, 1% L-glutamine and 1% penicillin/streptomycin. All experiments involving infectious SARS-CoV-2 were performed at the BSL3 facility of the Laboratory of Emerging Viruses (LEVE) located in the University of Campinas.
2.2 HTS Assay Design and Screening Process
2.2.1 HTS Assay
Medicine for Malaria Venture’s Pathogen Box is a compound library containing 400 compounds that were screened using survival against SARS-CoV-2 infection as primary criteria. 1.7x103 Vero CCL81 cells were plated in 45 µL per well in a 384 well plate at 37°C 5% CO2. After 24 h, 15 µL of a 0.5 mM compound stock solution were added to each well, resulting in a final concentration of 10 µM. During EC50 or CC50 assays, compounds were added in concentrations ranging from 40 µM to 0.156 µM. Plates were transported to the BSL3 laboratory where cells were infected with SARS-CoV-2 at a MOI of 0.1 in a volume of 15 µL and incubated at 37°C and 5% CO2 for 60 h. Supernatant was discarded and cells stained with 2 μM Hoechst-33342 and 100 nM MitoTracker Deep Red for 45 minutes at 37°C 5% CO2, and then fixed with paraformaldehyde (PFA) 4%.
2.2.3 HTS Data Acquisition and Processing
Data acquisition and analysis were performed using the Operetta High Content Imaging System (Perkin Elmer) and the Columbus server (Perkin Elmer), respectively. One central field position per well was acquired at 10x magnification. The number of Hoechst-stained healthy nuclei were counted, and the MitoTracker-stained mitochondrial mass/intensity was calculated in each field/image, plotted for each experimental condition and tested for statistical significance. Vehicle-treated non-infected wells and vehicle-treated SARS-CoV-2 infected wells were used as positive and negative controls for cell survival, respectively, and used to calculate the assay Z factor in each assay plate. Z-factors above 0.60 were considered valid and culminated in data analysis.
2.3 Hit Confirmation
2.3.1 Antiviral Assay
Antiviral assays were performed in Vero CCL81 cells using SARS-CoV-2 of MOI 0.01, and MOI 0.1 for HuH 7.5, Caco-2 and Calu-3 cells. Cell culture media was replaced by fresh media containing 20 µM of compound at 1h post infection. Supernatants were harvested after 48h of incubation at 37°C and 5% CO2. The viral load was measured in samples using viral plaque assay and 1-Step RT-qPCR.
2.3.2 Plaque Assay
Vero cells were seeded into 24-well plates and incubated with serially diluted samples (10-1 to 10-6) for 1h at 37°C and 5% CO2. After incubation, cells were overlaid with semi-solid medium (1% w/v carboxymethylcellulose in DMEM supplemented with 5% FBS) and kept in the incubator for 4 days. After removal of semi-solid medium, cells were fixed with PFA 4% and plaques were visualized after staining with 1% methylene blue. Viral lysis plaques were counted, and results were expressed as viral plaque forming units (PFU)/mL of sample. The limit of detection of the assay is 10 PFU/mL.
2.3.3 RT-qPCR
Viral RNA extraction was performed from 100 µl of cell culture supernatants with the Quick-RNA Viral Kit (cat. # R1035, Zymo Research, USA) according to the protocol provided by the manufacturer. The viral RNA was eluted in 50 µL of nuclease-free water. SARS-CoV-2 genome detection and quantification were performed with TaqMan Fast Virus 1-Step Master Mix System (cat. # 4444434, Applied Biosystems) using specific primers targeting SARS-CoV-2 E protein at 6 pmol (each) and probe at 5 pmol per reaction. Forward primer sequence: ACAGGTACGTTAATAGCGT; reverse primer: ATATTGCAGCAGTACGCACACA; and probe: FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ. The PCR mix containing Master Mix, primers and probe was incubated with 6 µL of an 1:50 (v/v) dilution of sample RNA. Cycling was conducted in Applied Biosystems 7500 Real-Time PCR System. The reactions were incubated at 50°C for 10 minutes for reverse transcription, and 95°C for 2 minutes, followed by incubation at 95°C for 5 seconds and 60°C for 30 seconds for data collection over 45 cycles. A plasmid containing SARS-CoV-2 E protein amplicon (kindly provided by Dr. Marcio Bajgelman) was used as standard curve in order to establish an absolute quantification of viral RNA copies per ml of supernatant.
2.3.4 Indirect Immunofluorescent Assay
2x104 Vero and 1x105 Calu-3 cell lines were seeded in 96-well plates in the presence of the 20 µM of compounds, then infected with SARS-CoV2 at the MOI 0.4 for 22 h of incubation at 37°C and 5% CO2. The cells were fixed with 4% paraformaldehyde for 2h, permeabilizated with 0,3% Triton X-100 for 15 min, and blocked with PBS containing 60% FBS plus 0,1% Triton X-100 for 30min. Subsequently, the cells were overnight incubated with PBS containing 1% BSA and the “SARS-CoV-2 Spike Protein (RBD) Chimeric Recombinant Rabbit Monoclonal Antibody (P06DHuRb) (Catalog # 703974, Invitrogen, Thermo Fisher) (dilution 100x). The cells were washed five times with PBS and stained with the secondary Alexa Fluor Goat anti-Rabbit 647 IgG (H + L) antibody (Invitrogen, Thermo Fisher) (dilution 1000x) for 2 h. Cell nuclei were counterstained with Hoechst 33342 and washed five times with PBS. Finally, images were acquired on the Operetta High Content Imaging System (Perkin Elmer) by using the Harmony software (Perkin Elmer). The data was updated in the Columbus server (Perkin Elmer) to further analyses. We evaluated the frequency of SARS-CoV-2 infected cells and the level of infection per cell by using the mean fluorescent intensity (MFI) on field positions acquired at 10x magnification. The representative images were selected from fields at 40x magnification.
2.4 Statistical Analysis
Viral plaque assays and RT-qPCR data were analyzed using non-parametric Kruskal-Wallis test followed by Dunn’s multiple comparison test. Group comparison was made between infected non-treated cells and infected treated cells. Each experiment was performed with technical triplicate and it was repeated at least twice. All tests were performed in GraphPad Prism v8.4.0 (GraphPad Software, La Jolla, CA, USA). P values < 0.05 were considered significant.
3 Results
3.1 HTS Assay Design and Screening Process
We developed a phenotypic cell culture-based HTS assay to screen 400 compounds included in the MMV Pathogen Box. The assay is based on the ability of SARS-CoV-2 to induce cytopathic effect (CPE) and death in Vero CCL81 cells, as a consequence of infection and viral replication. Thus, compounds in the Pathogen Box were initially screened for their cytoprotective properties as an indirect assessment of antiviral activity against SARS-CoV-2. According to the experimental pipeline depicted in Figure 1, selected hit compounds were resupplied for confirmation and validation of antiviral activity in low-throughput infection assays in cell lines relevant to SARS-CoV-2 biology and disease.
