Biology, Pathophysiological Role, and Clinical Implications of Exosomes: A Critical Appraisal
Abstract
:1. Introduction
2. Isolation and Characterization
3. Exosome Biogenesis
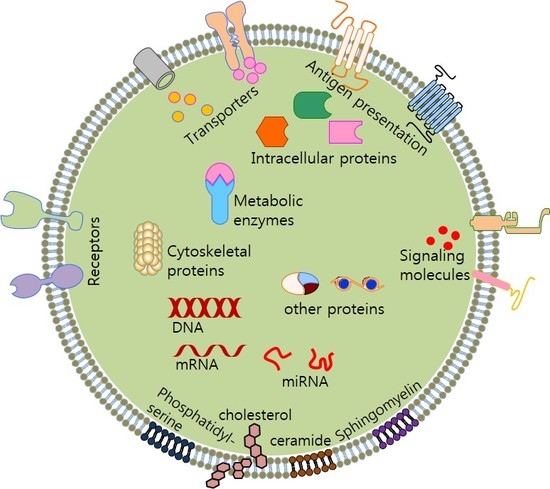
4. Exosome Composition
5. Exosome Secretion
6. Exosome Function
7. Prospective Applications
7.1. Exosomes in Diagnostics
7.2. Exosomes in Therapeutics
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [PubMed]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobrie, A.; Colombo, M.; Raposo, G.; Thery, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Naseri, Z.; Oskuee, R.K.; Jaafari, M.R.; Forouzandeh Moghadam, M. Exosome-mediated delivery of functionally active mirna-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. Int. J. Nanomed. 2018, 13, 7727–7747. [Google Scholar] [CrossRef]
- Fitzgerald, W.; Freeman, M.L.; Lederman, M.M.; Vasilieva, E.; Romero, R.; Margolis, L. A system of cytokines encapsulated in extracellular vesicles. Sci. Rep. 2018, 8, 8973. [Google Scholar] [CrossRef]
- Charoenviriyakul, C.; Takahashi, Y.; Morishita, M.; Nishikawa, M.; Takakura, Y. Role of extracellular vesicle surface proteins in the pharmacokinetics of extracellular vesicles. Mol. Pharm. 2018, 15, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Jesus, S.; Soares, E.; Cruz, M.T.; Borges, O. Exosomes as adjuvants for the recombinant hepatitis b antigen: First report. Eur. J. Pharm. Biopharm. 2018, 133, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Messenger, S.W.; Woo, S.S.; Sun, Z.; Martin, T.F.J. A ca(2+)-stimulated exosome release pathway in cancer cells is regulated by munc13-4. J. Cell Biol. 2018, 217, 2877–2890. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (ev): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef]
- Lamparski, H.G.; Metha-Damani, A.; Yao, J.Y.; Patel, S.; Hsu, D.H.; Ruegg, C.; Le Pecq, J.B. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods 2002, 270, 211–226. [Google Scholar] [CrossRef]
- Koga, K.; Matsumoto, K.; Akiyoshi, T.; Kubo, M.; Yamanaka, N.; Tasaki, A.; Nakashima, H.; Nakamura, M.; Kuroki, S.; Tanaka, M.; et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005, 25, 3703–3707. [Google Scholar]
- Clayton, A.; Court, J.; Navabi, H.; Adams, M.; Mason, M.D.; Hobot, J.A.; Newman, G.R.; Jasani, B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods 2001, 247, 163–174. [Google Scholar] [CrossRef]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.P.; Hole, P.; Carr, B.; Redman, C.W.G.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 780–788. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Varela, M.; de Menezes-Neto, A.; Perez-Zsolt, D.; Gamez-Valero, A.; Segui-Barber, J.; Izquierdo-Useros, N.; Martinez-Picado, J.; Fernandez-Becerra, C.; Del Portillo, H.A. Proteomics study of human cord blood reticulocyte-derived exosomes. Sci. Rep. 2018, 8, 14046. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Vidal, M.J.; Stahl, P.D. The small gtp-binding proteins rab4 and arf are associated with released exosomes during reticulocyte maturation. Eur. J. Cell Biol. 1993, 60, 261–267. [Google Scholar]
- Thery, C.; Boussac, M.; Veron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell-derived exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Savina, A.; Fader, C.M.; Damiani, M.T.; Colombo, M.I. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 2005, 6, 131–143. [Google Scholar] [CrossRef]
- Savina, A.; Vidal, M.; Colombo, M.I. The exosome pathway in k562 cells is regulated by rab11. J. Cell Sci. 2002, 115, 2505–2515. [Google Scholar]
- Hsu, C.; Morohashi, Y.; Yoshimura, S.; Manrique-Hoyos, N.; Jung, S.; Lauterbach, M.A.; Bakhti, M.; Gronborg, M.; Mobius, W.; Rhee, J.; et al. Regulation of exosome secretion by rab35 and its gtpase-activating proteins tbc1d10a-c. J. Cell Biol. 2010, 189, 223–232. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-alix regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Hosseini, H.M.; Fooladi, A.A.; Nourani, M.R.; Ghanezadeh, F. The role of exosomes in infectious diseases. Inflamm. Allergy Drug Targets 2013, 12, 29–37. [Google Scholar] [CrossRef]
- Bang, C.; Thum, T. Exosomes: New players in cell-cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064. [Google Scholar] [CrossRef] [PubMed]
- Babst, M.; Katzmann, D.J.; Estepa-Sabal, E.J.; Meerloo, T.; Emr, S.D. Escrt-iii: An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 2002, 3, 271–282. [Google Scholar] [CrossRef]
- McCullough, J.; Fisher, R.D.; Whitby, F.G.; Sundquist, W.I.; Hill, C.P. Alix-chmp4 interactions in the human escrt pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 7687–7691. [Google Scholar] [CrossRef] [PubMed]
