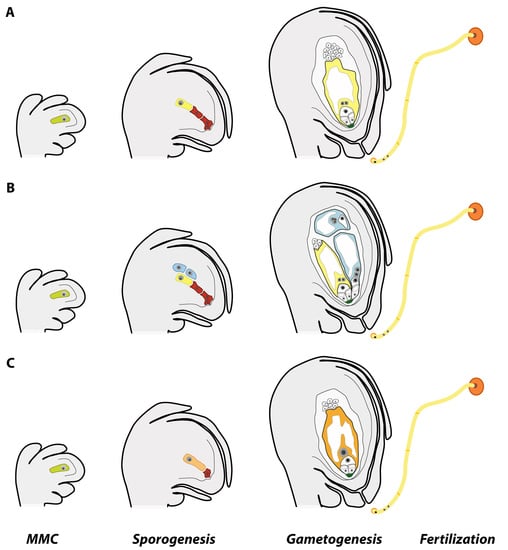
Apomixis Technology: Separating the Wheat from the Chaff
Abstract
:1. Introduction
2. Developmental Features of Apomixis
3. Genetic and Genomic Features of Apomixis
4. Molecular Control of Apomixis in Apomictic Plants
5. The Sexual Machinery and Apomixis-Like Phenotypes
5.1. Apomeiosis Mutants
5.2. Parthenogenetic Mutants
5.3. Mutants Associated with Endosperm Development
6. Overall Features, Hypotheses in Conflict, and the Quest for the Origin of Apomixis
7. Current View and Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- United Nations, Department of Economic and Social Affairs, P.D. World Population Prospects 2019. Volume II: Demographic Profiles; United Nations Press: New York, NY, USA, 2019. [Google Scholar]
- FAO. The State of Food and Agriculture; Food and Agriculture Organization of the United Nations Press: Rome, Italy, 2009; ISBN 978-92-5-106215-9. [Google Scholar]
- Sharma, H.C.; Crouch, J.H.; Sharma, K.K.; Seetharama, N.; Hash, C.T. Applications of biotechnology for crop improvement: Prospects and constraints. Plant Sci. 2002, 163, 381–395. [Google Scholar] [CrossRef] [Green Version]
- Acquaah, G. Principles of Plant Genetics and Breeding, 2nd ed.; Wiley- Blackwell: Chichester, UK, 2012; ISBN 978-0-470-66476-6. [Google Scholar]
- Van Dijk, P.J.; Rigola, D.; Schauer, S.E. Plant breeding: Surprisingly, less sex is better. Curr. Biol. 2016, 26, R122–R124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spillane, C.; Curtis, M.D.; Grossniklaus, U. Apomixis technology development—virgin births in farmers’ fields? Nat. Biotechnol. 2004, 22, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Sailer, C.; Schmid, B.; Grossniklaus, U. Apomixis allows the transgenerational fixation of phenotypes in hybrid plants. Curr. Biol. 2016, 26, 331–337. [Google Scholar] [CrossRef] [Green Version]
- Carman, J.G. Asynchonous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 1997, 61, 51–94. [Google Scholar] [CrossRef]
- Hojsgaard, D.; Klatt, S.; Baier, R.; Carman, J.G.; Hörandl, E. Taxonomy and biogeography of apomixis in angiosperms and associated biodiversity characteristics. CRC Crit. Rev. Plant Sci. 2014, 33, 414–427. [Google Scholar] [CrossRef] [Green Version]
- van Dijk, P.J.; Vijverberg, K. The significance of apomixis in the evolution of the angiosperms: A reappraisal. In Plant Species-Level Systematics: New Perspectives on Pattern & Process; Bakker, F.T., Chatrou, L.W., Gravendeel, B., Pelser, P.B., Eds.; Gantner Verlag: Ruggell, Lichtenstein, 2005; Volume 143, pp. 101–116. ISBN 0080-0694. [Google Scholar]
- Ray, D.T.; Coffelt, T.A.; Dierig, D.A. Breeding guayule for commercial production. Ind. Crops Prod. 2005, 22, 15–25. [Google Scholar] [CrossRef]
- Savidan, Y. Transfer of apomixis through wide crosses. In The Flowering of Apomixis: From Mechanisms to Genetic Engineering; Savidan, Y., Carman, J.G., Dresselhaus, T., Eds.; CIMMYT, IRD, European Commission OC VI (FAIR): Mexico City, Mexico, 2001; pp. 153–167. ISBN 970-648-074-9. [Google Scholar]
- Ozias-Akins, P.; Akiyama, Y.; Hanna, W.W. Molecular characterization of the genomic region linked with apomixis in Pennisetum/Cenchrus. Funct. Integr. Genomics 2003, 3, 94–104. [Google Scholar] [CrossRef]
- Ozias-Akins, P.; van Dijk, P.J. Mendelian genetics of apomixis in plants. Annu. Rev. Genet. 2007, 41, 509–537. [Google Scholar] [CrossRef]
- Barcaccia, G.; Albertini, E. Apomixis in plant reproduction: A novel perspective on an old dilemma. Plant Reprod. 2013, 26, 159–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardwell, J.H. A review of apomixis and differential expression analyses using microarrays; Utah State University Press: Logan, UT, USA, 2013. [Google Scholar]
- Grossniklaus, U.; Koltunow, A.; Van Lookeren Campagne, M. A bright future for apomixis. Trends Plant Sci. 1998, 3, 415–416. [Google Scholar] [CrossRef]