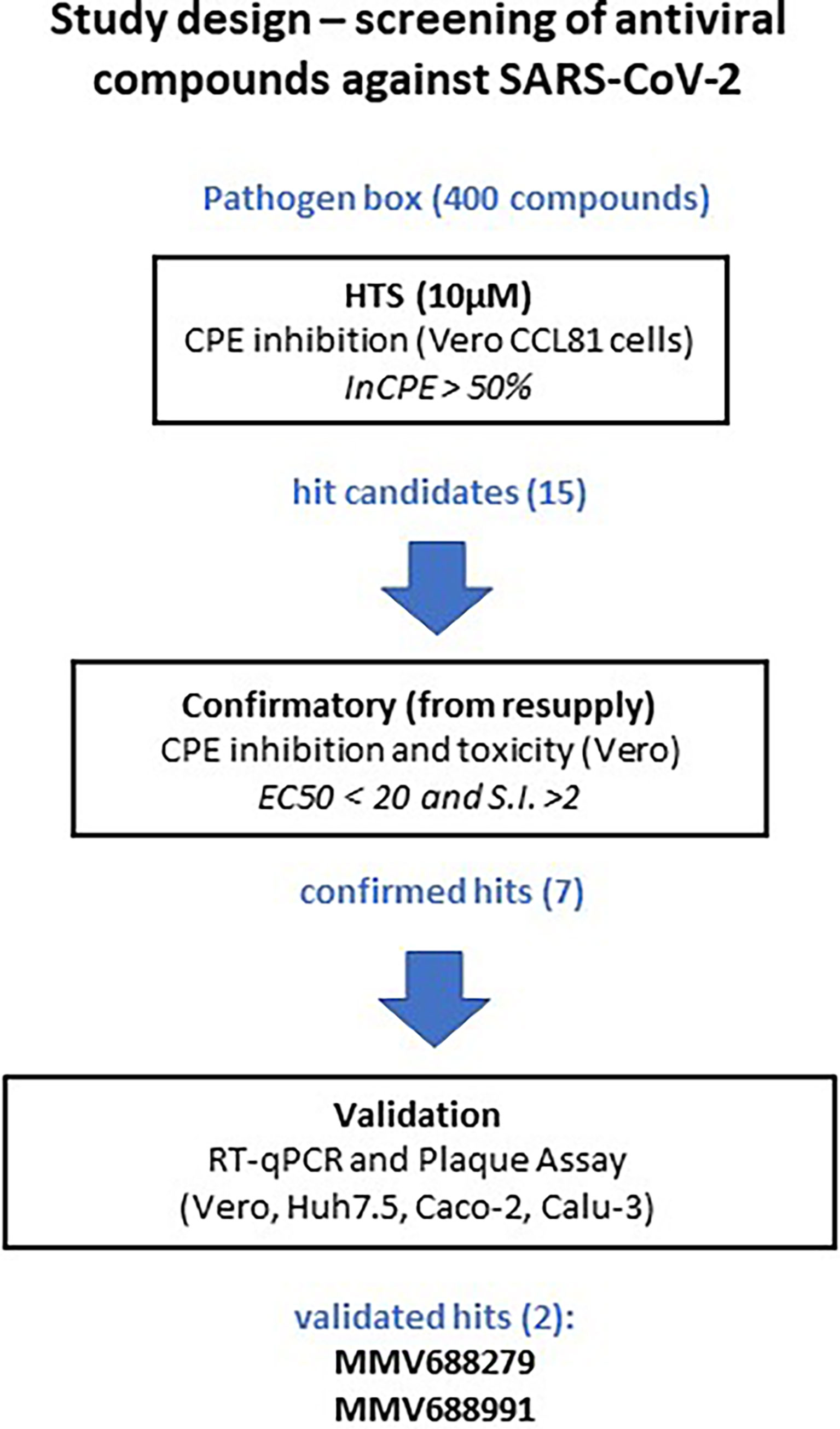
Figure 1 Study design for the high-throughput screening of MMV compound collections against SARS-CoV-2 in vitro. A total of 400 compounds present in MMV Pathogen Box were screened at 10 µM for the ability to inhibit virus-induced cytopathic effect (InCPE). Compounds that presented at least InCPE > 50% (15 compounds) were considered hits and progressed to confirmatory tests after resupply. Dose-response curves were performed and compounds that showed a half maximal effective concentration (EC50) below 20 µM and a selectivity index (SI) above 2 progressed to validation. The antiviral properties of 7 hit compounds were evaluated in different cell lines (Vero, HuH 7.5, Caco-2 and Calu-3) infected with SARS-CoV-2, leading to validation of MMV688279 and MMV688991.
Vero CCL81 cells were plated in a 384-well plate format and incubated with compounds from the MMV Pathogen Box at 10 µM. Assay plates were moved to BSL3 laboratory for infection with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.1 and kept in incubators at 37°C, 5% CO2 for 60 h. Assay plates were fixed and stained with Hoechst and MitoTracker to allow visualization of the cell nucleus and mitochondrial mass with the Operetta automated fluorescence microscope and calculation of virus-induced CPE. Control groups included DMSO-treated non-infected wells (positive control for healthy cells) and DMSO-treated SARS-CoV-2 infected wells (negative control, CPE-affected cells). Treatment with compounds that resulted in average 50% inhibition of SARS-CoV-2-induced CPE (InCPE > 50%), assessed by the number of Hoechst-stained nuclei, were considered hit candidates (Figure 2A). A total of 15 hit candidates were selected and resupplied from MMV, comprising 3.7% of all compounds in the Pathogen Box (Table S1). Our next step was to determine values for half maximal cytotoxic concentration (CC50) and half maximal effective concentration (EC50) for all 15 hit candidate compounds. Dose-response curves in Vero CCL81 were performed for each hit compound in the range of 40 µM to 0.156 µM, in the 384-well plate format (Figures S2A–H). SARS-CoV-2 infection, nuclei and mitochondrial staining and fixation procedures were performed as previously described for the HTS assays. Our criteria for hit confirmation were an EC50 lower than 20 µM and selectivity index (SI, given by CC50/EC50 ratio) over 2.0 (Figure 1). Seven compounds showed a concentration dependent cytoprotective effect against SARS-CoV-2 infection and fit our selection parameters (Figure 2B): MMV000063 (also known as sitamaquine), MMV688124, MMV688279 (dihydroquinazoline), MMV024829, MMV024937, MMV023953, MMV688991 (nitazoxanide) (Figure 2C). Importantly, none of the seven selected hit compounds caused an increase in mitochondrial fluorescence, which was observed in cells treated with chloroquine (Figures S2I–O). The most potent compound at this stage was MMV688279, with EC50 at 10 µM, while the most selective was MMV024829, with an SI at 4.0.