- Peters, P.J.; Geuze, H.J.; Van der Donk, H.A.; Slot, J.W.; Griffith, J.M.; Stam, N.J.; Clevers, H.C.; Borst, J. Molecules relevant for t cell-target cell interaction are present in cytolytic granules of human t lymphocytes. Eur. J. Immunol. 1989, 19, 1469–1475. [Google Scholar] [CrossRef]
- Raposo, G.; Tenza, D.; Mecheri, S.; Peronet, R.; Bonnerot, C.; Desaymard, C. Accumulation of major histocompatibility complex class ii molecules in mast cell secretory granules and their release upon degranulation. Mol. Biol. Cell 1997, 8, 2631–2645. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, H.F.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 1999, 94, 3791–3799. [Google Scholar] [PubMed]
- Cosme, J.; Liu, P.P.; Gramolini, A.O. The cardiovascular exosome: Current perspectives and potential. Proteom. 2013, 13, 1654–1659. [Google Scholar] [CrossRef]
- Schorey, J.S.; Bhatnagar, S. Exosome function: From tumor immunology to pathogen biology. Traffic 2008, 9, 871–881. [Google Scholar] [CrossRef]
- Fader, C.M.; Sanchez, D.G.; Mestre, M.B.; Colombo, M.I. Ti-vamp/vamp7 and vamp3/cellubrevin: Two v-snare proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta 2009, 1793, 1901–1916. [Google Scholar] [CrossRef]
- Gross, J.C.; Chaudhary, V.; Bartscherer, K.; Boutros, M. Active wnt proteins are secreted on exosomes. Nat. Cell Biol. 2012, 14, 1036–1045. [Google Scholar] [CrossRef]
- Marshansky, V.; Futai, M. The v-type h+-atpase in vesicular trafficking: Targeting, regulation and function. Curr. Opin. Cell Biol. 2008, 20, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Gong, A.Y.; Roth, A.L.; Huang, B.Q.; Ward, H.D.; Zhu, G.; Larusso, N.F.; Hanson, N.D.; Chen, X.M. Release of luminal exosomes contributes to tlr4-mediated epithelial antimicrobial defense. PLoS Pathog. 2013, 9, e1003261. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Schorey, J.S. Exosomes released from infected macrophages contain mycobacterium avium glycopeptidolipids and are proinflammatory. J. Biol. Chem. 2007, 282, 25779–25789. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ji, Q.; Yang, Y.; Li, Q.; Wang, Z. Exosome: Function and role in cancer metastasis and drug resistance. Technol. Cancer Res. Treat. 2018, 17, 1533033818763450. [Google Scholar] [CrossRef] [PubMed]
- Azimi, M.; Ghabaee, M.; Moghadasi, A.N.; Noorbakhsh, F.; Izad, M. Immunomodulatory function of treg-derived exosomes is impaired in patients with relapsing-remitting multiple sclerosis. Immunol. Res. 2018, 66, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Kalluri, R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 2013, 91, 431–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.G.; Grizzle, W.E. Exosomes and cancer: A newly described pathway of immune suppression. Clin. Cancer Res. 2011, 17, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Luo, J.D.; Jiang, H.; Duan, D.D. Tumor exosomes: A double-edged sword in cancer therapy. Acta Pharmacol. Sin. 2018, 39, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Gomari, H.; Forouzandeh Moghadam, M.; Soleimani, M. Targeted cancer therapy using engineered exosome as a natural drug delivery vehicle. Onco Targets Ther. 2018, 11, 5753–5762. [Google Scholar] [CrossRef] [PubMed]
- Muntasell, A.; Berger, A.C.; Roche, P.A. T cell-induced secretion of mhc class ii-peptide complexes on b cell exosomes. EMBO J. 2007, 26, 4263–4272. [Google Scholar] [CrossRef] [PubMed]
- Wahlund, C.J.E.; Gucluler, G.; Hiltbrunner, S.; Veerman, R.E.; Naslund, T.I.; Gabrielsson, S. Exosomes from antigen-pulsed dendritic cells induce stronger antigen-specific immune responses than microvesicles in vivo. Sci. Rep. 2017, 7, 17095. [Google Scholar] [CrossRef] [Green Version]
- Yin, W.; Ouyang, S.; Li, Y.; Xiao, B.; Yang, H. Immature dendritic cell-derived exosomes: A promise subcellular vaccine for autoimmunity. Inflammation 2013, 36, 232–240. [Google Scholar] [CrossRef]
- Van Niel, G.; Raposo, G.; Candalh, C.; Boussac, M.; Hershberg, R.; Cerf-Bensussan, N.; Heyman, M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 2001, 121, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Lachenal, G.; Pernet-Gallay, K.; Chivet, M.; Hemming, F.J.; Belly, A.; Bodon, G.; Blot, B.; Haase, G.; Goldberg, Y.; Sadoul, R. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell Neurosci. 2011, 46, 409–418. [Google Scholar] [CrossRef] [Green Version]
