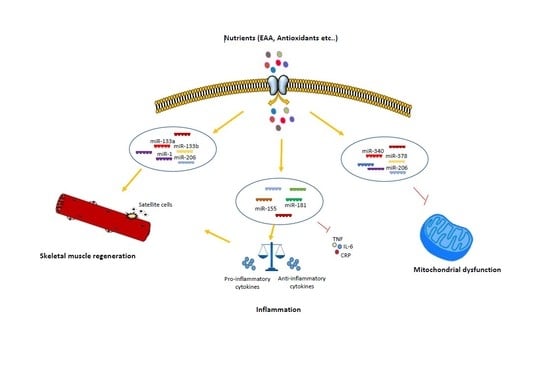
Nutrition and microRNAs: Novel Insights to Fight Sarcopenia
Abstract
:1. Introduction
2. Nutrition-Dependent microRNA Regulation of Skeletal Muscle Regeneration
3. Nutrition-Dependent microRNA Regulation of Inflammageing
4. Nutrition-Dependent microRNA Regulation of Mitochondrial Dysfunction
4.1. Autophagy
4.2. ROS Imbalance
5. High Fructose Diet Modulation of miRNAs Expression in Sarcopenia
6. Circulating miRNAs
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cruz-Jentoft, A.J.; Bahat, G.; Bauer, J.; Boirie, Y.; Bruyère, O.; Cederholm, T.; Cooper, C.; Landi, F.; Rolland, Y.; Sayer, A.A.; et al. Sarcopenia: Revised European consensus on definition and diagnosis. Age Ageing 2019, 48, 16–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmeli, E.; Reznick, A.Z.; Coleman, R.; Carmeli, V. Muscle strength and mass of lower extremities in relation to functional abilities in elderly adults. Gerontology 2000, 46, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Rolland, Y.; Czerwinski, S.; Van Kan, G.A.; Morley, J.E.; Cesari, M.; Onder, G.; Woo, J.; Baumgartner, R.; Pillard, F.; Boirie, Y.; et al. Sarcopenia: Its assessment, etiology, pathogenesis, consequences and future perspectives. J. Nutr. Health Aging 2008, 12, 433–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryall, J.G.; Schertzer, J.D.; Lynch, G.S. Cellular and molecular mechanisms underlying age-related skeletal muscle wasting and weakness. Biogerontology 2008, 9, 213–228. [Google Scholar] [CrossRef]
- Metter, E.J.; Talbot, L.A.; Schrager, M.; Conwit, R. Skeletal muscle strength as a predictor of all-cause mortality in healthy men. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2002, 57, B359–B365. [Google Scholar] [CrossRef]
- Marzetti, E.; Calvani, R.; Cesari, M.; Buford, T.W.; Lorenzi, M.; Behnke, B.J.; Leeuwenburgh, C. Mitochondrial dysfunction and sarcopenia of aging: From signaling pathways to clinical trials. Int. J. Biochem. Cell Biol. 2013, 45, 2288–2301. [Google Scholar] [CrossRef] [Green Version]
- Marzetti, E.; Hwang, J.C.Y.; Lees, H.A.; Wohlgemuth, S.E.; Dupont-Versteegden, E.E.; Carter, C.S.; Bernabei, R.; Leeuwenburgh, C. Mitochondrial death effectors: Relevance to sarcopenia and disuse muscle atrophy. Biochim. Biophys. Acta Gen. Subj. 2010, 1800, 235–244. [Google Scholar] [CrossRef] [Green Version]
- Alway, S.E.; Mohamed, J.S.; Myers, M.J. Mitochondria Initiate and Regulate Sarcopenia. Exerc. Sport Sci. Rev. 2017, 45, 58–69. [Google Scholar] [CrossRef]
- Dalle, S.; Rossmeislova, L.; Koppo, K. The role of inflammation in age-related sarcopenia. Front. Physiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Meng, S.J.; Yu, L.J. Oxidative stress, molecular inflammation and sarcopenia. Int. J. Mol. Sci. 2010, 11, 1509–1526. [Google Scholar] [CrossRef] [Green Version]
- Churchward-Venne, T.A.; Breen, L.; Phillips, S.M. Alterations in human muscle protein metabolism with aging: Protein and exercise as countermeasures to offset sarcopenia. BioFactors 2014, 40, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Scicchitano, B.M.; Sica, G. The Beneficial Effects of Taurine to Counteract Sarcopenia. Curr. Protein Pept. Sci. 2018, 19, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Short, K.R.; Nair, K.S. Muscle protein metabolism and the sarcopenia of aging. Int. J. Sport Nutr. Exerc. Metab. 2001, 11, S119–S127. [Google Scholar] [CrossRef] [PubMed]
- Welle, S.; Thornton, C.; Jozefowicz, R.; Statt, M. Myofibrillar protein synthesis in young and old men. Am. J. Physiol. 1993, 264, E693–E698. [Google Scholar] [CrossRef] [PubMed]
- Morton, R.W.; Traylor, D.A.; Weijs, P.J.M.; Phillips, S.M. Defining anabolic resistance: Implications for delivery of clinical care nutrition. Curr. Opin. Crit. Care 2018, 24, 124–130. [Google Scholar] [CrossRef]
- Haran, P.H.; Rivas, D.A.; Fielding, R.A. Role and potential mechanisms of anabolic resistance in sarcopenia. J. Cachexia. Sarcopenia Muscle 2012, 3, 157–162. [Google Scholar] [CrossRef] [Green Version]
- Dillon, E.L.; Durham, W.J.; Urban, R.J.; Sheffield-Moore, M. Hormone treatment and muscle anabolism during aging: Androgens. Clin. Nutr. 2010, 29, 697–700. [Google Scholar] [CrossRef] [Green Version]
- Basualto-Alarcón, C.; Varela, D.; Duran, J.; Maass, R.; Estrada, M. Sarcopenia and androgens: A link between pathology and treatment. Front. Endocrinol. (Lausanne) 2014, 5. [Google Scholar]
- Moore, D.R. Keeping Older Muscle “Young” through Dietary Protein and Physical Activity. Adv. Nutr. 2014, 5, 599S–607S. [Google Scholar] [CrossRef] [Green Version]
- Walker, D.K.; Dickinson, J.M.; Timmerman, K.L.; Drummond, M.J.; Reidy, P.T.; Fry, C.S.; Gundermann, D.M.; Rasmussen, B.B. Exercise, amino acids, and aging in the control of human muscle protein synthesis. Med. Sci. Sports Exerc. 2011, 43, 2249–2258. [Google Scholar] [CrossRef] [Green Version]
- Nieuwenhuizen, W.F.; Weenen, H.; Rigby, P.; Hetherington, M.M. Older adults and patients in need of nutritional support: Review of current treatment options and factors influencing nutritional intake. Clin. Nutr. 2010, 29, 160–169. [Google Scholar] [CrossRef]
- Robinson, S.; Granic, A.; Sayer, A.A. Nutrition and muscle strength, as the key component of sarcopenia: An overview of current evidence. Nutrients 2019, 11, 2942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Wienholds, E.; Koudijs, M.J.; van Eeden, F.J.M.; Cuppen, E.; Plasterk, R.H.A. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 2003, 35, 217–218. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Lee, R.C. Identification of microRNAs and Other Tiny Noncoding RNAs by cDNA Cloning. In RNA Interference, Editing, and Modification; Humana Press: Totowa, NJ, USA, 2004; Volume 265, pp. 131–158. [Google Scholar]
- Callis, T.E.; Deng, Z.; Chen, J.-F.; Wang, D.-Z. Muscling Through the microRNA World. Exp. Biol. Med. 2008, 233, 131–138. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, J.J.; Esser, K.A. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J. Appl. Physiol. 2007, 102, 306–313. [Google Scholar] [CrossRef]
- van Rooij, E.; Quiat, D.; Johnson, B.A.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Kelm, R.J.; Olson, E.N. A Family of microRNAs Encoded by Myosin Genes Governs Myosin Expression and Muscle Performance. Dev. Cell 2009, 17, 662–673. [Google Scholar] [CrossRef] [Green Version]
- Zacharewicz, E.; Lamon, S.; Russell, A.P. MicroRNAs in skeletal muscle and their regulation with exercise, ageing, and disease. Front. Physiol. 2013, 4, 266. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Nie, Q.; Zhang, X. MicroRNAs Involved in Skeletal Muscle Differentiation. J. Genet. Genom. 2013, 40, 107–116. [Google Scholar] [CrossRef]