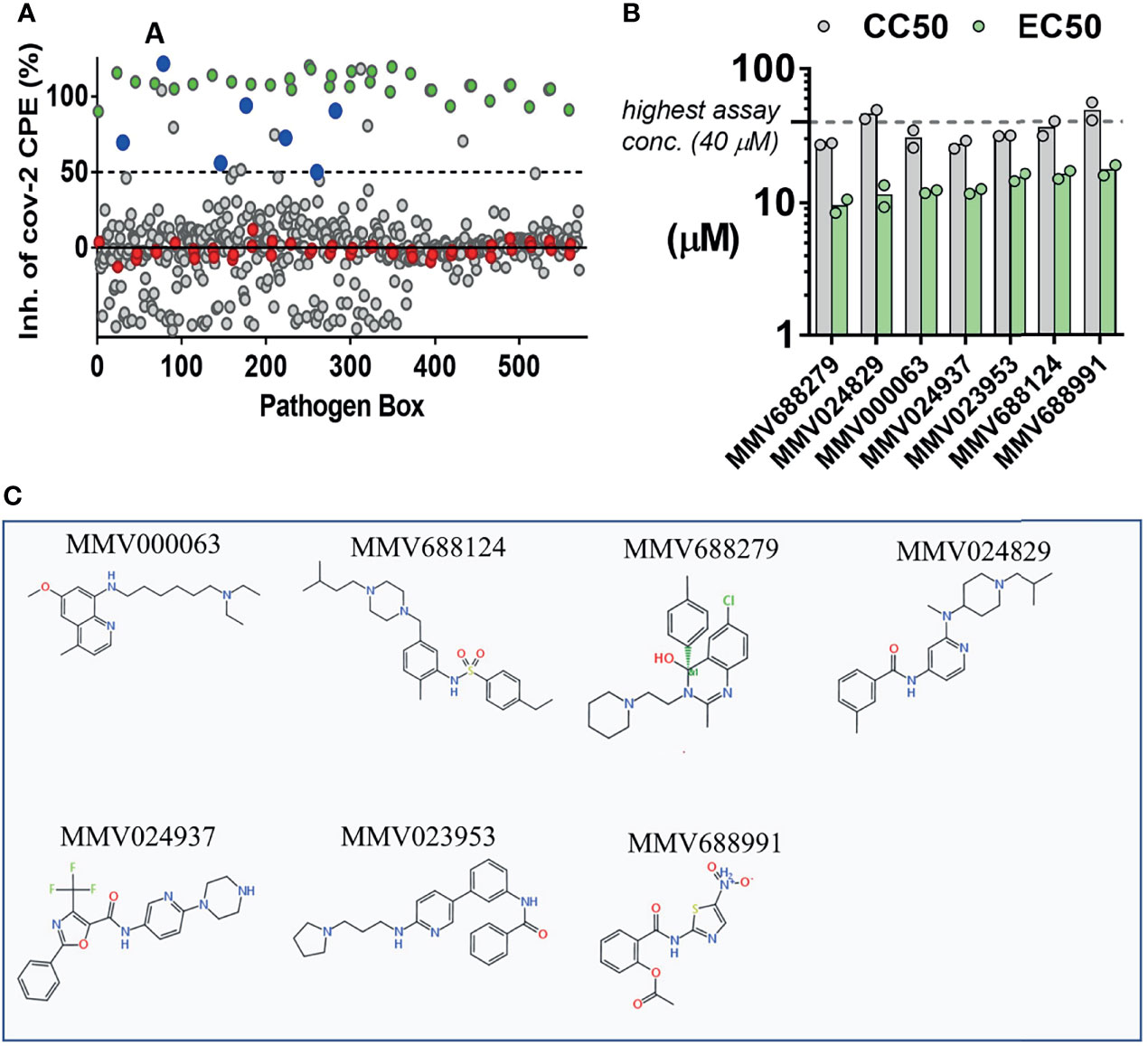
Figure 2 Screening of the MMV Pathogen Box for inhibitors of SARS-CoV-2-induced cytopathic effect (CPE) in Vero CCL81 cells. (A) 400 compounds (grey circles) were tested for the ability to inhibit virus-induced CPE and cell death. Non-infected wells (100%, green) and infected vehicle-treated (0%, red) were used as controls. Seven compounds (blue circles) that inhibited CPE > 50% were classified as hit candidates. (B) EC50/CC50 values for 7 hit compounds. Bars represent means of two independent experiments performed on different days. Dots represent mean values from independent concentration-response curves performed in triplicate. (C) Chemical structures of 7 selected hits.
3.2 Hit Confirmation
Seven hit compounds were tested for antiviral activity against SARS-CoV-2 in low-throughput confirmation tests at a final concentration of 20 µM, a concentration in which all 7 compounds were found to be non-toxic in Vero CCL81 cells (Figure S2). Compound anti-SARS-CoV-2 activity was evaluated in four different cell lines: Vero CCL81, HuH 7.5, Caco-2 and Calu-3, in the 24-wells plate format. Supernatant samples were collected 48h post infection (p.i.) to assess viral load by RT-qPCR and viral plaque assay (Figure 3; Figure S3). MMV688279 presented a potent antiviral effect against SARS-CoV-2 infection in all cell lines tested, resulting in 10 to 1000-fold reductions in viral RNA and up to 10000-fold (104) reduction in infectious viral progeny (Figures 3A, B). Of note, the most dramatic reductions in viral load caused by MMV688279 were observed in Calu-3 and Caco-2 human cell lines (Figure 3E), in which viral levels were below detection (less than 10 PFU/mL) in several samples (Figures 3B, D). Treatment with compound MMV688991 (nitazoxanide) also led to significant decreases in viral load in all cell types tested, which were of at least 10-fold in viral RNA and viral progeny (Figures 3C, D). MMV688991 was least effective in Vero CCL81 cells but caused up to a 1000-fold reduction in viral load in HuH 7.5, Caco-2 and Calu-3 cells. Compound MMV000063 caused a 10-fold reduction in viral load in HuH 7.5 and Caco-2 cells (Figures S3A, B). Treatment with MMV024937 led to at least 10-fold decrease in viral load in Vero CCL81 and Calu-3 (Figures S3G, H). MMV688124, MMV024829 and MMV023953 did not present an antiviral effect in any cell line tested, as assessed by RT-qPCR or plaque assay.