- Fevrier, B.; Vilette, D.; Archer, F.; Loew, D.; Faigle, W.; Vidal, M.; Laude, H.; Raposo, G. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. USA 2004, 101, 9683–9688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, B.A.; Abdelaziz, D.H.; Schatzl, H.M. Autophagy regulates exosomal release of prions in neuronal cells. J. Biol. Chem. 2018, 293, 8956–8968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Embade, N.; Gil, D.; Matthiesen, R.; Valle, M.; Elortza, F.; Lu, S.C.; Mato, J.M.; Falcon-Perez, J.M. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 2008, 7, 5157–5166. [Google Scholar] [CrossRef]
- Fonsato, V.; Collino, F.; Herrera, M.B.; Cavallari, C.; Deregibus, M.C.; Cisterna, B.; Bruno, S.; Romagnoli, R.; Salizzoni, M.; Tetta, C.; et al. Human liver stem cell-derived microvesicles inhibit hepatoma growth in scid mice by delivering antitumor micrornas. Stem Cells 2012, 30, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Masyuk, A.I.; Huang, B.Q.; Ward, C.J.; Gradilone, S.A.; Banales, J.M.; Masyuk, T.V.; Radtke, B.; Splinter, P.L.; LaRusso, N.F. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G990–G999. [Google Scholar] [CrossRef] [Green Version]
- Deng, F.; Magee, N.; Zhang, Y. Decoding the role of extracellular vesicles in liver diseases. Liver Res. 2017, 1, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Meng, F.; Venter, J.; Giang, T.; Glaser, S.; Alpini, G. Author correction: The role of the secretin/secretin receptor axis in inflammatory cholangiocyte communication via extracellular vesicles. Sci. Rep. 2018, 8, 11238. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Meng, F.; Venter, J.; Giang, T.; Glaser, S.; Alpini, G. The role of the secretin/secretin receptor axis in inflammatory cholangiocyte communication via extracellular vesicles. Sci. Rep. 2017, 7, 11183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Sinha, N.; Gerth, K.A.; Rahman, M.A.; Yallapu, M.M.; Midde, N.M. Specific packaging and circulation of cytochromes p450, especially 2e1 isozyme, in human plasma exosomes and their implications in cellular communications. Biochem. Biophys. Res. Commun. 2017, 491, 675–680. [Google Scholar] [CrossRef]
- Yang, S.J.; Wang, D.D.; Li, J.; Xu, H.Z.; Shen, H.Y.; Chen, X.; Zhou, S.Y.; Zhong, S.L.; Zhao, J.H.; Tang, J.H. Predictive role of gstp1-containing exosomes in chemotherapy-resistant breast cancer. Gene 2017, 623, 5–14. [Google Scholar] [CrossRef]
- Holmgren, L. Horizontal gene transfer: You are what you eat. Biochem. Biophys. Res. Commun. 2010, 396, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Garnier, D.; Magnus, N.; Lee, T.H.; Bentley, V.; Meehan, B.; Milsom, C.; Montermini, L.; Kislinger, T.; Rak, J. Cancer cells induced to express mesenchymal phenotype release exosome-like extracellular vesicles carrying tissue factor. J. Biol. Chem. 2012, 287, 43565–43572. [Google Scholar] [CrossRef]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor egfrviii by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Luhtala, N.; Aslanian, A.; Yates, J.R., 3rd; Hunter, T. Secreted glioblastoma nanovesicles contain intracellular signaling proteins and active ras incorporated in a farnesylation-dependent manner. J. Biol. Chem. 2017, 292, 611–628. [Google Scholar] [CrossRef] [PubMed]
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes mediate stromal mobilization of autocrine wnt-pcp signaling in breast cancer cell migration. Cell 2012, 151, 1542–1556. [Google Scholar] [CrossRef]
- Johnstone, R.M. The jeanne manery-fisher memorial lecture 1991. Maturation of reticulocytes: Formation of exosomes as a mechanism for shedding membrane proteins. Biochem. Cell Biol. 1992, 70, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell Proteom. 2010, 9, 1085–1099. [Google Scholar] [CrossRef]
- Kharaziha, P.; Chioureas, D.; Rutishauser, D.; Baltatzis, G.; Lennartsson, L.; Fonseca, P.; Azimi, A.; Hultenby, K.; Zubarev, R.; Ullen, A.; et al. Molecular profiling of prostate cancer derived exosomes may reveal a predictive signature for response to docetaxel. Oncotarget 2015, 6, 21740–21754. [Google Scholar] [CrossRef] [Green Version]
- Phuyal, S.; Skotland, T.; Hessvik, N.P.; Simolin, H.; Overbye, A.; Brech, A.; Parton, R.G.; Ekroos, K.; Sandvig, K.; Llorente, A. The ether lipid precursor hexadecylglycerol stimulates the release and changes the composition of exosomes derived from pc-3 cells. J. Biol. Chem. 2015, 290, 4225–4237. [Google Scholar] [CrossRef]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through met. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Faure, J.; Lachenal, G.; Court, M.; Hirrlinger, J.; Chatellard-Causse, C.; Blot, B.; Grange, J.; Schoehn, G.; Goldberg, Y.; Boyer, V.; et al. Exosomes are released by cultured cortical neurones. Mol. Cell Neurosci. 2006, 31, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Carayon, K.; Chaoui, K.; Ronzier, E.; Lazar, I.; Bertrand-Michel, J.; Roques, V.; Balor, S.; Terce, F.; Lopez, A.; Salome, L.; et al. Proteolipidic composition of exosomes changes during reticulocyte maturation. J. Biol. Chem. 2011, 286, 34426–34439. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Cossetti, C.; Iraci, N.; Mercer, T.R.; Leonardi, T.; Alpi, E.; Drago, D.; Alfaro-Cervello, C.; Saini, H.K.; Davis, M.P.; Schaeffer, J.; et al. Extracellular vesicles from neural stem cells transfer ifn-gamma via ifngr1 to activate stat1 signaling in target cells. Mol. Cell 2014, 56, 193–204. [Google Scholar] [CrossRef]