- Williams, A.H.; Liu, N.; van Rooij, E.; Olson, E.N. MicroRNA control of muscle development and disease. Curr. Opin. Cell Biol. 2009, 21, 461–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGregor, R.A.; Poppitt, S.D.; Cameron-Smith, D. Role of microRNAs in the age-related changes in skeletal muscle and diet or exercise interventions to promote healthy aging in humans. Ageing Res. Rev. 2014, 17, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Juvvuna, P.K.; Kukreti, H.; McFarlane, C. Mega roles of microRNAs in regulation of skeletal muscle health and disease. Front. Physiol. 2014, 5, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, J.; Qian, Z.; Chen, Y.; Li, Y.; Zhou, X. MicroRNA regulatory networks in the pathogenesis of sarcopenia. J. Cell. Mol. Med. 2020, 24, 4900–4912. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, B.; He, J.; Chen, D. From nutrient to MicroRNA: A novel insight into cell signaling involved in skeletal muscle development and disease. Int. J. Biol. Sci. 2016, 12, 1247–1261. [Google Scholar] [CrossRef] [Green Version]
- Barberi, L.; Scicchitano, B.M.; De Rossi, M.; Bigot, A.; Duguez, S.; Wielgosik, A.; Stewart, C.; McPhee, J.; Conte, M.; Narici, M.; et al. Age-dependent alteration in muscle regeneration: The critical role of tissue niche. Biogerontology 2013, 14. [Google Scholar] [CrossRef] [Green Version]
- Forcina, L.; Miano, C.; Pelosi, L.; Musarò, A. An Overview About the Biology of Skeletal Muscle Satellite Cells. Curr. Genom. 2019, 20, 24–37. [Google Scholar] [CrossRef] [Green Version]
- Sousa-Victor, P.; Gutarra, S.; García-Prat, L.; Rodriguez-Ubreva, J.; Ortet, L.; Ruiz-Bonilla, V.; Jardí, M.; Ballestar, E.; González, S.; Serrano, A.L.; et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 2014, 506, 316–321. [Google Scholar] [CrossRef]
- Zwetsloot, K.A.; Childs, T.E.; Gilpin, L.T.; Booth, F.W. Non-passaged muscle precursor cells from 32-month old rat skeletal muscle have delayed proliferation and differentiation. Cell Prolif. 2013, 46, 45–57. [Google Scholar] [CrossRef]
- Sousa-Victor, P.; García-Prat, L.; Serrano, A.L.; Perdiguero, E.; Muñoz-Cánoves, P. Muscle stem cell aging: Regulation and rejuvenation. Trends Endocrinol. Metab. 2015, 26, 287–296. [Google Scholar] [CrossRef]
- Scicchitano, B.M.; Sica, G.; Musarò, A. Stem Cells and Tissue Niche: Two Faces of the Same Coin of Muscle Regeneration. Eur. J. Transl. Myol. 2016, 26, 6125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conboy, I.M.; Conboy, M.J.; Wagers, A.J.; Girma, E.R.; Weismann, I.L.; Rando, T.A. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 2005, 433, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.; Thuret, S. The systemic milieu as a mediator of dietary influence on stem cell function during ageing. Ageing Res. Rev. 2015, 19, 53–64. [Google Scholar] [CrossRef]
- Brack, A.S.; Rando, T.A. Intrinsic changes and extrinsic influences of myogenic stem cell function during aging. Stem Cell Rev. 2007, 3, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Villeda, S.A.; Luo, J.; Mosher, K.I.; Zou, B.; Britschgi, M.; Bieri, G.; Stan, T.M.; Fainberg, N.; Ding, Z.; Eggel, A.; et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 2011, 477, 90–96. [Google Scholar] [CrossRef] [Green Version]
- Cheung, T.H.; Quach, N.L.; Charville, G.W.; Liu, L.; Park, L.; Edalati, A.; Yoo, B.; Hoang, P.; Rando, T.A. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 2012, 482, 524–528. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Hak, K.K.; Yong, S.L.; Sivaprasad, U.; Malhotra, A.; Dutta, A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006, 174, 677–687. [Google Scholar]
- Nakasa, T.; Ishikawa, M.; Shi, M.; Shibuya, H.; Adachi, N.; Ochi, M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J. Cell. Mol. Med. 2010, 14, 2495–2505. [Google Scholar] [CrossRef]
- Ling, Y.H.; Sui, M.H.; Zheng, Q.; Wang, K.Y.; Wu, H.; Li, W.Y.; Liu, Y.; Chu, M.X.; Fang, F.G.; Xu, L.N. MiR-27b regulates myogenic proliferation and differentiation by targeting Pax3 in goat. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Drummond, M.J.; Glynn, E.L.; Fry, C.S.; Dhanani, S.; Volpi, E.; Rasmussen, B.B. Essential Amino Acids Increase MicroRNA-499, -208b, and -23a and Downregulate Myostatin and Myocyte Enhancer Factor 2C mRNA Expression in Human Skeletal Muscle. J. Nutr. 2009, 139, 2279–2284. [Google Scholar] [CrossRef] [PubMed]
- Druz, A.; Betenbaugh, M.; Shiloach, J. Michael, S.J. Glucose Depletion Activates mmu-miR-466h-5p Expression Through Oxidative Stress and Inhibition of Histone Deacetylation. Nucleic Acids Res. 2012, 40, 7291–7302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dey, N.; Das, F.; Mariappan, M.M.; Mandal, C.C.; Ghosh-Choudhury, N.; Kasinath, B.S.; Choudhury, G.G. MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes. J. Biol. Chem. 2011, 286, 25586–25603. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Huang, Z.; Chen, D.; Yang, T.; Liu, G. MicroRNA-27a is induced by leucine and contributes to leucine-induced proliferation promotion in C2C12 cells. Int. J. Mol. Sci. 2013, 14, 14076–14084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, M.G.; Silva, M.T.; da Cunha, F.M.; Moriscot, A.S.; Aoki, M.S.; Miyabara, E.H. Leucine supplementation improves regeneration of skeletal muscles from old rats. Exp. Gerontol. 2015, 72, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Rogulska, A.; Kurasz, S. Regeneration of crushed skeletal muscles in experimental animals and the effect of leucine on the course of this process in white rat. Pol. Med. Sci. Hist. Bull. 1975, 15, 245–248. [Google Scholar] [PubMed]
- Iannone, F.; Montesanto, A.; Cione, E.; Crocco, P.; Caroleo, M.C.; Dato, S.; Rose, G.; Passarino, G. Expression patterns of muscle-specific miR-133b and miR-206 correlate with nutritional status and sarcopenia. Nutrients 2020, 12, 297. [Google Scholar] [CrossRef] [Green Version]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef]
- Ge, Y.; Chen, J. Mammalian target of rapamycin (mTOR) signaling network in skeletal myogenesis. J. Biol. Chem. 2012, 287, 43928–43935. [Google Scholar] [CrossRef] [Green Version]
- Calçada, D.; Vianello, D.; Giampieri, E.; Sala, C.; Castellani, G.; de Graaf, A.; Kremer, B.; van Ommen, B.; Feskens, E.; Santoro, A.; et al. The role of low-grade inflammation and metabolic flexibility in aging and nutritional modulation thereof: A systems biology approach. Mech. Ageing Dev. 2014, 136–137, 138–147. [Google Scholar] [CrossRef]