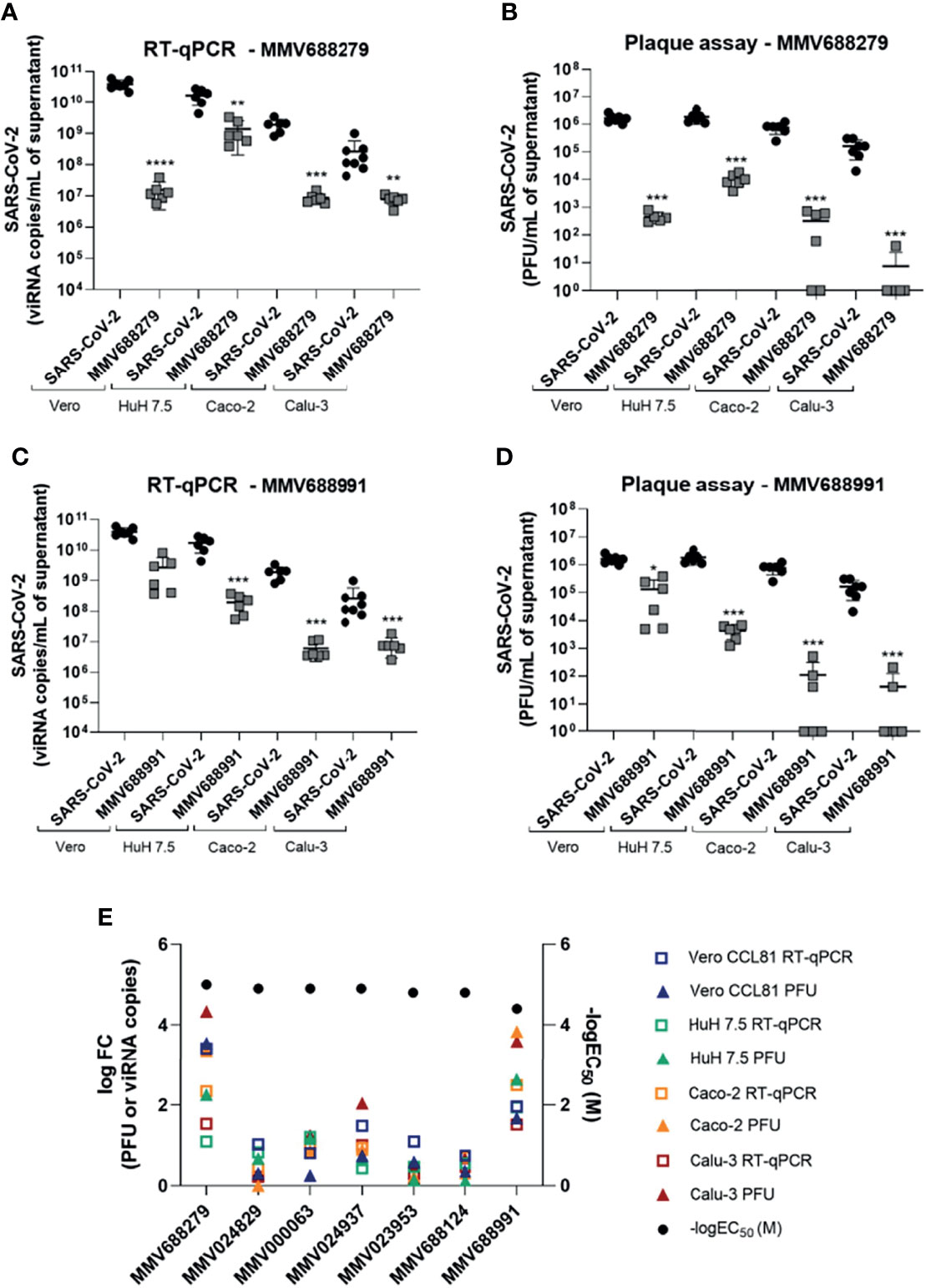
Figure 3 MMV688279 and MMV688991 have potent antiviral effects against SARS-CoV-2 in different cell lines. (A, C) Viral RNA measured by RT-qPCR in supernatant of cell cultures of different cell lines (Vero CCL81, HuH 7.5, Caco-2 and Calu-3) infected with SARS-CoV-2 and treated or not with MMV688279 (20 µM) and MMV688991 (20 µM). (B, D) Infectious viral particles measured in the supernatant of different cell lines infected with SARS-CoV-2 and treated or not with MMV688279 and MMV688991. *p<0.05, **p<0.01, ***p<0.001, ****P≤0.0001 relative to the virus-infected, vehicle-treated control group. Vehicle = DMSO (final concentration 0.4%). Data representative of 2 independent experiments (n=6). (E) Extracted potency parameters logFC RT-qPCR (squares), logFC plaque assay (triangle), -logEC50 (sphere) to each compound and each cell line. FC= fold change: FC= (average values of PFU/ml or viRNA copies/ml of untreated infected cell supernatants) over (average values of PFU/ml or viRNA copies/ml of treated cells supernatants). EC50: compound concentration needed to reduce 50% of the cytopathic effect induced by SARS-CoV-2 infection in Vero cells. -logEC50 (M) values are reported. The limit of detection of the assay is 10 PFU/mL.
Due to their antiviral effects, compounds MMV688279 and MMV688991 were selected for an extra validation step. We used an immunofluorescence assay to assess expression of the viral Spike protein, confirming the antiviral effect of both compounds in SARS-CoV-2-infected Vero CCL81 and Calu-3 cells (Figure 4). MMV688279 reduced the frequency of SARS-CoV2 infected cells (Figures 4B, E) and the mean fluorescent intensity (MFI) of infection per cell (Figures 4C, F) in both cell lines tested. In parallel, MMV688991 had the most preeminent effect on the frequency of infected cells from both lines.
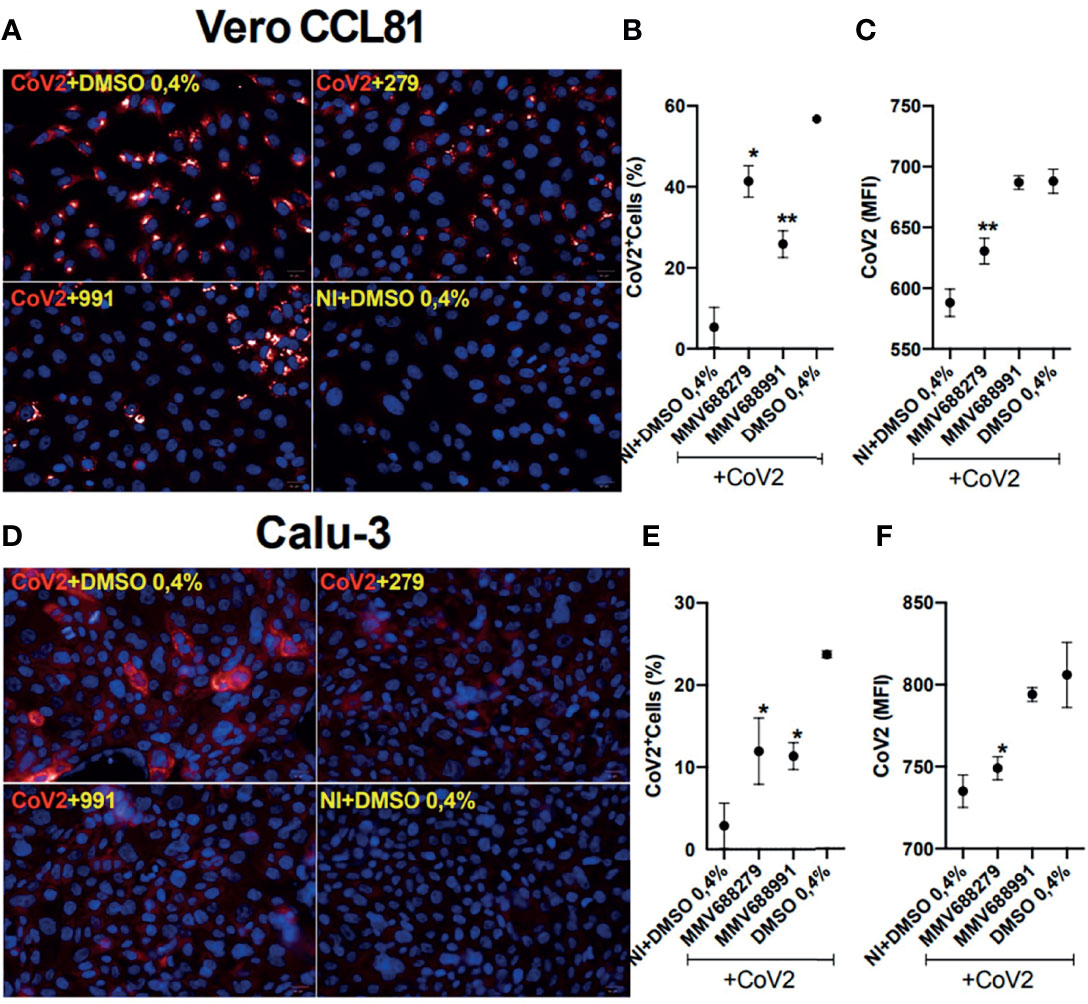
Figure 4 Indirect immunofluorescence (IFI) detection of SARS-CoV2 infecting Vero and Calu-3 cell lines after 22 h of infection. (A, D) The representative images show the presence of Spike-1 protein stained with the Alexa647-labeled antibody (red) and confirm the hit activity of the compounds MMV688279 and MMV688991. Cell nuclei were stained with Hoechst (blue); bar 20µm. (B, E) Frequency of CoV2+-infected cells and the (C, F) amount of infection per cell revealed by the mean fluorescent intensity (MFI). *p<0.05 and **p<0.01 relative to the virus-infected control group. Vehicle–DMSO 0.4%.