- Leblanc, O.; Grimanelli, D.; Hernandez-Rodriguez, M.; Galindo, P.A.; Soriano-Martinez, A.M.; Perotti, E. Seed development and inheritance studies in apomictic maize-Tripsacum hybrids reveal barriers for the transfer of apomixis into sexual crops. Int. J. Dev. Biol. 2009, 53, 585–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheben, A.; Hojsgaard, D.H. Can we use gene-editing to induce apomixis in sexual plants? Genes (Basel) 2020. submitted. [Google Scholar]
- Asker, S.E.; Jerling, L. Apomixis in plants; CRC Press: Boca Raton, FL, USA, 1992; ISBN 0-8493-4545-6. [Google Scholar]
- Naumova, T.N. Apomixis in angiosperms - nucellar and integumentary embryony; CRC Press: Boca Raton, FL, USA, 1993; ISBN 13: 978-1-315-89065-4. [Google Scholar]
- Nogler, G.A. Gametophytic apomixis. In Embryology of Angiosperms; Johri, B.M., Ed.; Springer-Verlag Press: Berlin/Heidelberg, Germany, 1984; pp. 475–518. ISBN 3-540-12739-9. [Google Scholar]
- Olsen, O.-A. (Ed.) Plant Cell Monographs; Plant Cell; Springer: Berlin/Heidelberg, Germany, 2007; Volume 8, ISBN 978-3-540-71234-3. [Google Scholar]
- Quarin, C.L. Effect of pollen source and pollen ploidy on endosperm formation and seed set in pseudogamous apomictic Paspalum notatum. Sex. Plant Rep. 1999, 11, 331–335. [Google Scholar] [CrossRef]
- Talent, N.; Dickinson, T.A. Endosperm formation in aposporous Crataegus (Rosaceae, Spiraeoideae, tribe Pyreae): Parallels to Ranunculaceae and Poaceae. New Phytol. 2007, 173, 231–249. [Google Scholar] [CrossRef]
- Köhler, C.; Mittelsten Scheid, O.; Erilova, A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 2010, 26, 142–148. [Google Scholar] [CrossRef]
- Schatlowski, N.; Wolff, P.; Santos-González, J.; Schoft, V.; Siretskiy, A.; Scott, R.; Tamaru, H.; Köhler, C. Hypomethylated pollen bypasses the interploidy hybridization barrier in Arabidopsis. Plant Cell 2014, 26, 3556–3568. [Google Scholar] [CrossRef] [Green Version]
- Hojsgaard, D.; Hörandl, E. Apomixis as a facilitator of range expansion and diversification in plants. In Evolutionary Biology: Biodiversification from Genotype to Phenotype; Pontarotti, P., Ed.; Springer International Press: Marseille, France, 2015; pp. 305–327. ISBN 10.1007/978-3-319-19932-0. [Google Scholar]
- Hojsgaard, D.; Hörandl, E. The rise of apomixis in natural plant populations. Front. Plant Sci. 2019, 10, 358. [Google Scholar] [CrossRef]
- Norrmann, G.A.; Quarin, C.L.; Burson, B.L. Cytogenetics and reproductive behavior of different chromosome races in six Paspalum species. J. Hered. 1989, 80, 24–28. [Google Scholar] [CrossRef]
- Naumova, N.; Osadtchiy, J.V.; Sharma, V.K.; Dijkhuis, P.; Ramulu, K.S. Apomixis in plants: Structural and functional aspects of diplospory in Poa nemoralis and P. palustris. Protoplasma 1999, 208, 186–195. [Google Scholar]
- Hojsgaard, D.; Schegg, E.; Valls, J.F.M.; Martínez, E.J.; Quarin, C.L. Sexuality, apomixis, ploidy levels, and genomic relationships among four Paspalum species of the subgenus Anachyris (Poaceae). Flora Morphol. Distrib. Funct. Ecol. Plants 2008, 203, 535–547. [Google Scholar] [CrossRef]
- Noyes, R.D.; Wagner, J.D. Dihaploidy yields diploid apomicts and parthenogens in Erigeron (Asteraceae). Am. J. Bot. 2014, 101, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Böcher, T. Cytological and embryological studies in the amphi– apomictic Arabis holboellii complex. Biol. Skr. 1951, 6, 1–58. [Google Scholar]
- Kantama, L.; Sharbel, T.F.; Schranz, M.E.; Mitchell-Olds, T.; de Vries, S.; de Jong, H. Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proc. Natl. Acad. Sci. USA 2007, 104, 14026–14031. [Google Scholar] [CrossRef] [Green Version]
- Tucker, M.R.; Koltunow, A.M. Traffic monitors at the cell periphery: The role of cell walls during early female reproductive cell differentiation in plants. Curr. Opin. Plant Biol. 2014, 17, 137–145. [Google Scholar] [CrossRef]
- Aliyu, O.M.; Schranz, M.E.; Sharbel, T.F. Quantitative variation for apomictic reproduction in the genus Boechera (Brassicaceae). Am. J. Bot. 2010, 97, 1719–1731. [Google Scholar] [CrossRef]