- Forcina, L.; Miano, C.; Scicchitano, B.M.; Rizzuto, E.; Berardinelli, M.G.; De Benedetti, F.; Pelosi, L.; Musarò, A. Increased Circulating Levels of Interleukin-6 Affect the Redox Balance in Skeletal Muscle. Oxid. Med. Cell. Longev. 2019, 2019. [Google Scholar] [CrossRef] [Green Version]
- Visser, M.; Pahor, M.; Taaffe, D.R.; Goodpaster, B.H.; Simonsick, E.M.; Newman, A.B.; Nevitt, M.; Harris, T.B. Relationship of interleukin-6 and tumor necrosis factor-α with muscle mass and muscle strength in elderly men and women: The health ABC study. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2002, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molanouri Shamsi, M.; Hassan, Z.H.; Gharakhanlou, R.; Quinn, L.S.; Azadmanesh, K.; Baghersad, L.; Isanejad, A.; Mahdavi, M. Expression of interleukin-15 and inflammatory cytokines in skeletal muscles of STZ-induced diabetic rats: Effect of resistance exercise training. Endocrine 2014, 46, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Bruunsgaard, H.; Ladelund, S.; Pedersen, A.N.; Schroll, M.; Jørgensen, T.; Pedersen, B.K. Predicting death from tumour necrosis factor-alpha and interleukin-6 in 80-year-old people. Clin. Exp. Immunol. 2003, 132, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Payette, H.; Roubenoff, R.; Jacques, P.F.; Dinarello, C.A.; Wilson, P.W.F.; Abad, L.W.; Harris, T. Insulin-like growth factor-1 and interleukin 6 predict sarcopenia in very old community-living men and women: The Framingham heart study. J. Am. Geriatr. Soc. 2003, 51, 1237–1243. [Google Scholar] [CrossRef]
- Reuben, D.B.; Cheh, A.I.; Harris, T.B.; Ferrucci, L.; Rowe, J.W.; Tracy, R.P.; Seeman, T.E. Peripheral blood markers of inflammation predict mortality and functional decline in high-functioning community-dwelling older persons. J. Am. Geriatr. Soc. 2002, 50, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, G.; Liang, X.; Zhu, M.; Du, M. Grape seed extract prevents skeletal muscle wasting in interleukin 10 knockout mice. BMC Complement. Altern. Med. 2014, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argilés, J.M.; Busquets, S.; Stemmler, B.; López-Soriano, F.J. Cachexia and sarcopenia: Mechanisms and potential targets for intervention. Curr. Opin. Pharmacol. 2015, 22, 100–106. [Google Scholar] [CrossRef]
- Beyer, I.; Mets, T.; Bautmans, I. Chronic low-grade inflammation and age-related sarcopenia. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 12–22. [Google Scholar] [CrossRef]
- Scicchitano, B.M.; Faraldi, M.; Musarò, A. The Proteolytic Systems of Muscle Wasting. Recent Adv. DNA Gene Seq. 2015, 9, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Scicchitano, B.M.; Dobrowolny, G.; Sica, G.; Musarò, A. Molecular Insights into Muscle Homeostasis, Atrophy and Wasting. Curr. Genom. 2018, 19, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Thoma, A.; Lightfoot, A.P. Nf-kb and inflammatory cytokine signalling: Role in skeletal muscle atrophy. In Advances in Experimental Medicine and Biology; Springer New York LLC: New York, NY, USA, 2018; Volume 1088, pp. 267–279. [Google Scholar]
- Li, H.; Malhotra, S.; Kumar, A. Nuclear factor-kappa B signaling in skeletal muscle atrophy. J. Mol. Med. 2008, 86, 1113–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandri, M.; Barberi, L.; Bijlsma, A.Y.; Blaauw, B.; Dyar, K.A.; Milan, G.; Mammucari, C.; Meskers, C.G.M.; Pallafacchina, G.; Paoli, A.; et al. Signalling pathways regulating muscle mass in ageing skeletal muscle. The role of the IGF1-Akt-mTOR-FoxO pathway. Biogerontology 2013, 14, 303–323. [Google Scholar] [CrossRef] [PubMed]
- Small, E.M.; O’Rourke, J.R.; Moresi, V.; Sutherland, L.B.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. USA 2010, 107, 4218–4223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.F.; Callis, T.E.; Wang, D.Z. microRNAs and muscle disorders. J. Cell Sci. 2009, 122, 13–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, M.J.; McCarthy, J.J.; Fry, C.S.; Esser, K.A.; Rasmussen, B.B. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am. J. Physiol. Endocrinol. Metab. 2008, 295. [Google Scholar] [CrossRef] [Green Version]
- Panguluri, S.K.; Bhatnagar, S.; Kumar, A.; McCarthy, J.J.; Srivastava, A.K.; Cooper, N.G.; Lundy, R.F.; Kumar, A. Genomic profiling of messenger RNAs and microRNAs reveals potential mechanisms of TWEAK-induced skeletal muscle wasting in mice. PLoS ONE 2010, 5, e8760. [Google Scholar] [CrossRef] [Green Version]
- Georgantas, R.W.; Streicher, K.; Greenberg, S.A.; Greenlees, L.M.; Zhu, W.; Brohawn, P.Z.; Higgs, B.W.; Czapiga, M.; Morehouse, C.A.; Amato, A.; et al. Inhibition of myogenic microRNAs 1, 133, and 206 by inflammatory cytokines links inflammation and muscle degeneration in adult inflammatory myopathies. Arthritis Rheumatol. 2014, 66, 1022–1033. [Google Scholar] [CrossRef]
- van Rooij, E.; Liu, N.; Olson, E.N. MicroRNAs flex their muscles. Trends Genet. 2008, 24, 159–166. [Google Scholar] [CrossRef]
- Baltimore, D.; Boldin, M.P.; O’Connell, R.M.; Rao, D.S.; Taganov, K.D. MicroRNAs: New regulators of immune cell development and function. Nat. Immunol. 2008, 9, 839–845. [Google Scholar] [CrossRef]
- Nie, Y.; Sato, Y.; Wang, C.; Yue, F.; Kuang, S.; Gavin, T.P. Impaired exercise tolerance, mitochondrial biogenesis, and muscle fiber maintenance in miR-133a-deficient mice. FASEB J. 2016, 30, 3745–3758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercken, E.M.; Majounie, E.; Ding, J.; Guo, R.; Kim, J.; Bernier, M.; Mattison, J.; Cookson, M.R.; Gorospe, M.; de Cabo, R.; et al. Age-associated miRNA alterations in skeletal muscle from rhesus monkeys reversed by caloric restriction. Aging (Albany NY) 2013, 5, 692–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, W.; Li, Z.; Li, M.; Xu, N.; Zhang, Y. MiR-181a and inflammation: MiRNA homeostasis response to inflammatory stimuli in vivo. Biochem. Biophys. Res. Commun. 2013, 430, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Keller, U. Nutritional Laboratory Markers in Malnutrition. J. Clin. Med. 2019, 8, 775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moen, I.W.; Bergholdt, H.K.M.; Mandrup-Poulsen, T.; Nordestgaard, B.G.; Ellervik, C. Increased plasma ferritin concentration and low-grade inflammation—a mendelian randomization study. Clin. Chem. 2018, 64, 374–385. [Google Scholar] [CrossRef] [PubMed]
- Soeters, P.B.; Wolfe, R.R.; Shenkin, A. Hypoalbuminemia: Pathogenesis and Clinical Significance. J. Parenter. Enter. Nutr. 2019, 43, 181–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C.; Klionsky, D.J. Regulation Mechanisms and Signaling Pathways of Autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [Green Version]