In summary, four out of seven hit compounds (57%) were validated in our assays as compounds with antiviral activity against SARS-CoV-2 in vitro, at a concentration of 20 µM. Among these compounds, MMV688279 and MMV688991 were the most potent, causing a significant reduction in viral load in all cell lines tested, assessed either by viral RNA levels, release of infectious viral progeny and spike protein expression.
3.3 Screening of a Compound Series Derived From MMV688279 and Identification of MMV1578396
A series of 40 compounds derived from MMV688279 were obtained and screened in 384 well plates against SARS-CoV-2 infection in Vero CCL81 cells. We included resupplied MMV688279, nitazoxanide and chloroquine as controls. As expected, all control compounds protected Vero CCL81 cells against SARS-CoV-2-induced CPE in the low micromolar range, while vehicle-treated cells were mostly destroyed. We considered two criteria for hit compound selection: 1) EC50 value lower than 20 µM; 2) SI greater than 4.0 (the SI of MMV688279), which led us to select MMV1578396. MMV1578396 presented an average EC50 of 7.5 µM and a CC50 of 39.1 µM, which resulted in an SI of 5.2.
The antiviral properties of MMV1578396 were validated in infection assays in Vero CCL81 and Calu-3 cells (Figure 5). In Vero CCL81 cells, MMV1578396 caused a 42-fold reduction in infectious viral titers while MMV688279 caused a 380-fold reduction, both at a concentration of 20 µM (Figure 5A). The antiviral effect of MMV1578396 against SARS-CoV-2 was more pronounced in Calu-3 cells, in which a 500-fold reduction in infectious viral load was observed in cell cultures treated with the compound at 20 µM. While treatment with MMV688279 or MMV1578396 resulted in comparable reductions in infectious viruses in cell culture at 20 µM, treatment with MMV1578396 at 2 µM caused a 50-fold reduction in viral load in Calu-3 cells (Figure 5B). Analysis of viral RNA levels indicated that treatment with MMV1578396 had no effect on viral load in Vero CCL81 cells (Figure 5C) but caused a 10-fold reduction in viral load in Calu-3 cells at 20 µM concentration (Figure 5D). The effects of MMV688279 and MMV1578396 on viral RNA levels in Calu-3 cells were mostly similar. Compounds MMV688279 and MMV1578396 were not cytotoxic in the concentrations tested in both Vero CCL81 and Calu-3 cells (Figures 5E, F).
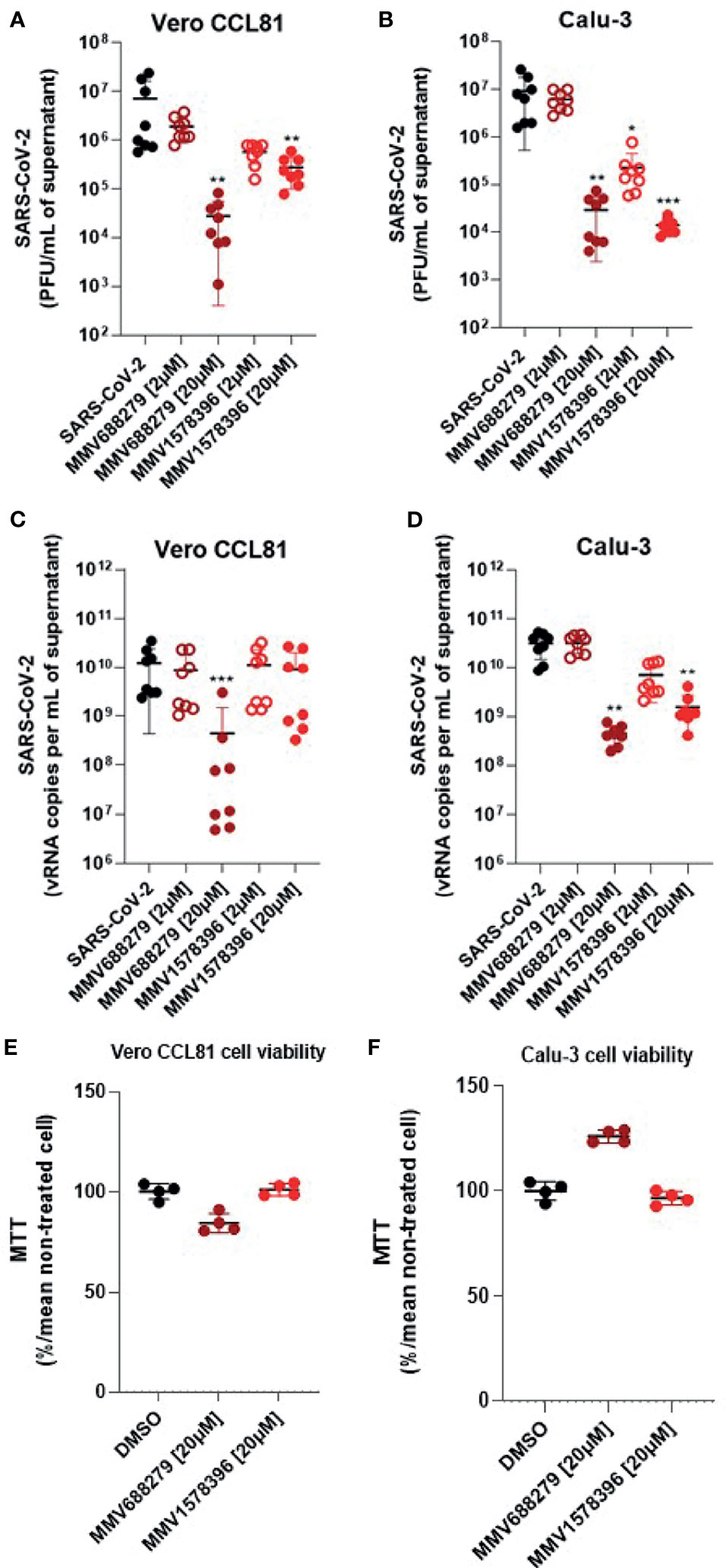
Figure 5 MMV1578396 has potent antiviral activity against SARS-CoV-2 in cell culture. (A, C) Viral RNA measured by RT-qPCR in supernatant of cell cultures of different cell lines (Vero CCL81 and Calu-3) infected with SARS-CoV-2 and treated or not with MMV688279 (20 and 2 µM) and MMV1578396 (20 and 2 µM). (B, D) Infectious viral particles measured in the supernatant of different cell lines infected with SARS-CoV-2 and treated or not with MMV688279 and MMV1578396. (E, F) Cell viability measured by MTT assay in Vero CCL81 and Calu-3 non-infected cells treated with DMSO (control) or MMV688279 (20 µM) and MMV1578396 (20 µM). Viability expressed in percentage (%). *p<0.05, **p<0.01, ***p<0.001 relative to the virus-infected, vehicle-treated control group. Vehicle = DMSO (final concentration 0.4%). Data representative of 2 independent experiments (n=8). The limit of detection of the assay is 10 PFU/mL.