- Rebozzio, R.N.; Sartor, M.E.; Quarin, C.L.; Espinoza, F. Residual sexuality and its seasonal variation in natural apomictic Paspalum notatum accessions. Biol. Plant. 2011, 55, 391–395. [Google Scholar] [CrossRef]
- Hojsgaard, D.H.; Martínez, E.J.; Quarin, C.L. Competition between meiotic and apomictic pathways during ovule and seed development results in clonality. New Phytol. 2013, 197, 336–347. [Google Scholar] [CrossRef]
- Hand, M.L.; Vít, P.; Krahulcová, A.; Johnson, S.D.; Oelkers, K.; Siddons, H.; Chrtek, J.; Fehrer, J.; Koltunow, A.M.G. Evolution of apomixis loci in Pilosella and Hieracium (Asteraceae) inferred from the conservation of apomixis-linked markers in natural and experimental populations. Heredity (Edinb.) 2015, 114, 17–26. [Google Scholar] [CrossRef]
- Pessino, S.C.; Ortiz, J.P.A.; Leblanc, O.; do Valle, C.B.; Evans, C.; Hayward, M.D. Identification of a maize linkage group related to apomixis in Brachiaria. Theor. Appl. Genet. 1997, 94, 439–444. [Google Scholar] [CrossRef]
- Pessino, S.; Evans, C.; Ortiz, J.; Armstead, I.; do Valle, C.; Hayward, M. A genetic map of the apospory- region in Brachiaria hybrids: Identification of two markers closely associated with the trait. Hereditas 1998, 128, 153–158. [Google Scholar] [CrossRef]
- Gustine, D.; Sherwood, R.; Huff, D. Apospory-linked molecular markers in buffelgrass. Crop Sci. 1997, 37, 947–951. [Google Scholar] [CrossRef]
- Ozias-Akins, P.; Roche, D.; Hanna, W. Tight clustering and hemizygosity of apomixis-linked markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc. Natl. Acad. Sci. USA 1998, 95, 5127–5132. [Google Scholar] [CrossRef] [Green Version]
- Noyes, R.D.; Rieseberg, L.H. Two independent loci control agamospermy (apomixis) in the triploid flowering plant Erigeron annuus. Genetics 2000, 155, 379–390. [Google Scholar]
- Catanach, A.S.; Erasmuson, S.K.; Podivinsky, E.; Jordan, B.R.; Bicknell, R. Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc. Natl. Acad. Sci. USA 2006, 103, 18650–18655. [Google Scholar] [CrossRef] [Green Version]
- Schallau, A.; Arzenton, F.; Johnston, A.J.; Hähnel, U.; Koszegi, D.; Blattner, F.R.; Altschmied, L.; Haberer, G.; Barcaccia, G.; Bäumlein, H. Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. Plant J. 2010, 62, 773–784. [Google Scholar] [CrossRef]
- Labombarda, P.; Busti, A.; Caceres, M.E.; Pupilli, F.; Arcioni, S. An AFLP marker tightly linked to apomixis reveals hemizygosity in a portion of the apomixis-controlling locus in Paspalum simplex. Genome 2002, 45, 513–519. [Google Scholar] [CrossRef]
- Pupilli, F.; Martinez, E.J.; Busti, A.; Calderini, O.; Quarin, C.L.; Arcioni, S. Comparative mapping reveals partial conservation of synteny at the apomixis locus in Paspalum spp. Mol. Genet. Genomics 2004, 270, 539–548. [Google Scholar] [CrossRef]
- Stein, J.; Quarin, C.L.; Martínez, E.J.; Pessino, S.C.; Ortiz, J.P.A. Tetraploid races of Paspalum notatum show polysomic inheritance and preferential chromosome pairing around the apospory-controlling locus. Theor. Appl. Genet. 2004, 109, 186–191. [Google Scholar] [CrossRef]
- Hojsgaard, D.H.; Martínez, E.J.; Acuña, C.A.; Quarin, C.L.; Pupilli, F. A molecular map of the apomixis-control locus in Paspalum procurrens and its comparative analysis with other species of Paspalum. Theor. Appl. Genet. 2011, 123, 959–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogler, G.A. Genetics of apospory in apomictic Ranunculus auricomus. V. Conclusion. Bot. Helvet. 1984, 94, 411–422. [Google Scholar]
- Van Dijk, P.J.; Bakx-Schotman, J.M.T. Formation of unreduced megaspores (diplospory) in apomictic dandelions (Taraxacum officinale, s.l.) is controlled by a sex-specific dominant locus. Genetics 2004, 166, 483–492. [Google Scholar] [CrossRef] [Green Version]