- Hassani, K.; Olivier, M. Immunomodulatory impact of leishmania-induced macrophage exosomes: A comparative proteomic and functional analysis. PLoS Negl. Trop. Dis. 2013, 7, e2185. [Google Scholar] [CrossRef]
- Lee, J.E.; Moon, P.G.; Lee, I.K.; Baek, M.C. Proteomic analysis of extracellular vesicles released by adipocytes of otsuka long-evans tokushima fatty (oletf) rats. Protein J. 2015, 34, 220–235. [Google Scholar] [CrossRef]
- Graner, M.W.; Alzate, O.; Dechkovskaia, A.M.; Keene, J.D.; Sampson, J.H.; Mitchell, D.A.; Bigner, D.D. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009, 23, 1541–1557. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, T.A.; Sacco, R.E.; Nonnecke, B.J.; Lippolis, J.D. Bovine milk proteome: Quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from staphylococcus aureus mastitis. J. Proteom. 2013, 82, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Buschow, S.I.; van Balkom, B.W.; Aalberts, M.; Heck, A.J.; Wauben, M.; Stoorvogel, W. Mhc class ii-associated proteins in b-cell exosomes and potential functional implications for exosome biogenesis. Immunol. Cell Biol. 2010, 88, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.S.; Lee, J.M.; Park, G.W.; Lim, H.W.; Bang, J.Y.; Kim, Y.K.; Kwon, K.H.; Kwon, H.J.; Kim, K.P.; Gho, Y.S. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J. Proteome Res. 2007, 6, 4646–4655. [Google Scholar] [CrossRef]
- Xu, R.; Greening, D.W.; Rai, A.; Ji, H.; Simpson, R.J. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line lim1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 2015, 87, 11–25. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Qin, H.; Poon, T.C.; Sze, S.C.; Ding, X.; Co, N.N.; Ngai, S.M.; Chan, T.F.; Wong, N. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and rnas. Carcinogenesis 2015, 36, 1008–1018. [Google Scholar] [CrossRef] [PubMed]
- Pisitkun, T.; Shen, R.F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potolicchio, I.; Carven, G.J.; Xu, X.; Stipp, C.; Riese, R.J.; Stern, L.J.; Santambrogio, L. Proteomic analysis of microglia-derived exosomes: Metabolic role of the aminopeptidase cd13 in neuropeptide catabolism. J. Immunol. 2005, 175, 2237–2243. [Google Scholar] [CrossRef]
- Stamer, W.D.; Hoffman, E.A.; Luther, J.M.; Hachey, D.L.; Schey, K.L. Protein profile of exosomes from trabecular meshwork cells. J. Proteom. 2011, 74, 796–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staubach, S.; Razawi, H.; Hanisch, F.G. Proteomics of muc1-containing lipid rafts from plasma membranes and exosomes of human breast carcinoma cells mcf-7. Proteomics 2009, 9, 2820–2835. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Munoz, C.; Kilani, R.T.; Ghahary, A. Profile of exosomes related proteins released by differentiated and undifferentiated human keratinocytes. J. Cell Physiol. 2009, 221, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Jeong, J.; Lee, K.J. Characterization of vesicles secreted from insulinoma nit-1 cells. J. Proteome Res. 2009, 8, 2851–2862. [Google Scholar] [CrossRef]
- Ochieng, J.; Pratap, S.; Khatua, A.K.; Sakwe, A.M. Anchorage-independent growth of breast carcinoma cells is mediated by serum exosomes. Exp. Cell Res. 2009, 315, 1875–1888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demory Beckler, M.; Higginbotham, J.N.; Franklin, J.L.; Ham, A.J.; Halvey, P.J.; Imasuen, I.E.; Whitwell, C.; Li, M.; Liebler, D.C.; Coffey, R.J. Proteomic analysis of exosomes from mutant kras colon cancer cells identifies intercellular transfer of mutant kras. Mol. Cell Proteom. 2013, 12, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Hosseini-Beheshti, E.; Pham, S.; Adomat, H.; Li, N.; Tomlinson Guns, E.S. Exosomes as biomarker enriched microvesicles: Characterization of exosomal proteins derived from a panel of prostate cell lines with distinct ar phenotypes. Mol. Cell Proteom. 2012, 11, 863–885. [Google Scholar] [CrossRef]