In summary, screening of a compound series derived from MMV688279 resulted in the identification of MMV1578396, which has a comparable selectivity index and was also validated in antiviral assays against SARS-CoV-2.
4 Discussion
The MMV Pathogen Box is a library containing 400 drug-like molecules to be used for drug discovery against neglected and new diseases. It contains reference compounds approved for human use, and new compounds that have shown inhibitory activity against the causative agents of malaria, tuberculosis, toxoplasmosis, Chagas disease, cryptosporidiosis and dengue. Using these compounds for drug discovery in the context of a pandemic is a faster strategy to find bioactive compounds against SARS-CoV-2. All compounds in the Pathogen Box were previously tested for toxicity in vitro by MMV and found to be adequate for initial drug discovery assays.
In the initial phase of our screening campaign, we identified seven compounds that reduced virus-induced CPE, of which four compounds had confirmed antiviral activity against SARS-CoV-2: MMV000063, MMV024937, MMV688279 and MMV688991. Interestingly, there are compounds that protect Vero CCL81 cells from virus-induced cell death without affecting viral replication (Figure 3E), thus being solely cytoprotective in the context of infection. Compounds MMV688214, MMV024829 and MMV023953 may increase Vero CCL81 cell survival or cell proliferation, leading to their identification as hits in a cell culture-based screening campaign. We could not ascertain whether such cytoprotective compounds would be beneficial to treat COVID-19 patients. Regarding the cells lines, Caco-2 is a human epithelial cell line used for assays of drug absorption, intestinal permeability and drug transport (23), and thus frequently used in initial phases of drug discovery. HuH 7.5 is a human hepatocyte cell line traditionally used for hepatitis C virus research and found to express angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2), which are critical for SARS-CoV-2 infection (24). Our data indicates that HuH 7.5 is a suitable model for antiviral testing against SARS-CoV-2 in vitro. Calu-3 is also a human epithelial cell line derived from lung and a recurrent in vitro model used in COVID-19 studies. The use of different cell lines to validate our findings is based on the dependence of SARS-CoV-2 on host proteases to initiate infection. SARS-CoV-2 Spike protein is cleaved by the pH-dependent cysteine protease cathepsin L (CTSL) in Vero CCL81 cells, whereas the same Spike protein is cleaved by the pH-independent TMPRSS2 protease for SARS-CoV-2 entry in Calu-3 cells (25).
MMV000063 and MMV688991 are reference compounds within the Pathogen Box. MMV000063 is a synthetic compound commonly known as sitamaquine that has been used in the treatment of leishmaniasis (26). Conversely, MMV688991 (nitazoxanide) has already been described as an inhibitor of SARS-CoV-2 replication in vitro (27). Nitazoxanide is a broad-spectrum antiviral compound and has been recommended as a possible treatment against MERS-CoV and SARS-CoV-2 (28, 29). Recent studies indicate that treatment with nitazoxanide reduces viral load in moderate COVID-19 patients, though amelioration of symptoms seems dependent on early treatment administration (28, 30, 31). To the best of our knowledge, we are first to report that MMV688279 and MMV024937 (Figure 1) are active against SARS-CoV-2. In vitro ADME and mouse PK data are available and can be accessed in MMV portal (32). MMV688279 is a dihydroquinazoline compound originally tested against kinetoplastids. MMV688279 acts by inhibiting trypanothione reductase, an enzyme involved in the antioxidant system of Trypanosoma brucei that is essential for parasite growth in cell culture (33). MMV024937 is a heterocyclic compound based on a pyridine skeleton and is part of the anti-malarial compound set.
According to our initial results, compounds MMV688279 and MMV688991 were the most promising. Both compounds caused a significant reduction of viral load in all cell lines tested. Thus, these compounds are likely to act on conserved cellular mechanisms to promote the antiviral effect against SARS-CoV-2, which suggests that they may also be effective in multicellular infection models and in vivo. Early PK studies show poor oral bioavailability (F% = 20%) in mice, which could be attributed to its high logD, rapid metabolization, low Cmax and high volume of distribution, although the latter parameter could potentially suggest distribution to the target tissue, the lungs. However, with an IC50 below 20 µM in a CYP2D6 inhibition assay, at this stage, MMV688279 could lead to potential drug-drug interactions. Therefore, further structure-activity and structure-property analyses should be undertaken for this chemical class in order to achieve better DMPK and safety profiles.
We identified compound MMV1578396 from a series of 40 compounds inspired in compound MMV688279. MMV1578396 performed similarly to compound MMV688279 in screening and validation assays in Vero CCL81 cells and reduced infectious SARS-CoV-2 levels by 50-fold at a concentration of 2 μM in Calu-3 cells (Figure 5B). Calu-3 cells are the cell culture gold standard for compound testing in the context of COVID-19. Compounds that effectively inhibit SARS-CoV-2 in this cell type would be more likely to be effective in vivo, and eventually in clinical trials in patients.