- Grimanelli, D.; Leblanc, O.; Espinosa, E.; Perotti, E.; Gonzalez de Leon, D.; Savidan, Y. Mapping diplosporous apomixis in tetraploid Tripsacum: One gene or several genes? Heredity (Edinb) 1998, 80, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Goel, S.; Chen, Z.; Conner, J.A.; Akiyama, Y.; Hanna, W.W.; Ozias-Akins, P. Delineation by fluorescence in situ hybridization of a single hemizygous chromosomal region associated with aposporous embryo sac formation in Pennisetum squamulatum and Cenchrus ciliaris. Genetics 2003, 163, 1069–1082. [Google Scholar] [PubMed]
- Calderini, O.; Chang, S.B.; De Jong, H.; Busti, A.; Paolocci, F.; Arcioni, S.; De Vries, S.C.; Abma-Henkens, M.H.C.; Lankhorst, R.M.K.; Donnison, I.S.; et al. Molecular cytogenetics and DNA sequence analysis of an apomixis-linked BAC in Paspalum simplex reveal a non pericentromere location and partial microcolinearity with rice. Theor. Appl. Genet. 2006, 112, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Ito, K.; Johnson, S.D.; Oelkers, K.; Suzuki, G.; Houben, A.; Mukai, Y.; Koltunow, A.M. Chromosomes carrying meiotic avoidance loci in three apomictic eudicot Hieracium subgenus Pilosella species share structural features. Plant Physiol. 2011, 157, 1327–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worthington, M.; Ebina, M.; Yamanaka, N.; Heffelfinger, C.; Quintero, C.; Zapata, Y.P.; Perez, J.G.; Selvaraj, M.; Ishitani, M.; Duitama, J.; et al. Translocation of a parthenogenesis gene candidate to an alternate carrier chromosome in apomictic Brachiaria humidicola. BMC Genomics 2019, 20, 1–18. [Google Scholar] [CrossRef]
- Galla, G.; Siena, L.A.; Ortiz, J.P.A.; Baumlein, H.; Barcaccia, G.; Pessino, S.C.; Bellucci, M.; Pupilli, F. A Portion of the apomixis locus of Paspalum simplex is microsyntenic with an unstable chromosome segment highly conserved among Poaceae. Sci. Rep. 2019, 9, 3271. [Google Scholar] [CrossRef] [Green Version]
- Bowers, J.E.; Arias, M.A.; Asher, R.; Avise, J.A.; Ball, R.T.; Brewer, G.A.; Buss, R.W.; Chen, A.H.; Edwards, T.M.; Estill, J.C.; et al. Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc. Natl. Acad. Sci. USA 2005, 102, 13206–13211. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, Y.; Hanna, W.W.; Ozias-Akins, P. High-resolution physical mapping reveals that the apospory-specific genomic region (ASGR) in Cenchrus ciliaris is located on a heterochromatic and hemizygous region of a single chromosome. Theor. Appl. Genet. 2005, 111, 1042–1051. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.A.; Goel, S.; Gunawan, G.; Cordonnier-Pratt, M.M.; Johnson, V.E.; Liang, C.; Wang, H.; Pratt, L.H.; Mullet, J.E.; DeBarry, J.; et al. Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 2008, 147, 1396–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siena, L.A.; Ortiz, J.P.A.; Calderini, O.; Paolocci, F.; Cáceres, M.E.; Kaushal, P.; Grisan, S.; Pessino, S.C.; Pupilli, F. An apomixis-linked ORC3-like pseudogene is associated with silencing of its functional homolog in apomictic Paspalum simplex. J. Exp. Bot. 2016, 67, 1965–1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, H.J. The relation of recombination to mutational advance. Mutat. Res. Mol. Mech. Mutagen. 1964, 1, 2–9. [Google Scholar] [CrossRef]
- Kondrashov, A.S. Selection against harmful mutations in large sexual and asexual populations. Genet. Res. 1982, 40, 325–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlesworth, D. Evolution of recombination rates between sex chromosomes. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pupilli, F.; Barcaccia, G. Cloning plants by seeds: Inheritance models and candidate genes to increase fundamental knowledge for engineering apomixis in sexual crops. J. Biotechnol. 2012, 159, 291–311. [Google Scholar] [CrossRef]
- Vielle Calzada, J.P.; Craneand, C.F.; Stelly, D.M. Apomixis: The asexual revolution. Science (80-) 1996, 274, 1322–1323. [Google Scholar] [CrossRef]
- Singh, M.; Burson, B.L.; Finlayson, S.A. Isolation of candidate genes for apomictic development in buffelgrass (Pennisetum ciliare). Plant Mol. Biol. 2007, 64, 673–682. [Google Scholar] [CrossRef]