- Beckett, K.; Monier, S.; Palmer, L.; Alexandre, C.; Green, H.; Bonneil, E.; Raposo, G.; Thibault, P.; Le Borgne, R.; Vincent, J.P. Drosophila s2 cells secrete wingless on exosome-like vesicles but the wingless gradient forms independently of exosomes. Traffic 2013, 14, 82–96. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microrna-loaded exosomes from t cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- Lim, J.W.; Mathias, R.A.; Kapp, E.A.; Layton, M.J.; Faux, M.C.; Burgess, A.W.; Ji, H.; Simpson, R.J. Restoration of full-length apc protein in sw480 colon cancer cells induces exosome-mediated secretion of dkk-4. Electrophoresis 2012, 33, 1873–1880. [Google Scholar] [CrossRef]
- Ohshima, K.; Inoue, K.; Fujiwara, A.; Hatakeyama, K.; Kanto, K.; Watanabe, Y.; Muramatsu, K.; Fukuda, Y.; Ogura, S.; Yamaguchi, K.; et al. Let-7 microrna family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE 2010, 5, e13247. [Google Scholar] [CrossRef] [PubMed]
- Welton, J.L.; Khanna, S.; Giles, P.J.; Brennan, P.; Brewis, I.A.; Staffurth, J.; Mason, M.D.; Clayton, A. Proteomics analysis of bladder cancer exosomes. Mol. Cell Proteom. 2010, 9, 1324–1338. [Google Scholar] [CrossRef]
- Ji, H.; Erfani, N.; Tauro, B.J.; Kapp, E.A.; Zhu, H.J.; Moritz, R.L.; Lim, J.W.; Simpson, R.J. Difference gel electrophoresis analysis of ras-transformed fibroblast cell-derived exosomes. Electrophoresis 2008, 29, 2660–2671. [Google Scholar] [CrossRef]
- Lazar, I.; Clement, E.; Ducoux-Petit, M.; Denat, L.; Soldan, V.; Dauvillier, S.; Balor, S.; Burlet-Schiltz, O.; Larue, L.; Muller, C.; et al. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment Cell Melanoma Res. 2015, 28, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Peng, P.; Chen, S.; Li, L.; Zhang, M.; Cao, D.; Yang, J.; Li, H.; Gui, T.; Li, X.; et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J. Proteom. 2013, 80, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zoller, M. Cell surface tetraspanin tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010, 70, 1668–1678. [Google Scholar] [CrossRef]
- Van Balkom, B.W.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small rnas in human endothelial cells and exosomes provides insights into localized rna processing, degradation and sorting. J. Extracell. Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef]
- Pienimaeki-Roemer, A.; Kuhlmann, K.; Bottcher, A.; Konovalova, T.; Black, A.; Orso, E.; Liebisch, G.; Ahrens, M.; Eisenacher, M.; Meyer, H.E.; et al. Lipidomic and proteomic characterization of platelet extracellular vesicle subfractions from senescent platelets. Transfusion 2015, 55, 507–521. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Begne, M.; Lu, B.; Han, X.; Hagen, F.K.; Hand, A.R.; Melvin, J.E.; Yates, J.R. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (mudpit). J. Proteome Res. 2009, 8, 1304–1314. [Google Scholar] [CrossRef]
- Skogberg, G.; Gudmundsdottir, J.; van der Post, S.; Sandstrom, K.; Bruhn, S.; Benson, M.; Mincheva-Nilsson, L.; Baranov, V.; Telemo, E.; Ekwall, O. Characterization of human thymic exosomes. PLoS ONE 2013, 8, e67554. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Konig, A.K.; Marme, F.; Runz, S.; Wolterink, S.; Koensgen, D.; Mustea, A.; Sehouli, J.; Altevogt, P. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 2009, 278, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Gonzalez, E.; Berisa, A.; Gil, D.; Embade, N.; Valle, M.; Luka, Z.; Elortza, F.; Wagner, C.; et al. Candidate biomarkers in exosome-like vesicles purified from rat and mouse urine samples. Proteom. Clin. Appl. 2010, 4, 416–425. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Wu, Y.; Duan, J.; Ma, Y.; Shen, Z.; Wei, L.; Cui, X.; Zhang, J.; Xie, Y.; Liu, J. Quantitative proteomic analysis of exosome protein content changes induced by hepatitis b virus in huh-7 cells using silac labeling and lc-ms/ms. J. Proteome Res. 2014, 13, 5391–5402. [Google Scholar] [CrossRef] [PubMed]
- Raj, D.A.; Fiume, I.; Capasso, G.; Pocsfalvi, G. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. 2012, 81, 1263–1272. [Google Scholar] [CrossRef] [Green Version]
- Looze, C.; Yui, D.; Leung, L.; Ingham, M.; Kaler, M.; Yao, X.; Wu, W.W.; Shen, R.F.; Daniels, M.P.; Levine, S.J. Proteomic profiling of human plasma exosomes identifies ppargamma as an exosome-associated protein. Biochem. Biophys. Res. Commun. 2009, 378, 433–438. [Google Scholar] [CrossRef]
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 2009, 20, 363–379. [Google Scholar] [CrossRef]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mrna and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef]
- Kim, Y.S.; Ahn, J.S.; Kim, S.; Kim, H.J.; Kim, S.H.; Kang, J.S. The potential theragnostic (diagnostic+therapeutic) application of exosomes in diverse biomedical fields. Korean J. Physiol. Pharmacol. 2018, 22, 113–125. [Google Scholar] [CrossRef] [Green Version]