The experimental assessment of antiviral activity included the use of both RT-qPCR and plaque assays. Quantification of copies of viral RNA by RT-qPCR, although overly sensitive, does not discriminate between infectious and non-infectious particles. On the other hand, plaque assay titration reveals infectious viral particles. Cell lines treated with MMV688279, MMV688991 or MMV1578396 showed a more pronounced reduction in viral load as indicated by plaque assays than by RT-qPCR (Figure 3E). This result indicates that such compounds are likely interfering with middle-to-late phases of viral replication, after viral RNA replication takes place. Moreover, the mechanism of action by which MMV688279 and MMV1578396 inhibit SARS-CoV-2 replication is presently unknown. Although none of these compounds are potent in the nanomolar range, the discovery of their mechanisms of action may inspire the development of more potent compounds and new antiviral strategies.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
LC, AB, MF, HG, AN, JFS and KB-D-S performed experiments. PO, KF, MB and DT contributed resources and funding. KS represents the partnership with MMV. FG and JP-M provided access and supported activities at the BSL3 laboratory. LC, AB, MF, HG, AN, DT, JHCS, AC and RM analyzed and interpreted data. LC, AB, MF, HG, AN, AC and RM wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We thank RedeVirus MCTI (Brazilian Ministry of Sciences, Technology and Innovation), CNPq: Brazilian National Council for Scientific and Technological Development (grant #403709/2020-2), Finep (Brazilian Funding Authority for Studies and Projects) and FNDCT (Brazilian National Fund for Scientific and Technological Development) for the financial support (grant #01.20.0003.00). We also thank FAPESP (#2016/00194-8, #2020/04558-0, #2019/06459-1, #2020/02159-0) and FAEPEX-UNICAMP (#2266/20) for supporting this study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Mr. Valber Ferreira and Ms. Tereza Silva for all technical support. We thank Dr. Celso Benedetti for critically reviewing our manuscript. Graphical abstract was in created BioRender.com. Finally, we thank MMV for providing the Pathogen box.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2022.854363/full#supplementary-material
Supplementary Figure 1 | HTS assay quality assessment. 400 molecules from the PathogenBox (from MMV) were redistributed in two 384-wells microplates, named P1 and P2, with 32 positive (+) and negative (-) controls per plate. (A) plots and statistical tables for the number of cells in control wells of HTS run-5 and run-6, two independent replicates. (B) Z-factor values for plates P1 and P2 from HTS run-5 and run-6. (C) Correlation plot between the normalized Inhibition of Sars-Cov-2 mediated cytopathic effect (%InCPE) obtained from HTS run-5 and run-6. Negative controls (not infected), positive controls (infected and untreated), tested samples and hit candidates are colored green, red, gray and blue, respectively.
Supplementary Figure 2 | Dose-response curves for seven hit compounds from MMV Pathogen box. Vero CCL81 cells were infected with SARS-CoV-2 (MOI 0,1) and incubated with compounds for 60h. (A–H) Dose-response curves for compounds calculated on CPE inhibition (red) and compound toxicity (blue) to calculate EC50 and CC50 values. (I–O) Evaluation of Mitotracker-stained mitochondria fluorescence in Vero CCL81 cells incubated with seven hit compounds. *Chloroquine used as reference compound.
Supplementary Figure 3 | Assessment of antiviral activity of confirmed hit compounds against SARS-CoV-2 in different cell lines. (A, C, E, G, I) Infectious viral load measured in the supernatant of different cell lines (Vero CCL81, HuH 7.5, Caco-2 and Calu-3) infected with SARS-CoV-2 and treated or not with MMV000063, MMV688124, MMV024829, MMV024937 and MMV023953. (B, D, F, H, J) Viral RNA measured by RT-qPCR in supernatant of Vero, Huh 7.5, Caco-2 and Calu-3 cells infected with SARS-CoV-2 and treated or not with compounds. *p<0.05, **p<0.01, ***p<0.001 relative to the virus-infected control group. Data representative of 2 independent experiments (n=6). Vehicle – DMSO 0.4%. The limit of detection of the assay is 10 PFU/mL
Supplementary Figure 4 | Screening of a compound series derived from MMV688279 identified the hit compound MMV1578396 (A) 6-point dose-response curve of MMV1578396 used to calculate EC50/CC50 values. (B) Chemical structure of MMV1578396.
References
1. Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N Engl J Med (2020) 382:1199–207. doi: 10.1056/nejmoa2001316
2. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus From Patients With Pneumonia in China 2019. N Engl J Med (2020) 382:727–33. doi: 10.1056/nejmoa2001017
3. Khalili A, Petersen E, Koopmans M, Go U, Hamer DH, Petrosillo N, et al. Personal View Comparing SARS-CoV-2 With SARS-CoV and Influenza Pandemics. Lancet Infect Dis (2020) 20:e238–44. doi: 10.1016/S1473-3099(20)30484-9
4. WHO. Coronavirus Disease (COVID-19) Dashboard | WHO Coronavirus Disease (COVID-19) Dashboard. Available at: https://covid19.who.int/ (Accessed 2.20.21).
5. Guan W, Ni Z, Hu Yu, Liang W, Ou C, He J, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med (2020) 382:1708–20. doi: 10.1056/nejmoa2002032
6. Matricardi PM, Dal Negro RW, Nisini R. The First, Holistic Immunological Model of COVID-19: Implications for Prevention, Diagnosis, and Public Health Measures. Pediatr Allergy Immunol (2020) 31:454–70. doi: 10.1111/pai.13271
7. Yang J, Zheng Y, Gou X, Pu K, Chen Z, Guo Q, et al. Prevalence of Comorbidities and its Effects in Patients Infected With SARS-CoV-2: A Systematic Review and Meta-Analysis. Int J Infect Dis (2020) 94:91–5. doi: 10.1016/j.ijid.2020.03.017
8. Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, et al. Dexamethasone in Hospitalized Patients With Covid-19. N Engl J Med (2021) 384:693–704. doi: 10.1056/NEJMoa2021436
9. Katz LM. (A Little) Clarity on Convalescent Plasma for Covid-19. N Engl J Med (2021) 384:666–8. doi: 10.1056/nejme2035678
10. Pallotta AM, Kim C, Gordon SM, Kim A. Monoclonal Antibodies for Treating COVID-19. Cleve. Clin J Med (2021) 1:1–5. doi: 10.3949/ccjm.88a.ccc074
11. Pan H, Peto R, Henao-Restrepo A-M, Preziosi M-P, Sathi-yamoorthy V, Abdool Karim Q, et al. Repurposed Antiviral Drugs for Covid-19 — Interim WHO Solidarity Trial Results. N Engl J Med (2021) 384:497–511. doi: 10.1056/NEJMoa2023184
12. Lokhande AS, Devarajan PV. A Review on Possible Mechanistic Insights of Nitazoxanide for Repurposing in COVID-19. Eur J Pharmacol (2021) 891:1–15. doi: 10.1016/j.ejphar.2020.173748
13. Forni G, Mantovani A, Forni G, Mantovani A, Moretta L, Rappuoli R, et al. COVID-19 Vaccines: Where We Stand and Challenges Ahead. Cell Death Differ (2021) 28:626–39. doi: 10.1038/s41418-020-00720-9
14. de Souza WM, Amorim MR, Sesti-Costa R, Coimbra LD, Toledo-Teixeira DA, de, Parise PL, et al. Levels of SARS-CoV-2 Lineage P.1 Neutralization by Antibodies Elicited After Natural Infection and Vaccination. SSRN Electron J (2021) 2:E527–35. doi: 10.2139/ssrn.3793486
15. Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, et al. Neutralization of SARS-CoV-2 Spike 69/70 Deletion, E484K and N501Y Variants by BNT162b2 Vaccine-Elicited Sera. Nat Med (2021) 4:620–21 1–2. doi: 10.1038/s41591-021-01270-4
16. Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, et al. Discovery of SARS-CoV-2 Antiviral Drugs Through Large-Scale Compound Repurposing. Nature (2020) 586:113–9. doi: 10.1038/s41586-020-2577-1
17. Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, et al. A Trial of Lopinavir–Ritonavir in Adults Hospitalized With Severe Covid-19. N Engl J Med (2020) 382:1787–99. doi: 10.1056/nejmoa2001282
18. Lythgoe MP, Middleton P. Ongoing Clinical Trials for the Management of the COVID-19 Pandemic, Trends in Pharmacological. Sciences (2020) 41:363–82. doi: 10.1016/j.tips.2020.03.006
19. Mayr LM, Bojanic D. Novel Trends in High-Throughput Screening. Curr Opin Pharmacol (2009) 9:580–8. doi: 10.1016/j.coph.2009.08.004
20. Pawar AY. Combating Devastating COVID-19 by Drug Repurposing. Int J Antimicrob Agents (2020) 56:1–4. doi: 10.1016/j.ijantimicag.2020.105984
21. Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat Rev Drug Discovery (2018) 18:41–58. doi: 10.1038/nrd.2018.168
22. Veale CGL. Unpacking the Pathogen Box—An Open Source Tool for Fighting Neglected Tropical Disease. ChemMedChem (2019) 14:386–453. doi: 10.1002/cmdc.201800755
23. Kumar KKV, Karnati S, Reddy MB, Chandramouli R. Caco-2 Cell Lines in Drug Discovery- an Updated Perspective. J Basic Clin Pharm (2010) 1:63–9.