- Leblanc, O.; Armstead, I.; Pessino, S.; Ortiz, J.P.A.; Evans, C.; do Valle, C.; Hayward, M.D. Non-radioactive mRNA fingerprinting to visualise gene expression in mature ovaries of Brachiaria hybrids derived from B. brizantha, an apomictic tropical forage. Plant Sci. 1997, 126, 49–58. [Google Scholar] [CrossRef]
- Rodrigues, J.C.M.; Cabral, G.B.; Dusi, D.M.A.; de Mello, L.V.; Rigden, D.J.; Carneiro, V.T.C. Identification of differentially expressed cDNA sequences in ovaries of sexual and apomictic plants of Brachiaria brizantha. Plant Mol. Biol. 2003, 53, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Miyazaki, C.; Kojimai, A.; Saito, A.; Adachi, T. Isolation and characterization of a gene expressed during early embryo sac development in apomictic Guinea grass (Panicum maximum). J. Plant Physiol. 1999, 154, 55–62. [Google Scholar] [CrossRef]
- Yamada-Akiyama, H.; Akiyama, Y.; Ebina, M.; Xu, Q.; Tsuruta, S.; Yazaki, J.; Kishimoto, N.; Kikuchi, S.; Takahara, M.; Takamizo, T.; et al. Analysis of expressed sequence tags in apomictic guineagrass (Panicum maximum). J. Plant Physiol. 2009, 166, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Albertini, E.; Marconi, G.; Barcaccia, G.; Raggi, L.; Falcinelli, M. Isolation of candidate genes for apomixis in Poa pratensis L. Plant Mol. Biol. 2004, 56, 879–894. [Google Scholar] [CrossRef] [PubMed]
- Cervigni, G.D.L.; Paniego, N.; Díaz, M.; Selva, J.P.; Zappacosta, D.; Zanazzi, D.; Landerreche, I.; Martelotto, L.; Felitti, S.; Pessino, S.; et al. Expressed sequence tag analysis and development of gene associated markers in a near-isogenic plant system of Eragrostis curvula. Plant Mol. Biol. 2008, 67, 1–10. [Google Scholar] [CrossRef]
- Rodrigo, J.M.; Zappacosta, D.C.; Selva, J.P.; Garbus, I.; Albertini, E.; Echenique, V. Apomixis frequency under stress conditions in weeping lovegrass (Eragrostis curvula). PLoS ONE 2017, 12, 1–17. [Google Scholar] [CrossRef]
- Garbus, I.; Romero, J.R.; Selva, J.P.; Pasten, M.C.; Chinestra, C.; Carballo, J.; Zappacosta, D.C.; Echenique, V. De novo transcriptome sequencing and assembly from apomictic and sexual Eragrostis curvula genotypes. PLoS ONE 2017, 12, 1–22. [Google Scholar] [CrossRef]
- Laspina, N.V.; Vega, T.; Seijo, J.G.; González, A.M.; Martelotto, L.G.; Stein, J.; Podio, M.; Ortiz, J.P.A.; Echenique, V.C.; Quarin, C.L.; et al. Gene expression analysis at the onset of aposporous apomixis in Paspalum notatum. Plant Mol. Biol. 2008, 67, 615–628. [Google Scholar] [CrossRef]
- Pessino, S.C.; Espinoza, F.; Martínez, E.J.; Ortiz, J.P.A.; Valle, E.M.; Quarin, C.L. Isolation of cDNA clones differentially expressed in flowers of apomictic and sexual Paspalum notatum. Hereditas 2001, 134, 35–42. [Google Scholar] [CrossRef]
- De Oliveira, F.A.; Vigna, B.B.Z.; Da Silva, C.C.; Fávero, A.P.; De Matta, F.P.; Azevedo, A.L.S.; De Souza, A.P. Coexpression and transcriptome analyses identify active apomixis-related genes in Paspalum notatum leaves. BMC Genomics 2020, 21, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Guerin, J.; Rossel, J.B.; Robert, S.; Tsuchiya, T.; Koltunow, A. A DEFICIENS homologue is down-regulated during apomictic initiation in ovules of Hieracium. Planta 2000, 210, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Bräuning, S.; Catanach, A.; Lord, J.M.; Bicknell, R.; Macknight, R.C. Comparative transcriptome analysis of the wild-type model apomict Hieracium praealtum and its loss of parthenogenesis (lop) mutant. BMC Plant Biol. 2018, 18, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galla, G.; Vogel, H.; Sharbel, T.F.; Barcaccia, G. De novo sequencing of the Hypericum perforatum L. flower transcriptome to identify potential genes that are related to plant reproduction sensu lato. BMC Genomics 2015, 16, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galla, G.; Zenoni, S.; Avesani, L.; Altschmied, L.; Rizzo, P.; Sharbel, T.F.; Barcaccia, G. Pistil transcriptome analysis to disclose genes and gene products related to aposporous apomixis in Hypericum perforatum L. Front. Plant Sci. 2017, 8, 79. [Google Scholar] [CrossRef] [Green Version]