- Keller, S.; Rupp, C.; Stoeck, A.; Runz, S.; Fogel, M.; Lugert, S.; Hager, H.D.; Abdel-Bakky, M.S.; Gutwein, P.; Altevogt, P. Cd24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007, 72, 1095–1102. [Google Scholar] [CrossRef]
- Kim, H.K.; Song, K.S.; Park, Y.S.; Kang, Y.H.; Lee, Y.J.; Lee, K.R.; Ryu, K.W.; Bae, J.M.; Kim, S. Elevated levels of circulating platelet microparticles, vegf, il-6 and rantes in patients with gastric cancer: Possible role of a metastasis predictor. Eur. J. Cancer 2003, 39, 184–191. [Google Scholar] [CrossRef]
- Lasser, C.; Alikhani, V.S.; Ekstrom, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjostrand, M.; Gabrielsson, S.; Lotvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain rna: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of micrornas in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Li, X.; Gong, S.; Yuan, H.; Jiang, Y.; Huang, W.; Sun, X.; Dang, X. Serum exosomes contain ecrg4 mrna that suppresses tumor growth via inhibition of genes involved in inflammation, cell proliferation, and angiogenesis. Cancer Gene Ther. 2018, 25, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Rubio, D.; Martin-Burriel, I.; Gil, A.; Cubero, P.; Forner, M.; Khalyfa, A.; Marin, J.M. Stability of circulating exosomal mirnas in healthy subjects. Sci. Rep. 2018, 8, 10306. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, X.; Fu, X.; Li, W.; Xing, S.; Yang, Y. Identification of common differentially-expressed mirnas in ovarian cancer cells and their exosomes compared with normal ovarian surface epithelial cell cells. Oncol. Lett. 2018, 16, 2391–2401. [Google Scholar] [CrossRef]
- Leblanc, P.; Alais, S.; Porto-Carreiro, I.; Lehmann, S.; Grassi, J.; Raposo, G.; Darlix, J.L. Retrovirus infection strongly enhances scrapie infectivity release in cell culture. EMBO J. 2006, 25, 2674–2685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, F.; Jiang, W.; Zhou, L.; Chen, Z. Circulating exosomal mir-17-5p and mir-92a-3p predict pathologic stage and grade of colorectal cancer. Transl. Oncol. 2018, 11, 221–232. [Google Scholar] [CrossRef]
- Poon, K.S.; Palanisamy, K.; Chang, S.S.; Sun, K.T.; Chen, K.B.; Li, P.C.; Lin, T.C.; Li, C.Y. Plasma exosomal mir-223 expression regulates inflammatory responses during cardiac surgery with cardiopulmonary bypass. Sci. Rep. 2017, 7, 10807. [Google Scholar] [CrossRef]
- Casadei, L.; Calore, F.; Creighton, C.J.; Guescini, M.; Batte, K.; Iwenofu, O.H.; Zewdu, A.; Braggio, D.A.; Bill, K.L.; Fadda, P.; et al. Exosome-derived mir-25-3p and mir-92a-3p stimulate liposarcoma progression. Cancer Res. 2017, 77, 3846–3856. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating micrornas as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Li, Z.; Ma, Y.Y.; Wang, J.; Zeng, X.F.; Li, R.; Kang, W.; Hao, X.K. Exosomal microrna-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 2016, 9, 139–148. [Google Scholar] [PubMed]
- Palanisamy, V.; Sharma, S.; Deshpande, A.; Zhou, H.; Gimzewski, J.; Wong, D.T. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS ONE 2010, 5, e8577. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.; Bajracharya, S.D.; Yuen, P.S.; Zhou, H.; Star, R.A.; Illei, G.G.; Alevizos, I. Exosomes from human saliva as a source of microrna biomarkers. Oral. Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef]
- Li, C.Y.; Li, H.J.; Zhang, T.; Gao, H.S.; Chang, J.W.; Men, X.L.; Wu, J.; Li, J.M. Significance of apolipoprotein a1 as biomarker for early diagnosis and classification of bladder urothelial carcinoma. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 2013, 31, 266–270. [Google Scholar] [PubMed]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Dorval, T.; Chaput, N.; André, F.; Caby, M.-P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (dc) derived-exosomes: Results of thefirst phase i clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Morse, M.A.; Garst, J.; Osada, T.; Khan, S.; Hobeika, A.; Clay, T.M.; Valente, N.; Shreeniwas, R.; Sutton, M.A.; Delcayre, A.; et al. A phase i study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Chaput, N.; Flament, C.; Viaud, S.; Taieb, J.; Roux, S.; Spatz, A.; Andre, F.; LePecq, J.B.; Boussac, M.; Garin, J.; et al. Dendritic cell derived-exosomes: Biology and clinical implementations. J. Leukoc Biol. 2006, 80, 471–478. [Google Scholar] [CrossRef]
- Viaud, S.; Thery, C.; Ploix, S.; Tursz, T.; Lapierre, V.; Lantz, O.; Zitvogel, L.; Chaput, N. Dendritic cell-derived exosomes for cancer immunotherapy: What’s next? Cancer Res. 2010, 70, 1281–1285. [Google Scholar] [CrossRef]