24. Hoffmann H-H, Sá Nchez-Rivera FJ, Schneider WM, Macdonald MR, Poirier JT, Rice CM. Functional Interrogation of a SARS-CoV-2 Host Protein Interactome Identifies Unique and Shared Coronavirus Host Factors. Cell Host Microbe (2021) 29:267–80. doi: 10.1016/j.chom.2020.12.009
25. Hoffmann M, Mösbauer K, Hofmann-Winkler H, Kaul A, Kleine-Weber H, Krüger N, et al. Chloroquine Does Not Inhibit Infection of Human Lung Cells With SARS-CoV-2. Nature (2020) 585:588–90. doi: 10.1038/s41586-020-2575-3
26. Hussain H, Al-Harrasi A, Al-Rawahi A, Green IR, Gibbons S. Fruitful Decade for Antileishmanial Compounds From 2002 to Late 2011. Chem Rev (2014) 114:10369–428. doi: 10.1021/cr400552x
27. Son J, Huang S, Zeng Q, Bricker TL, Case JB, Zhou J, et al. Nitazoxanide and JIB-04 Have Broad-Spectrum Antiviral Activity and Inhibit SARS-CoV-2 Replication in Cell Culture and Coronavirus Pathogenesis in a Pig Model. bioRxiv (2020) 1:1–41. doi: 10.1101/2020.09.24.312165
28. Rocco PRM, Silva PL, Cruz FF, Junior MACM, Tierno P.F.G.M.M., Moura MA, et al. Early Use of Nitazoxanide in Mild Covid-19 Disease: Randomised, Placebo-Controlled Trial. Eur Respir J (2021) 58:1–10. doi: 10.1183/13993003.03725-2020
29. Rossignol JF. Nitazoxanide, a New Drug Candidate for the Treatment of Middle East Respiratory Syndrome Coronavirus. J Infect Public Health (2016) 9:227–30. doi: 10.1016/j.jiph.2016.04.001
30. Blum VF, Cimerman S, Hunter JR, Tierno P, Lacerda A, Soeiro A, et al. Nitazoxanide In Vitro Efficacy Against SARS CoV-2 and In Vivo Superiority to Placebo to Treat Moderate COVID-19 – A Phase 2 Randomized Double-Blind Clinical Trial. SSRN Electron J (2021) 1:1–30. doi: 10.2139/ssrn.3763773
31. Silva M, Espejo A, Pereyra ML, Lynch M, Thompson M, Taconelli H, et al. Efficacy of Nitazoxanide in Reducing the Viral Load in COVID-19 Patients. Randomized, Placebo-Controlled, Single-Blinded, Parallel Group, Pilot Study. medRxiv (2021), 1–17. doi: 10.1101/2021.03.03.21252509
32. About the Pathogen Box | Medicines for Malaria Venture . Available at: https://www.mmv.org/mmv-open/pathogen-box/about-pathogen-box (Accessed 3.12.21). WWW Document.
33. Patterson S, Alphey MS, Jones DC, Shanks EJ, Street IP, Frearson JA, et al. Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography. J Med Chem (2011) 54:6514–30. doi: 10.1021/jm200312v
Keywords: SARS-CoV-2, biosafety level 3, MMV pathogen box, antivirals, drug repositioning, high-throughput screening
Citation: Coimbra LD, Borin A, Fontoura M, Gravina HD, Nagai A, Shimizu JF, Bispo-dos-Santos K, Granja F, Oliveira PSL, Franchini KG, Samby K, Bruder M, Proença-Módena JL, Trivella DBB, Smetana JHC, Cordeiro AT and Marques RE (2022) Identification of Compounds With Antiviral Activity Against SARS-CoV-2 in the MMV Pathogen Box Using a Phenotypic High-Throughput Screening Assay. Front.Virol. 2:854363. doi: 10.3389/fviro.2022.854363
Received: 19 January 2022; Accepted: 17 March 2022;
Published: 14 April 2022.
Edited by:
Siew Pheng Lim, Denka Life Innovation Research (DLIR), SingaporeReviewed by:
Xuping Xie, University of Texas Medical Branch at Galveston, United StatesAmelia K. Pinto, Saint Louis University, United States
Copyright © 2022 Coimbra, Borin, Fontoura, Gravina, Nagai, Shimizu, Bispo-dos-Santos, Granja, Oliveira, Franchini, Samby, Bruder, Proença-Módena, Trivella, Smetana, Cordeiro and Marques. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rafael Elias Marques, rafael.marques@lnbio.cnpem.br
†These authors have contributed equally to this work
 Laís D. Coimbra
Laís D. Coimbra Alexandre Borin
Alexandre Borin Marina Fontoura
Marina Fontoura Humberto D. Gravina
Humberto D. Gravina Alice Nagai1
Alice Nagai1  Karina Bispo-dos-Santos
Karina Bispo-dos-Santos Paulo S. L. Oliveira
Paulo S. L. Oliveira Kleber G. Franchini
Kleber G. Franchini Marjorie Bruder
Marjorie Bruder José Luiz Proença-Módena
José Luiz Proença-Módena Daniela B. B. Trivella
Daniela B. B. Trivella Juliana H. C. Smetana
Juliana H. C. Smetana Rafael Elias Marques
Rafael Elias Marques