- Sharbel, T.F.; Voigt, M.-L.; Corral, J.M.; Thiel, T.; Varshney, A.; Kumlehn, J.; Vogel, H.; Rotter, B. Molecular signatures of apomictic and sexual ovules in the Boechera holboellii complex. Plant J. 2009, 58, 870–882. [Google Scholar] [CrossRef]
- Sharbel, T.F.; Voigt, M.-L.; Corral, J.M.; Galla, G.; Kumlehn, J.; Klukas, C.; Schreiber, F.; Vogel, H.; Rotter, B. Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell 2010, 22, 655–671. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, A.; Schmid, M.W.; Klostermeier, U.C.; Qi, W.; Guthörl, D.; Sailer, C.; Waller, M.; Rosenstiel, P.; Grossniklaus, U. Apomictic and sexual germline development differ with respect to cell cycle, transcriptional, hormonal and epigenetic regulation. PLoS Genet. 2014, 10, e1004476. [Google Scholar] [CrossRef] [Green Version]
- Zühl, L.; Volkert, C.; Ibberson, D.; Schmidt, A.; Wilson, Z. Differential activity of F-box genes and E3 ligases distinguishes sexual versus apomictic germline specification in Boechera. J. Exp. Bot. 2019, 70, 5643–5657. [Google Scholar] [CrossRef] [Green Version]
- Polegri, L.; Calderini, O.; Arcioni, S.; Pupilli, F. Specific expression of apomixis-linked alleles revealed by comparative transcriptomic analysis of sexual and apomictic Paspalum simplex Morong flowers. J. Exp. Bot. 2010, 61, 1869–1883. [Google Scholar] [CrossRef] [Green Version]
- Ortiz, J.P.A.; Revale, S.; Siena, L.A.; Podio, M.; Delgado, L.; Stein, J.; Leblanc, O.; Pessino, S.C. A reference floral transcriptome of sexual and apomictic Paspalum notatum. BMC Genomics 2017, 18, 318. [Google Scholar] [CrossRef] [Green Version]
- Sahu, P.P.; Gupta, S.; Malaviya, D.R.; Roy, A.K.; Kaushal, P.; Prasad, M. Transcriptome analysis of differentially expressed genes during embryo sac development in apomeiotic non-parthenogenetic interspecific hybrid of Pennisetum glaucum. Mol. Biotechnol. 2012, 51, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Hu, Y.; Tucker, M.R.; Taylor, J.M.; Johnson, S.D.; Spriggs, A.; Tsuchiya, T.; Oelkers, K.; Rodrigues, J.C.M.; Koltunow, A.M.G. Enlarging cells initiating apomixis in Hieracium praealtum transition to an embryo sac program prior to entering mitosis. Plant Physiol. 2013, 163, 216–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabiger, D.S.; Taylor, J.M.; Spriggs, A.; Hand, M.L.; Henderson, S.T.; Johnson, S.D.; Oelkers, K.; Hrmova, M.; Saito, K.; Suzuki, G.; et al. Generation of an integrated Hieracium genomic and transcriptomic resource enables exploration of small RNA pathways during apomixis initiation. BMC Biol. 2016, 14, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Galla, G.; Basso, A.; Grisan, S.; Bellucci, M.; Pupilli, F.; Barcaccia, G. Ovule gene expression analysis in sexual and aposporous apomictic Hypericum perforatum L. (Hypericaceae) accessions. Front. Plant Sci. 2019, 10, 654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellino, M. Gene Expression Analysis and Transcriptome Evolution in Apomicts: A Case Study in Boechera and Ranunculus; Heidelberg University: Heidelberg, Germany, 2015. [Google Scholar]
- Pellino, M.; Hojsgaard, D.; Hörandl, E.; Sharbel, T. Chasing the apomictic factors in the Ranunculus auricomus complex: The effects of hybridization, parent of origin and ploidy in comparative ovule specific gene expression analysis. Genes (Basel) 2020. submitted. [Google Scholar]
- Grimanelli, D.; García, M.; Kaszas, E.; Perotti, E.; Leblanc, O. Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics 2003, 165, 1521–1531. [Google Scholar]
- Hojsgaard, D.; Greilhuber, J.; Pellino, M.; Paun, O.; Sharbel, T.F.; Hörandl, E. Emergence of apospory and bypass of meiosis via apomixis after sexual hybridisation and polyploidisation. New Phytol. 2014, 204, 1000–1012. [Google Scholar] [CrossRef] [Green Version]