- Cuervo, A.M.; Bergamini, E.; Brunk, U.T.; Dröge, W.; Ffrench, M.; Terman, A. Autophagy and aging: The importance of maintaining “clean” cells. Autophagy 2005, 1, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Maiuri, M.; Galluzzi, L.; Vicencio, J.; Kepp, O.; Tasdemir, E.; Kroemer, G. To Die or Not to Die: That is the Autophagic Question. Curr. Mol. Med. 2008, 8, 78–91. [Google Scholar] [CrossRef]
- Marzetti, E.; Wohlgemuth, S.E.; Lees, H.A.; Chung, H.Y.; Giovannini, S.; Leeuwenburgh, C. Age-related activation of mitochondrial caspase-independent apoptotic signaling in rat gastrocnemius muscle. Mech. Ageing Dev. 2008, 129, 542–549. [Google Scholar] [CrossRef] [Green Version]
- Terman, A.; Brunk, U.T. Oxidative stress, accumulation of biological “garbage”, and aging. Antioxidants Redox Signal. 2006, 8, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Petrovski, G.; Das, D.K. Does autophagy take a front seat in lifespan extension? J. Cell. Mol. Med. 2010, 14, 2543–2551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, J.; Kou, X.; Jia, S.; Yang, X.; Yang, Y.; Chen, N. Autophagy as a Potential Target for Sarcopenia. J. Cell. Physiol. 2016, 231, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Ro, S.H.; Cao, J.; Otto, N.M.; Kim, D.H. MTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, T.; Hall, M.N. The TOR signalling pathway controls nuclear localization of nutrient- regulated transcription factors. Nature 1999, 402, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Martina, J.A.; Chen, Y.; Gucek, M.; Puertollano, R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012, 8, 903–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peña-Llopis, S.; Vega-Rubin-De-Celis, S.; Schwartz, J.C.; Wolff, N.C.; Tran, T.A.T.; Zou, L.; Xie, X.J.; Corey, D.R.; Brugarolas, J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011, 30, 3242–3258. [Google Scholar] [CrossRef] [Green Version]
- Roczniak-Ferguson, A.; Petit, C.S.; Froehlich, F.; Qian, S.; Ky, J.; Angarola, B.; Walther, T.C.; Ferguson, S.M. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 2012, 5. [Google Scholar] [CrossRef] [Green Version]
- Settembre, C.; Zoncu, R.; Medina, D.L.; Vetrini, F.; Erdin, S.; Erdin, S.; Huynh, T.; Ferron, M.; Karsenty, G.; Vellard, M.C.; et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012, 31, 1095–1108. [Google Scholar] [CrossRef] [Green Version]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Arencibia, M.G.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science (80-) 2011, 332, 1429–1433. [Google Scholar] [CrossRef] [Green Version]
- Settembre, C.; Ballabio, A. TFEB regulates autophagy: An integrated coordination of cellular degradation and recycling processes. Autophagy 2011, 7, 1379–1381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R.; Montefusco, S.; Scotto-Rosato, A.; Prezioso, C.; Forrester, A.; et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapierre, L.R.; De Magalhaes Filho, C.D.; McQuary, P.R.; Chu, C.C.; Visvikis, O.; Chang, J.T.; Gelino, S.; Ong, B.; Davis, A.E.; Irazoqui, J.E.; et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Settembre, C.; De Cegli, R.; Mansueto, G.; Saha, P.K.; Vetrini, F.; Visvikis, O.; Huynh, T.; Carissimo, A.; Palmer, D.; Jürgen Klisch, T.; et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013, 15, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Mammucari, C.; Milan, G.; Romanello, V.; Masiero, E.; Rudolf, R.; Del Piccolo, P.; Burden, S.J.; Di Lisi, R.; Sandri, C.; Zhao, J.; et al. FoxO3 Controls Autophagy in Skeletal Muscle In Vivo. Cell Metab. 2007, 6, 458–471. [Google Scholar] [CrossRef]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef]
- Murphy, C.T.; McCarroll, S.A.; Bargmann, C.I.; Fraser, A.; Kamath, R.S.; Ahringer, J.; Li, H.; Kenyon, C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 2003, 424, 277–284. [Google Scholar] [CrossRef]
- Murphy, C.T. The search for DAF-16/FOXO transcriptional targets: Approaches and discoveries. Exp. Gerontol. 2006, 41, 910–921. [Google Scholar] [CrossRef]
- Brown, D.M.; Goljanek-Whysall, K. MicroRNAs: Modulators of the underlying pathophysiology of sarcopenia? Ageing Res. Rev. 2015, 24, 263–273. [Google Scholar] [CrossRef]
- Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13421–13426. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Li, Q.J.; Gong, Z.B.; Zhou, L.; You, N.; Wang, S.; Li, X.L.; Li, J.J.; An, J.Z.; Wang, D.S.; et al. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol. Cancer Res. Treat. 2014, 13, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, D.; He, Y.; Meléndez, A.; Feng, Z.; Hong, Q.; Bai, X.; Li, Q.; Cai, G.; Wang, J.; et al. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Omaha) 2013, 35, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Jiang, J.; Liu, W.; Wang, H.; Zhao, L.; Liu, S.; Li, P.; Zhang, S.; Sun, C.; Wu, Y.; et al. MicroRNA-378 promotes autophagy and inhibits apoptosis in skeletal muscle. Proc. Natl. Acad. Sci. USA 2018, 115, E10849–E10858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abramov, A.Y.; Canevari, L.; Duchen, M.R. Calcium signals induced by amyloid β peptide and their consequences in neurons and astrocytes in culture. Biochim. Biophys. Acta 2004, 1742, 81–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galluzzi, L.; Kepp, O.; Trojel-Hansen, C.; Kroemer, G. Mitochondrial control of cellular life, stress, and death. Circ. Res. 2012, 111, 1198–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fariss, M.W.; Chan, C.B.; Patel, M.; Van Houten, B.; Orrenius, S. Role of mitochondria in toxic oxidative stress. Mol. Interv. 2005, 5, 94–111. [Google Scholar] [CrossRef]
- Gemma, C.; Vila, J.; Bachstetter, A.; Bickford, P.C. Oxidative Stress and the Aging Brain: From Theory to Prevention. In Brain Aging; CRC Press: Boca Raton, FL, USA, 2019; pp. 353–374. ISBN 0849338182. [Google Scholar]
- Fanò, G.; Mecocci, P.; Vecchiet, J.; Belia, S.; Fulle, S.; Polidori, M.C.; Felzani, G.; Senin, U.; Vecchiet, L.; Beal, M.F. Age and sex influence on oxidative damage and functional status in human skeletal muscle. J. Muscle Res. Cell Motil. 2001, 22, 345–351. [Google Scholar] [CrossRef]
- Pansarasa, O.; Castagna, L.; Colombi, B.; Vecchiet, J.; Felzani, G.; Marzatico, F. Age and sex differences in human skeletal muscle: Role of reactive oxygen species. Free Radic. Res. 2000, 33, 287–293. [Google Scholar] [CrossRef]
- Damiano, S.; Muscariello, E.; La Rosa, G.; Di Maro, M.; Mondola, P.; Santillo, M. Dual role of reactive oxygen species in muscle function: Can antioxidant dietary supplements counteract age-related sarcopenia? Int. J. Mol. Sci. 2019, 20, 3815. [Google Scholar] [CrossRef] [Green Version]