- Lee, R.H.; Kim, B.; Choi, I.; Kim, H.; Choi, H.S.; Suh, K.; Bae, Y.C.; Jung, J.S. Characterization and expression analysis of mesenchymal stem cells from human bone marrow and adipose tissue. Cell Physiol. Biochem. 2004, 14, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Bruno, S.; Grange, C.; Deregibus, M.C.; Calogero, R.A.; Saviozzi, S.; Collino, F.; Morando, L.; Busca, A.; Falda, M.; Bussolati, B.; et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J. Am. Soc. Nephrol. 2009, 20, 1053–1067. [Google Scholar] [CrossRef]
- Yan, Y.; Jiang, W.; Tan, Y.; Zou, S.; Zhang, H.; Mao, F.; Gong, A.; Qian, H.; Xu, W. Hucmsc exosome-derived gpx1 is required for the recovery of hepatic oxidant injury. Mol. Ther. 2017, 25, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Janowska-Wieczorek, A.; Majka, M.; Kijowski, J.; Baj-Krzyworzeka, M.; Reca, R.; Turner, A.R.; Ratajczak, J.; Emerson, S.G.; Kowalska, M.A.; Ratajczak, M.Z. Platelet-derived microparticles bind to hematopoietic stem/progenitor cells and enhance their engraftment. Blood 2001, 98, 3143–3149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beltrami, C.; Besnier, M.; Shantikumar, S.; Shearn, A.I.; Rajakaruna, C.; Laftah, A.; Sessa, F.; Spinetti, G.; Petretto, E.; Angelini, G.D.; et al. Human pericardial fluid contains exosomes enriched with cardiovascular-expressed micrornas and promotes therapeutic angiogenesis. Mol. Ther. 2017, 25, 679–693. [Google Scholar] [CrossRef] [PubMed]
- Makkar, R.R.; Smith, R.R.; Cheng, K.; Malliaras, K.; Thomson, L.E.; Berman, D.; Czer, L.S.; Marban, L.; Mendizabal, A.; Johnston, P.V.; et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (caduceus): A prospective, randomised phase 1 trial. Lancet 2012, 379, 895–904. [Google Scholar] [CrossRef]
- Ibrahim, A.G.; Cheng, K.; Marban, E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Rep. 2014, 2, 606–619. [Google Scholar] [CrossRef]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport rna and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Fruhbeis, C.; Frohlich, D.; Kramer-Albers, E.M. Emerging roles of exosomes in neuron-glia communication. Front. Physiol. 2012, 3, 119. [Google Scholar] [CrossRef]
- Marcus, M.E.; Leonard, J.N. Fedexosomes: Engineering therapeutic biological nanoparticles that truly deliver. Pharmaceuticals 2013, 6, 659–680. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of sirna to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Kooijmans, S.A.; Aleza, C.G.; Roffler, S.R.; van Solinge, W.W.; Vader, P.; Schiffelers, R.M. Display of gpi-anchored anti-egfr nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. Vesicles 2016, 5, 31053. [Google Scholar] [CrossRef] [PubMed]
- Kooijmans, S.A.; Stremersch, S.; Braeckmans, K.; de Smedt, S.C.; Hendrix, A.; Wood, M.J.; Schiffelers, R.M.; Raemdonck, K.; Vader, P. Electroporation-induced sirna precipitation obscures the efficiency of sirna loading into extracellular vesicles. J. Control Release 2013, 172, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Yue, S.; Stadel, D.; Zoller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef]
- El-Andaloussi, S.; Lee, Y.; Lakhal-Littleton, S.; Li, J.; Seow, Y.; Gardiner, C.; Alvarez-Erviti, L.; Sargent, I.L.; Wood, M.J. Exosome-mediated delivery of sirna in vitro and in vivo. Nat. Protoc. 2012, 7, 2112–2126. [Google Scholar] [CrossRef] [PubMed]
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212. [Google Scholar] [CrossRef]


| No. | Proteins (Species Found) | Symbol | Tissue/Cell/Body Fluids | Identification Methods | References |
|---|---|---|---|---|---|
| 1. | Programmed cell death 6 interacting protein (R, M, H) | PDCD6IP | Pancreatic, colorectal, breast, ovarian, liver, and brain cancer cells, hepatocytes, neural stem cells, embryonic fibroblast, dendritic cells, platelets, macrophages, reticulocytes, urine, serum, saliva | Mass spectrometry, Western blotting | [72,73,74,75,76,77,78,79,80] |
| 2. | Heat-shock protein 8 (R, M, H, B) | HSPA8 | Prostate, colorectal, and brain cancer cells, macrophages, mast cells, adipocytes, reticulocytes, platelets, urine, milk, serum, saliva | Mass spectrometry, Western blotting, RNA sequencing | [70,77,81,82,83,84,85,86,87,88] |
| 3. | Annexin A2 (R, M, H, B) | ANXA2 | Ovarian, colorectal, and breast cancer cells, microglia, dendritic cells, macrophages, fibroblasts, hepatocytes, adipocytes, reticulocytes, platelets, thymus, urine, milk, saliva | Mass spectrometry, Western blotting, RNA sequencing | [70,77,80,81,83,84,86,87,89,90,91,92] |