- Quarin, C.L. Seasonal changes in the incidence of apomixis of diploid, triploid, and tetraploid plants of Paspalum cromyorrhizon. Euphytica 1986, 35, 515–522. [Google Scholar] [CrossRef]
- Quarin, C.L.; Espinoza, F.; Martinez, E.J.; Pessino, S.C.; Bovo, O.A. A rise of ploidy level induces the expression of apomixis in Paspalum notatum. Sex. Plant Reprod. 2001, 13, 243–249. [Google Scholar] [CrossRef]
- Rios, E.F.; Blount, A.; Kenworthy, K.E.; Acuña, C.A.; Quesenberry, K.H. Seasonal expression of apospory in bahiagrass. Trop. Grasslands 2013, 1, 116. [Google Scholar] [CrossRef]
- Shah, J.N.; Kirioukhova, O.; Pawar, P.; Tayyab, M.; Mateo, J.L.; Johnston, A.J. Depletion of key meiotic genes and transcriptome-wide abiotic stress reprogramming mark early preparatory events ahead of apomeiotic transition. Front. Plant Sci. 2016, 7, 1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karunarathne, P.; Reutemann, A.V.; Schedler, M.; Gluecksberg, A.; Martinez, E.J.; Honfi, A.I.; Hojsgaard, D.H. Sexual modulation in a polyploid grass: a reproductive contest between environmentally inducible sexual and genetically dominant apomictic pathways. Sci. Rep. 2020. under review. [Google Scholar]
- Klatt, S.; Hadacek, F.; Hodač, L.; Brinkmann, G.; Eilerts, M.; Hojsgaard, D.; Hörandl, E. Photoperiod extension enhances sexual megaspore formation and triggers metabolic reprogramming in facultative apomictic Ranunculus auricomus. Front. Plant Sci. 2016, 7, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Guan, L.; Seo, M.; Hoffmann, F.; Adachi, T. Developmental expression of ASG-1 during gametogenesis in apomictic guinea grass (Panicum maximum). J. Plant Physiol. 2005, 162, 1141–1148. [Google Scholar] [CrossRef]
- Albertini, E.; Marconi, G.; Reale, L.; Barcaccia, G.; Porceddu, A.; Ferranti, F.; Falcinelli, M. SERK and APOSTART. Candidate genes for apomixis in Poa pratensis. Plant Physiol. 2005, 138, 2185–2199. [Google Scholar] [CrossRef] [Green Version]
- Corral, J.M.; Vogel, H.; Aliyu, O.M.; Hensel, G.; Thiel, T.; Kumlehn, J.; Sharbel, T.F. A conserved apomixis-specific polymorphism is correlated with exclusive exonuclease expression in premeiotic ovules of apomictic Boechera species. Plant Physiol. 2013, 163, 1660–1672. [Google Scholar] [CrossRef] [Green Version]
- Mau, M.; Corral, J.M.; Vogel, H.; Melzer, M.; Fuchs, J.; Kuhlmann, M.; de Storme, N.; Geelen, D.; Sharbel, T.F. The conserved chimeric transcript UPGRADE2 is associated with unreduced pollen formation and is exclusively found in apomictic Boechera species. Plant Physiol. 2013, 163, 1640–1659. [Google Scholar] [CrossRef] [Green Version]
- Mancini, M.; Permingeat, H.; Colono, C.; Siena, L.; Pupilli, F.; Azzaro, C.; de Alencar Dusi, D.M.; de Campos Carneiro, V.T.; Podio, M.; Seijo, J.G.; et al. The MAP3K-coding QUI-GON JINN (QGJ) gene is essential to the formation of unreduced embryo sacs in Paspalum. Front. Plant Sci. 2018, 871, 1547. [Google Scholar] [CrossRef] [Green Version]
- Krasileva, K.V.; Buffalo, V.; Bailey, P.; Pearce, S.; Ayling, S.; Tabbita, F.; Soria, M.; Wang, S.; Akhunov, E.; Uauy, C.; et al. Separating homeologs by phasing in the tetraploid wheat transcriptome. Genome Biol. 2013, 14, R66. [Google Scholar] [CrossRef] [Green Version]
- Vijverberg, K.; Ozias-Akins, P.; Schranz, M.E. Identifying and engineering genes for parthenogenesis in plants. Front. Plant Sci. 2019, 10, 128. [Google Scholar] [CrossRef] [Green Version]
- Ozias-Akins, P.; Conner, J.A. Clonal reproduction through seeds in sight for crops. Trends Genet. 2020, 36, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A. Controlling apomixis: Shared features and distinct characteristics of gene regulation. Genes (Basel) 2020, 11, 329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siena, L.A.; Ortiz, J.P.A.; Leblanc, O.; Pessino, S. PnTgs1-like expression during reproductive development supports a role for RNA methyltransferases in the aposporous pathway. BMC Plant Biol. 2014, 14, 297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colono, C.; Ortiz, J.P.A.; Permingeat, H.R.; Bicknell, R.A. A Plant-Specific TGS1 homolog influences gametophyte development in sexual tetraploid Paspalum notatum ovules. Front. Plant Sci. 2019, 10, 1566. [Google Scholar] [CrossRef] [PubMed]