- Pelosi, L.; Forcina, L.; Nicoletti, C.; Scicchitano, B.M.; Musarò, A. Increased Circulating Levels of Interleukin-6 Induce Perturbation in Redox-Regulated Signaling Cascades in Muscle of Dystrophic Mice. Oxid. Med. Cell. Longev. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Scicchitano, B.M.; Pelosi, L.; Sica, G.; Musarò, A. The physiopathologic role of oxidative stress in skeletal muscle. Mech. Ageing Dev. 2018, 170, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, V.; Cornelius, C.; Dinkova-Kostova, A.T.; Calabrese, E.J.; Mattson, M.P. Cellular stress responses, the hormesis paradigm, and vitagenes: Novel targets for therapeutic intervention in neurodegenerative disorders. Antioxid. Redox Signal. 2010, 13, 1763–1811. [Google Scholar] [CrossRef] [PubMed]
- Fougère, B.; van Kan, G.A.; Vellas, B.; Cesari, M. Redox Systems, Antioxidants and Sarcopenia. Curr. Protein Pept. Sci. 2018, 19, 643–648. [Google Scholar] [CrossRef]
- Leri, M.; Scuto, M.; Ontario, M.L.; Calabrese, V.; Calabrese, E.J.; Bucciantini, M.; Stefani, M. Healthy effects of plant polyphenols: Molecular mechanisms. Int. J. Mol. Sci. 2020, 21, 1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, A.; Wendt, S.; Benstoem, C.; Neubauer, C.; Meybohm, P.; Langlois, P.; Adhikari, N.K.J.; Heyland, D.K.; Stoppe, C. Vitamin C to improve organ dysfunction in cardiac surgery patients—Review and pragmatic approach. Nutrients 2018, 10, 974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerullo, F.; Gambassi, G.; Cesari, M. Rationale for antioxidant supplementation in sarcopenia. J. Aging Res. 2012, 2012, 316943. [Google Scholar] [CrossRef] [Green Version]
- Jakemanl, P.; Maxwell, S. Effect of antioxidant vitamin supplementation on muscle function after eccentric exercise. Eur. J. Appl. Physiol. Occup. Physiol. 1993, 67, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Shafat, A.; Butler, P.; Jensen, R.L.; Donnelly, A.E. Effects of dietary supplementation with vitamins C and E on muscle function during and after eccentric contractions in humans. Eur. J. Appl. Physiol. 2004, 93, 196–202. [Google Scholar] [CrossRef]
- Dobrowolny, G.; Aucello, M.; Rizzuto, E.; Beccafico, S.; Mammucari, C.; Boncompagni, S.; Belia, S.; Wannenes, F.; Nicoletti, C.; Del Prete, Z.; et al. Skeletal Muscle Is a Primary Target of SOD1G93A-Mediated Toxicity. Cell Metab. 2008, 8, 425–436. [Google Scholar] [CrossRef]
- Marzani, B.; Balage, M.; Vénien, A.; Astruc, T.; Papet, I.; Dardevet, D.; Mosoni, L. Antioxidant supplementation restores defective leucine stimulation of protein synthesis in skeletal muscle from old rats. J. Nutr. 2008, 138, 2205–2211. [Google Scholar] [CrossRef] [Green Version]
- Stefani, M.; Rigacci, S. Beneficial properties of natural phenols: Highlight on protection against pathological conditions associated with amyloid aggregation. BioFactors 2014, 40, 482–493. [Google Scholar] [CrossRef] [PubMed]
- Murase, T.; Haramizu, S.; Ota, N.; Hase, T. Suppression of the aging-associated decline in physical performance by a combination of resveratrol intake and habitual exercise in senescence-accelerated mice. Biogerontology 2009, 10, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Sahin, K.; Pala, R.; Tuzcu, M.; Ozdemir, O.; Orhan, C.; Sahin, N.; Juturu, V. Curcumin prevents muscle damage by regulating NF-κB and Nrf2 pathways and improves performance: An in vivo model. J. Inflamm. Res. 2016, 9, 147–154. [Google Scholar] [PubMed] [Green Version]
- Charles, A.-L.; Meyer, A.; Dal-Ros, S.; Auger, C.; Keller, N.; Ramamoorthy, T.G.; Zoll, J.; Metzger, D.; Schini-Kerth, V.; Geny, B. Polyphenols prevent ageing-related impairment in skeletal muscle mitochondrial function through decreased reactive oxygen species production. Exp. Physiol. 2013, 98, 536–545. [Google Scholar] [CrossRef]
- Teixeira, V.H.; Valente, H.F.; Casal, S.I.; Marques, A.F.; Moreira, P.A. Antioxidants do not prevent postexercise peroxidation and may delay muscle recovery. Med. Sci. Sports Exerc. 2009, 41, 1752–1760. [Google Scholar] [CrossRef]
- Peternelj, T.T.; Coombes, J.S. Antioxidant supplementation during exercise training: Beneficial or detrimental? Sport. Med. 2011, 41, 1043–1069. [Google Scholar] [CrossRef]
- Rowlands, D.S.; Pearce, E.; Aboud, A.; Gillen, J.B.; Gibala, M.J.; Donato, S.; Waddington, J.M.; Green, J.G.; Tarnopolsky, M.A. Oxidative stress, inflammation, and muscle soreness in an 894-km relay trail run. Eur. J. Appl. Physiol. 2012, 112, 1839–1848. [Google Scholar] [CrossRef]
- Gomez-Cabrera, M.C.; Domenech, E.; Romagnoli, M.; Arduini, A.; Borras, C.; Pallardo, F.V.; Sastre, J.; Viña, J. Oral administration of vitamin C decreases muscle mitochondrial biogenesis and hampers training-induced adaptations in endurance performance. Am. J. Clin. Nutr. 2008, 87, 142–149. [Google Scholar] [CrossRef] [Green Version]
- Croft, K.D. Dietary polyphenols: Antioxidants or not? Arch. Biochem. Biophys. 2016, 595, 120–124. [Google Scholar] [CrossRef] [Green Version]
- Surai, P.F. Polyphenol compounds in the chicken/animal diet: From the past to the future. J. Anim. Physiol. Anim. Nutr. (Berl.) 2014, 98, 19–31. [Google Scholar] [CrossRef]
- Calabrese, V.; Cornelius, C.; Dinkova-Kostova, A.T.; Calabrese, E.J. Vitagenes, cellular stress response, and acetylcarnitine: Relevance to hormesis. BioFactors 2009, 35, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, V.; Cornelius, C.; Mancuso, C.; Barone, E.; Calafato, S.; Bates, T.; Rizzarelli, E.; Kostova, A.T.D. Vitagenes, dietary antioxidants and neuroprotection in neurodegenerative diseases. Front. Biosci. 2009, 14, 376–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calabrese, V.; Cornelius, C.; Mancuso, C.; Pennisi, G.; Calafato, S.; Bellia, F.; Bates, T.E.; Giuffrida Stella, A.M.; Schapira, T.; Dinkova Kostova, A.T.; et al. Cellular stress response: A novel target for chemoprevention and nutritional neuroprotection in aging, neurodegenerative disorders and longevity. Neurochem. Res. 2008, 33, 2444–2471. [Google Scholar] [CrossRef]
- Calabrese, V.; Mancuso, C.; Calvani, M.; Rizzarelli, E.; Butterfield, D.A.; Giuffrida Stella, A.M. Nitric oxide in the central nervous system: Neuroprotection versus neurotoxicity. Nat. Rev. Neurosci. 2007, 8, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, V.; Cornelius, C.; Rizzarelli, E.; Owen, J.B.; Dinkova-Kostova, A.T.; Butterfield, D.A. Nitric Oxide in Cell Survival: A Janus Molecule. Antioxid. Redox Signal. 2009, 11, 2717–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surai, P.F. Silymarin as a natural antioxidant: An overview of the current evidence and perspectives. Antioxidants 2015, 4, 204–247. [Google Scholar] [CrossRef] [Green Version]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Kauppinen, A.; Suuronen, T.; Ojala, J.; Kaarniranta, K.; Salminen, A. Antagonistic crosstalk between NF-κB and SIRT1 in the regulation of inflammation and metabolic disorders. Cell. Signal. 2013, 25, 1939–1948. [Google Scholar] [CrossRef]