| 4. | Syndecan-binding protein (R, M, H, B) | SDCBP | Pancreatic, ovarian, colorectal, prostate, and brain cancer cells, dendritic cells, macrophages, mast cells, reticulocytes, hepatocytes, platelets, fibroblasts, urine, milk, saliva | Mass spectrometry, Western blotting | [24,77,82,83,84,85,87,93] |
| 5. | Heat-shock protein 90 alpha class A member 1 (R, M, H, B) | HSP90AA1 | Ovarian, colorectal, prostate, and bladder cancer cells, neural stem cells, macrophages, mast cells, adipocytes, reticulocytes, hepatocytes, pancreatic cells, platelets, fibroblasts, urine, milk, serum, saliva | Mass spectrometry, Western blotting, microarray | [70,77,80,81,83,84,86,94,95,96,97,98] |
| 6. | Tumor susceptibility gene 101 (R, M, H, B, D) | TSG101 | Colorectal, liver, prostate, and bladder cancer cells, neural stem cells, dendritic cells, macrophages, reticulocytes, hepatocytes, platelets, urine, milk, Drosophila s2 cells. | Mass spectrometry, Western blotting | [24,58,76,77,80,83,87,97,99,100,101,102] |
| 7. | Eukaryotic translation elongation factor 1 alpha 1 (R, M, H, B) | EEF1A1 | Ovarian, colorectal, prostate, and bladder cancer cells, mast cells, adipocytes, hepatocytes, reticulocytes, dendritic cells, platelets, macrophages, pancreatic cells, fibroblasts, thymus, urine, saliva, milk | Mass spectrometry, RNA sequencing, microarray | [24,58,70,77,80,81,83,84,86,87,93,94,103] |
| 8. | Tyrosine-3-monooxygenase/ tryptophan-5-monooxygenase activation protein, zeta (R, M, H, B) | YWHAZ | Ovarian, colorectal, prostate, and bladder cancer cells, mast cells, hepatocytes, reticulocytes, dendritic cells, platelets, pancreatic cells, fibroblasts, thymus, urine, saliva, milk | Mass spectrometry, microarray | [24,70,77,83,84,94,95,104,105,106] |
| 9. | Eukaryotic translation elongation factor 2 (R, M, H, B) | EEF2 | Ovarian, pancreatic, bladder, colorectal, prostate, and breast cancer cells, adipocytes, hepatocytes, reticulocytes, platelets, thymus, saliva, urine, milk | Mass spectrometry, RNA sequencing, microarray | [58,70,77,80,81,83,85,86,87,93,94,95,98,103,106,107,108,109,110,111] |
| 10. | Heat-shock protein 90 alpha class B member 1 (R, M, H, B) | HSP90AB1 | Ovarian, pancreatic, bladder, colorectal, and prostate cancer cells, dendritic cells, macrophages, mast cells, neural stem, cells, pancreatic cells, adipocytes, hepatocytes, reticulocytes, platelets, thymus, urine, milk, serum | Mass spectrometry, Western blotting, | [24,58,77,79,80,81,83,85,86,88,94,95,96,98,103,106,109,111] |
| 11. | Annexin 5 (R, M, H) | ANXA5 | Ovarian, bladder, colorectal, and prostate cancer cells, adipocytes, hepatocytes, reticulocytes, platelets, thymus, urine, serum, dendritic cells, fibroblast, macrophages, mast cells, pancreatic cells, urine | Mass spectrometry, Western blotting, fluorescence-activated cell sorying | [24,58,70,73,77,80,81,85,87,88,94,95,103,109,110,111,112,113] |
| 12. | Fatty-acid synthase (R, M, H, B) | FASN | Adipocytes, hepatocytes, reticulocytes, fibroblast, pancreatic cells, milk, breast milk, bladder, colorectal, ovarian, and prostrate cancer cells, platelets, serum, thymus, urine | Mass spectroscopy, Western blotting, RNA sequencing | [58,70,73,77,81,83,85,87,93,95,103,106,109,111,114,115] |
| 13. | Tyrosine-3-monooxygenase/ tryptophan-5-monooxygenase activation protein, epsilon (R, M, H, B) | YWHAE | Adipocytes, hepatocytes, reticulocytes, urine, milk, mast cells, pancreatic cells, bladder, colorectal, ovarian, and prostrate cancer cells, platelets, saliva, thymus, urine | Mass spectroscopy | [58,73,77,81,83,85,88,94,95,103,106,109,110,111,113] |
| 14. | Clathrin heavy chain (Hc) (R, M, H) | CLTC | Adipocytes, hepatocytes, reticulocytes, fibroblast, macrophages, bladder, colorectal, ovarian and prostate cancer cells, plasma, platelets, saliva, thymus, urine | Mass Spectroscopy, RNA Sequencing | [58,70,73,77,80,81,85,87,103,106,109,110,111,116,117] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jan, A.T.; Rahman, S.; Khan, S.; Tasduq, S.A.; Choi, I. Biology, Pathophysiological Role, and Clinical Implications of Exosomes: A Critical Appraisal. Cells 2019, 8, 99. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8020099
Jan AT, Rahman S, Khan S, Tasduq SA, Choi I. Biology, Pathophysiological Role, and Clinical Implications of Exosomes: A Critical Appraisal. Cells. 2019; 8(2):99. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8020099
Chicago/Turabian StyleJan, Arif Tasleem, Safikur Rahman, Shahanavaj Khan, Sheikh Abdullah Tasduq, and Inho Choi. 2019. "Biology, Pathophysiological Role, and Clinical Implications of Exosomes: A Critical Appraisal" Cells 8, no. 2: 99. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8020099