- Zhang, X.; Zuo, X.; Yang, B.; Li, Z.; Xue, Y.; Zhou, Y.; Huang, J.; Zhao, X.; Zhou, J.; Yan, Y.; et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 2014, 158, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Khatri, B.; Seo, D.; Shouse, S.; Pan, J.H.; Hudson, N.J.; Kim, J.K.; Bottje, W.; Kong, B.C. MicroRNA profiling associated with muscle growth in modern broilers compared to an unselected chicken breed. BMC Genom. 2018, 19. [Google Scholar] [CrossRef]
- Mei, T.; Liu, Y.; Wang, J.; Zhang, Y. miR-340-5p: A potential direct regulator of Nrf2 expression in the post-exercise skeletal muscle of mice. Mol. Med. Rep. 2019, 19, 1340–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, S.; Hagiwara, Y.; Tsuchiya, M.; Shinoda, M.; Koide, M.; Hatakeyama, H.; Chaweewannakorn, C.; Suzuki, K.; Yano, T.; Sogi, Y.; et al. Involvement of inflammasome activation via elevation of uric acid level in nociception in a mouse model of muscle pain. Mol. Pain 2019, 15. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, N.; Maurya, C.K.; Pandey, J.; Rai, A.K.; Tamrakar, A.K. Fructose-induced ROS generation impairs glucose utilization in L6 skeletal muscle cells. Free Radic. Res. 2015, 49, 1055–1068. [Google Scholar] [CrossRef] [PubMed]
- Lanaspa, M.A.; Tapia, E.; Soto, V.; Sautin, Y.; Sánchez-Lozada, L.G. Uric Acid and Fructose: Potential Biological Mechanisms. Semin. Nephrol. 2011, 31, 426–432. [Google Scholar] [CrossRef]
- Lanaspa, M.A.; Sanchez-Lozada, L.G.; Cicerchi, C.; Li, N.; Roncal-Jimenez, C.A.; Ishimoto, T.; Le, M.; Garcia, G.E.; Thomas, J.B.; Rivard, C.J.; et al. Uric Acid Stimulates Fructokinase and Accelerates Fructose Metabolism in the Development of Fatty Liver. PLoS ONE 2012, 7, e47948. [Google Scholar] [CrossRef] [Green Version]
- Hokayem, M.; Blond, E.; Vidal, H.; Lambert, K.; Meugnier, E.; Feillet-Coudray, C.; Coudray, C.; Pesenti, S.; Luyton, C.; Lambert-Porcheron, S.; et al. Grape polyphenols prevent fructose-induced oxidative stress and insulin resistance in first-degree relatives of type 2 diabetic patients. Diabetes Care 2013, 36, 1454–1461. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.C.; Bleich, S.N.; Gortmaker, S.L. Increasing caloric contribution from sugar- sweetened beverages and 100% fruit juices among US children and adolescents, 1988-2004. Pediatrics 2008, 121. [Google Scholar] [CrossRef]
- Hanover, L.M.; White, J.S. Manufacturing, composition, and applications of fructose. Am. J. Clin. Nutr. 1993, 58. [Google Scholar] [CrossRef]
- Sievenpiper, J.L.; Chiavaroli, L.; De Souza, R.J.; Mirrahimi, A.; Cozma, A.I.; Ha, V.; Wang, D.D.; Yu, M.E.; Carleton, A.J.; Beyene, J.; et al. “Catalytic” doses of fructose may benefit glycaemic control without harming cardiometabolic risk factors: A small meta-analysis of randomised controlled feeding trials. Br. J. Nutr. 2012, 108, 418–423. [Google Scholar] [CrossRef] [Green Version]
- Madero, M.; Rodríguez Castellanos, F.E.; Jalal, D.; Villalobos-Martín, M.; Salazar, J.; Vazquez-Rangel, A.; Johnson, R.J.; Sanchez-Lozada, L.G. A pilot study on the impact of a low fructose diet and allopurinol on clinic blood pressure among overweight and prehypertensive subjects: A randomized placebo controlled trial. J. Am. Soc. Hypertens. 2015, 9, 837–844. [Google Scholar] [CrossRef]
- Busserolles, J.; Rock, E.; Gueux, E.; Mazur, A.; Grolier, P.; Rayssiguier, Y. Short-term consumption of a high-sucrose diet has a pro-oxidant effect in rats. Br. J. Nutr. 2002, 87, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Coffee, E.M.; Yerkes, L.; Ewen, E.P.; Zee, T.; Tolan, D.R. Increased prevalence of mutant null alleles that cause hereditary fructose intolerance in the American population. J. Inherit. Metab. Dis. 2010, 33, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Delbosc, S.; Paizanis, E.; Magous, R.; Araiz, C.; Dimo, T.; Cristol, J.P.; Cros, G.; Azay, J. Involvement of oxidative stress and NADPH oxidase activation in the development of cardiovascular complications in a model of insulin resistance, the fructose-fed rat. Atherosclerosis 2005, 179, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Kunde, S.S.; Roede, J.R.; Vos, M.B.; Orr, M.L.; Go, Y.M.; Park, Y.; Ziegler, T.R.; Jones, D.P. Hepatic oxidative stress in fructose-induced fatty liver is not caused by sulfur amino acid insufficiency. Nutrients 2011, 3, 987–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellor, K.; Ritchie, R.H.; Meredith, G.; Woodman, O.L.; Morris, M.J.; Delbridge, L.M.D. High-fructose diet elevates myocardial superoxide generation in mice in the absence of cardiac hypertrophy. Nutrition 2010, 26, 842–848. [Google Scholar] [CrossRef]
- Nyby, M.D.; Abedi, K.; Smutko, V.; Eslami, P.; Tuck, M.L. Vascular angiotensin type 1 receptor expression is associated with vascular dysfunction, oxidative stress and inflammation in fructose-fed rats. Hypertens. Res. 2007, 30, 451–457. [Google Scholar] [CrossRef] [Green Version]
- Brosh, S.; Boer, P.; Sperling, O. Effects of fructose on synthesis and degradation of purine nucleotides in isolated rat hepatocytes. BBA Gen. Subj. 1982, 717, 459–464. [Google Scholar] [CrossRef]
- Johnson, R.J.; Nakagawa, T.; Sanchez-Lozada, L.G.; Shafiu, M.; Sundaram, S.; Le, M.; Ishimoto, T.; Sautin, Y.Y.; Lanaspa, M.A. Sugar, uric acid, and the etiology of diabetes and obesity. Diabetes 2013, 62, 3307–3315. [Google Scholar]
- Hecker, L.; Vittal, R.; Jones, T.; Jagirdar, R.; Luckhardt, T.R.; Horowitz, J.C.; Pennathur, S.; Martinez, F.J.; Thannickal, V.J. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat. Med. 2009, 15, 1077–1081. [Google Scholar] [CrossRef] [Green Version]
- Faeh, D.; Minehira, K.; Schwarz, J.M.; Periasami, R.; Seongsu, P.; Tappy, L. Effect of fructose overfeeding and fish oil administration on hepatic de novo lipogenesis and insulin sensitivity in healthy men. Diabetes 2005, 54, 1907–1913. [Google Scholar] [CrossRef] [Green Version]
- Lam, Y.Y.; Ha, C.W.Y.; Campbell, C.R.; Mitchell, A.J.; Dinudom, A.; Oscarsson, J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS ONE 2012, 7, e34233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDevitt, R.M.; Bott, S.J.; Harding, M.; Coward, W.A.; Bluck, L.J.; Prentice, A.M. De novo lipogenesis during controlled overfeeding with sucrose or glucose in lean and obese women. Am. J. Clin. Nutr. 2001, 74, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Samuel, V.T. Fructose induced lipogenesis: From sugar to fat to insulin resistance. Trends Endocrinol. Metab. 2011, 22, 60–65. [Google Scholar] [PubMed]
- Choi, H.K.; Atkinson, K.; Karlson, E.W.; Willett, W.; Curhan, G. Purine-Rich Foods, Daily and Protein Intake, and the Risk of Gout in Men. N. Engl. J. Med. 2004, 350, 1093–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.W.J.; Ford, E.S.; Gao, X.; Choi, H.K. Sugar-sweetened soft drinks, diet soft drinks, and serum uric acid level: The Third National Health and Nutrition Examination Survey. Arthritis Care Res. 2008, 59, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Jegatheesan, P.; De Bandt, J.P. Fructose and NAFLD: The multifaceted aspects of fructose metabolism. Nutrients 2017, 9, 230. [Google Scholar] [CrossRef] [Green Version]
- Rippe, J.M.; Angelopoulos, T.J. Sucrose, high-fructose corn syrup, and fructose, their metabolism and potential health effects: What do we really know? Adv. Nutr. 2013, 4, 236–245. [Google Scholar] [CrossRef]
- Lê, K.A. Consommation de fructose: Facteur déclenchant du syndrome métabolique? Obesite 2008, 3, 276–279. [Google Scholar] [CrossRef]
- Fernández-Novell, J.M.; Ramió-Lluch, L.; Orozco, A.; Gómez-Foix, A.M.; Guinovart, J.J.; Rodríguez-Gil, J.E. Glucose and fructose have sugar-specific effects in both liver and skeletal muscle in vivo: A role for liver fructokinase. PLoS ONE 2014, 9, e109726. [Google Scholar] [CrossRef]
- Jegatheesan, P.; Beutheu, S.; Freese, K.; Waligora-Dupriet, A.J.; Nubret, E.; Butel, M.J.; Bergheim, I.; De Bandt, J.P. Preventive effects of citrulline on Western diet-induced non-alcoholic fatty liver disease in rats. Br. J. Nutr. 2016, 116, 191–203. [Google Scholar] [CrossRef] [Green Version]
- Cabrera, D.; Ruiz, A.; Cabello-Verrugio, C.; Brandan, E.; Estrada, L.; Pizarro, M.; Solis, N.; Torres, J.; Barrera, F.; Arrese, M. Diet-Induced Nonalcoholic Fatty Liver Disease Is Associated with Sarcopenia and Decreased Serum Insulin-Like Growth Factor-1. Dig. Dis. Sci. 2016, 61, 3190–3198. [Google Scholar] [CrossRef] [PubMed]
- Caliceti, C.; Calabria, D.; Roda, A.; Cicero, A.F.G. Fructose intake, serum uric acid, and cardiometabolic disorders: A critical review. Nutrients 2017, 9, 395. [Google Scholar] [CrossRef] [PubMed]
- Hallfrisch, J. Metabolic effects of dietary fructose. FASEB J. 1990, 4, 2652–2660. [Google Scholar] [CrossRef] [PubMed]
- Emmerson, B.T. Effect of oral fructose on urate production. Ann. Rheum. Dis. 1974, 33, 276–280. [Google Scholar] [CrossRef] [Green Version]
- Perez-Ruiz, F.; Calabozo, M.; García Erauskin, G.; Ruibal, A.; Herrero-Beites, A.M. Renal underexcretion of uric acid is present in patients with apparent high urinary uric acid output. Arthritis Care Res. 2002, 47, 610–613. [Google Scholar] [CrossRef]
- Kaneko, C.; Ogura, J.; Sasaki, S.; Okamoto, K.; Kobayashi, M.; Kuwayama, K.; Narumi, K.; Iseki, K. Fructose suppresses uric acid excretion to the intestinal lumen as a result of the induction of oxidative stress by NADPH oxidase activation. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 559–566. [Google Scholar] [CrossRef] [Green Version]
- Madlala, H.P.; Van Heerden, F.R.; Mubagwa, K.; Musabayane, C.T. Changes in renal function and oxidative status associated with the hypotensive effects of oleanolic acid and related synthetic derivatives in experimental animals. PLoS ONE 2015, 10, e0128192. [Google Scholar] [CrossRef] [Green Version]
- Lambertucci, R.H.; Hirabara, S.M.; Silveira, L.D.R.; Levada-Pires, A.C.; Curi, R.; Pithon-Curi, T.C. Palmitate increases superoxide production through mitochondrial electron transport chain and NADPH oxidase activity in skeletal muscle cells. J. Cell. Physiol. 2008, 216, 796–804. [Google Scholar] [CrossRef]
- Duval, C.; Müller, M.; Kersten, S. PPARα and dyslipidemia. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 961–971. [Google Scholar] [CrossRef]
- Carvalho-Filho, M.A.; Ueno, M.; Hirabara, S.M.; Seabra, A.B.; Carvalheira, J.B.C.; De Oliveira, M.G.; Velloso, L.A.; Curi, R.; Saad, M.J.A. S-nitrosation of the insulin receptor, insulin receptor substrate 1, and protein kinase B/Akt: A novel mechanism of insulin resistance. Diabetes 2005, 54, 959–967. [Google Scholar] [CrossRef] [Green Version]
- Kahn, B.B.; Flier, J.S. Obesity and insulin resistance. J. Clin. Investig. 2000, 106, 473–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabio, G.; Das, M.; Mora, A.; Zhang, Z.; Jun, J.Y.; Hwi, J.K.; Barrett, T.; Kim, J.K.; Davis, R.J. A stress signaling pathway in adipose tissue regulates hepatic insulin resistance. Science (80-) 2008, 322, 1539–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schenk, S.; Saberi, M.; Olefsky, J.M. Insulin sensitivity: Modulation by nutrients and inflammation. J. Clin. Investig. 2008, 118, 2992–3002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landrier, J.F.; Derghal, A.; Mounien, L. MicroRNAs in Obesity and Related Metabolic Disorders. Cells 2019, 8, 859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintanilha, B.J.; Reis, B.Z.; Silva Duarte, G.B.; Cozzolino, S.M.F.; Rogero, M.M. Nutrimiromics: Role of micrornas and nutrition in modulating inflammation and chronic diseases. Nutrients 2017, 9, 1168. [Google Scholar] [CrossRef] [PubMed]
- Sud, N.; Zhang, H.; Pan, K.; Cheng, X.; Cui, J.; Su, Q. Aberrant expression of microRNA induced by high-fructose diet: Implications in the pathogenesis of hyperlipidemia and hepatic insulin resistance. J. Nutr. Biochem. 2017, 43, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Zhan, S.; Wang, Y.; Wang, L.; Zhong, T.; Li, L.; Fan, J.; Xiong, C.; Wang, Y.; Zhang, H. Role of microRNA-101a in the regulation of goat skeletal muscle satellite cell proliferation and differentiation. Gene 2015, 572, 198–204. [Google Scholar] [CrossRef]
- Guess, M.G.; Barthel, K.K.B.; Harrison, B.C.; Leinwand, L.A. miR-30 family microRNAs regulate myogenic differentiation and provide negative feedback on the microRNA pathway. PLoS ONE 2015, 10, e0118229. [Google Scholar] [CrossRef]
- Taniguchi, C.M.; Ueki, K.; Kahn, C.R. Erratum: Complementary roles of IRS-1 and IRS-2 in the hepatic regulation of metabolism. J. Clin. Investig. 2005, 115, 1388. [Google Scholar]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. MicroRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1. [Google Scholar] [CrossRef] [Green Version]
- Gautam, A.; Kumar, R.; Dimitrov, G.; Hoke, A.; Hammamieh, R.; Jett, M. Identification of extracellular miRNA in archived serum samples by next-generation sequencing from RNA extracted using multiple methods. Mol. Biol. Rep. 2016, 43, 1165–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [Green Version]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef]
- Zernecke, A.; Bidzhekov, K.; Noels, H.; Shagdarsuren, E.; Gan, L.; Denecke, B.; Hristov, M.; Köppel, T.; Jahantigh, M.N.; Lutgens, E.; et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2009, 2. [Google Scholar] [CrossRef]
- Ai, J.; Zhang, R.; Li, Y.; Pu, J.; Lu, Y.; Jiao, J.; Li, K.; Yu, B.; Li, Z.; Wang, R.; et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem. Biophys. Res. Commun. 2010, 391, 73–77. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Fitzpatrick, M.; Wood, W.H.; De, S.; Ejiogu, N.; Zhang, Y.; Mattison, J.A.; Becker, K.G.; Zonderman, A.B.; Evans, M.K. Age-related changes in microRNA levels in serum. Aging (Albany NY) 2013, 5, 725–740. [Google Scholar] [CrossRef] [Green Version]
- Olivieri, F.; Spazzafumo, L.; Santini, G.; Lazzarini, R.; Albertini, M.C.; Rippo, M.R.; Galeazzi, R.; Abbatecola, A.M.; Marcheselli, F.; Monti, D.; et al. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mech. Ageing Dev. 2012, 133, 675–685. [Google Scholar] [CrossRef]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma MicroRNA profiling reveals loss of endothelial MiR-126 and other MicroRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, D.; Chen, X.; Li, J.; Li, L.; Bian, Z.; Sun, F.; Lu, J.; Yin, Y.; Cai, X.; et al. Secreted Monocytic miR-150 Enhances Targeted Endothelial Cell Migration. Mol. Cell 2010, 39, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.Z.Y.; Ong, K.L.; Seeher, K.; Armstrong, N.J.; Thalamuthu, A.; Brodaty, H.; Sachdev, P.; Mather, K. Circulating microRNAs as biomarkers of alzheimer’s disease: A systematic review. J. Alzheimer’s Dis. 2015, 49, 755–766. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Zhang, Y.L.; Zhang, Y.; Feng, B.; Zheng, Z.; Wang, D.; Zhang, S.; Guo, Q.; Ye, H. Circulating MicroRNAs in Plasma Decrease in Response to Sarcopenia in the Elderly. Front. Genet. 2020, 11, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margolis, L.M.; Rivas, D.A.; Pasiakos, S.M.; McClung, J.P.; Ceglia, L.; Fielding, R.A. Upregulation of circulating myomiR following short-term energy restriction is inversely associated with whole body protein synthesis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 313, R298–R304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cong, L.; Lukiw, W.J. Plant and Animal microRNAs (miRNAs) and Their Potential for Inter-kingdom Communication. Cell. Mol. Neurobiol. 2018, 38, 133–140. [Google Scholar] [CrossRef]
- Witwer, K.W. XenomiRs and miRNA homeostasis in health and disease: Evidence that diet and dietary miRNAs directly and indirectly influence circulating miRNA profiles. RNA Biol. 2012, 9, 1147–1154. [Google Scholar] [CrossRef]
- Fan, Y.; Habib, M.; Xia, J. Xeno-mirNet: A comprehensive database and analytics platform to explore xeno-miRNAs and their potential targets. PeerJ 2018, 2018. [Google Scholar] [CrossRef]
- Wolf, T.; Baier, S.R.; Zempleni, J. The Intestinal Transport of Bovine Milk Exosomes Is Mediated by Endocytosis in Human Colon Carcinoma Caco-2 Cells and Rat Small Intestinal IEC-6 Cells1-3. J. Nutr. 2015, 145, 2201–2206. [Google Scholar] [CrossRef] [Green Version]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granjon, A.; Gustin, M.P.; Rieusset, J.; Lefai, E.; Meugnier, E.; Güller, I.; Cerutti, C.; Paultre, C.; Disse, E.; Rabasa-Lhoret, R.; et al. The microRNA signature in response to insulin reveals its implication in the transcriptional action of insulin in human skeletal muscle and the role of a sterol regulatory element-binding protein-1c/myocyte enhancer factor 2C pathway. Diabetes 2009, 58, 2555–2564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaminski, J.; Lançon, A.; Aires, V.; Limagne, E.; Tili, E.; Michaille, J.J.; Latruffe, N. Resveratrol initiates differentiation of mouse skeletal muscle-derived C2C12 myoblasts. Biochem. Pharmacol. 2012, 84, 1251–1259. [Google Scholar] [CrossRef] [PubMed]
- Lançon, A.; Kaminski, J.; Tili, E.; Michaille, J.J.; Latruffe, N. Control of microRNA expression as a new way for resveratrol to deliver its beneficial effects. Proc. J. Agric. Food Chem. Am. Chem. Soc. 2012, 60, 8783–8789. [Google Scholar]
- Yang, W.M.; Jeong, H.J.; Park, S.Y.; Lee, W. Induction of miR-29a by saturated fatty acids impairs insulin signaling and glucose uptake through translational repression of IRS-1 in myocytes. FEBS Lett. 2014, 588, 2170–2176. [Google Scholar] [CrossRef] [Green Version]
- Seldeen, K.L.; Pang, M.; Leiker, M.M.; Bard, J.E.; Rodríguez-Gonzalez, M.; Hernandez, M.; Sheridan, Z.; Nowak, N.; Troen, B.R. Chronic vitamin D insufficiency impairs physical performance in C57BL/6J mice. Aging (Albany NY) 2018, 10, 1338–1355. [Google Scholar] [CrossRef]



| Nutraceuticals | Final Effect | References | ||
|---|---|---|---|---|
| EAAs | miR-1, miR-23a, miR-208b, miR-499, miR-27a | Skeletal muscle regeneration, proliferation and differentiation | [52,55] | |
| Serum iron | miR-133b | Skeletal muscle differentiation | [58] | |
| Albumin | miR-133b, miR-206 | Skeletal muscle regeneration and differentiation | [58] | |
| Ferritin | miR-133b, miR-206 | Downregulation of skeletal muscle regeneration and differentiation | [58] | |
| Insulin | miR-1, miR-133a, miR-206, miR-29a, miR-29c | Downregulation of skeletal muscle regeneration, differentiation and insulin resistance | [225] | |
| Resveratrol | miR-21, miR-27b | miR-133b, miR-20b, miR-149 | Modulation of skeletal muscle differentiation | [198,226,227] |
| Palmitic Acid | miR-29a | Insulin resistance and diabetes | [228] | |
| Vitamin D | miR-26a | Skeletal muscle regeneration and differentiation | [229] | |
| Fructose | miR-101, miR-30a | miR-128a | Skeletal muscle proliferation and differentiation, insulin signaling, insulin resistance | [199] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbiera, A.; Pelosi, L.; Sica, G.; Scicchitano, B.M. Nutrition and microRNAs: Novel Insights to Fight Sarcopenia. Antioxidants 2020, 9, 951. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox9100951
Barbiera A, Pelosi L, Sica G, Scicchitano BM. Nutrition and microRNAs: Novel Insights to Fight Sarcopenia. Antioxidants. 2020; 9(10):951. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox9100951
Chicago/Turabian StyleBarbiera, Alessandra, Laura Pelosi, Gigliola Sica, and Bianca Maria Scicchitano. 2020. "Nutrition and microRNAs: Novel Insights to Fight Sarcopenia" Antioxidants 9, no. 10: 951. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox9100951