- Ochogavía, A.; Galla, G.; Seijo, J.G.; González, A.M.; Bellucci, M.; Pupilli, F.; Barcaccia, G.; Albertini, E.; Pessino, S. Structure, target-specificity and expression of PN_LNC_N13, a long non-coding RNA differentially expressed in apomictic and sexual Paspalum notatum. Plant Mol. Biol. 2018, 96, 53–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, J.P.A.; Leblanc, O.; Rohr, C.; Grisolia, M.; Siena, L.A.; Podio, M.; Colono, C.; Azzaro, C.; Pessino, S.C. Small RNA-seq reveals novel regulatory components for apomixis in Paspalum notatum. BMC Genom. 2019, 20, 487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amiteye, S.; Corral, J.M.; Vogel, H.; Sharbel, T.F. Analysis of conserved microRNAs in floral tissues of sexual and apomictic Boechera species. BMC Genomics 2011, 12, 500. [Google Scholar] [CrossRef] [Green Version]
- Amiteye, S.; Corral, J.M.; Vogel, H.; Kuhlmann, M.; Mette, M.F.; Sharbel, T.F. Novel microRNAs and microsatellite-like small RNAs in sexual and apomictic Boechera species. MicroRNA 2013, 2, 45–62. [Google Scholar] [CrossRef]
- Selva, J.P.; Siena, L.; Rodrigo, J.M.; Garbus, I.; Zappacosta, D.; Romero, J.R.; Ortiz, J.P.A.; Pessino, S.C.; Leblanc, O.; Echenique, V. Temporal and spatial expression of genes involved in DNA methylation during reproductive development of sexual and apomictic Eragrostis curvula. Sci. Rep. 2017, 7, 15092. [Google Scholar] [CrossRef]
- Garbus, I.; Selva, J.P.; Pasten, M.C.; Bellido, A.M.; Carballo, J.; Albertini, E.; Echenique, V. Characterization and discovery of miRNA and miRNA targets from apomictic and sexual genotypes of Eragrostis curvula. BMC Genomics 2019, 20, 839. [Google Scholar] [CrossRef] [Green Version]
- Juranić, M.; Tucker, M.R.; Schultz, C.J.; Shirley, N.J.; Taylor, J.M.; Spriggs, A.; Johnson, S.D.; Bulone, V.; Koltunow, A.M. Asexual female gametogenesis involves contact with a sexually fated megaspore in apomictic Hieracium. Plant Physiol. 2018, 177, 1027–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bocchini, M.; Galla, G.; Pupilli, F.; Bellucci, M.; Barcaccia, G.; Ortiz, J.P.A.; Pessino, S.C.; Albertini, E. The vesicle trafficking regulator PN-SCD1 is demethylated and overexpressed in florets of apomictic Paspalum notatum genotypes. Sci. Rep. 2018, 8, 3030. [Google Scholar] [CrossRef] [PubMed]
- Podio, M.; Cáceres, M.E.; Samoluk, S.S.; Seijo, J.G.; Pessino, S.C.; Ortiz, J.P.A.; Pupilli, F. A methylation status analysis of the apomixis-specific region in Paspalum spp. suggests an epigenetic control of parthenogenesis. J. Exp. Bot. 2014, 65, 6411–6424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirioukhova, O.; Shah, J.N.; Larsen, D.S.; Tayyab, M.; Mueller, N.E.; Govind, G.; Baroux, C.; Federer, M.; Gheyselinck, J.; Barrell, P.J.; et al. Aberrant imprinting may underlie evolution of parthenogenesis. Sci. Rep. 2018, 8, 10626. [Google Scholar] [CrossRef]
- Thomas, C.; Meyer, D.; Himber, C.; Steinmetz, A. Spatial expression of a sunflower SERK gene during induction of somatic embryogenesis and shoot organogenesis. Plant Physiol. Biochem. 2004, 42, 35–42. [Google Scholar] [CrossRef]
- Podio, M.; Felitti, S.A.; Siena, L.A.; Delgado, L.; Mancini, M.; Seijo, J.G.; González, A.M.; Pessino, S.C.; Ortiz, J.P.A. Characterization and expression analysis of SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) genes in sexual and apomictic Paspalum notatum. Plant Mol. Biol. 2014, 84, 479–495. [Google Scholar] [CrossRef]
- Conner, J.A.; Mookkan, M.; Huo, H.; Chae, K.; Ozias-Akins, P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. USA 2015, 112, 11205–11210. [Google Scholar] [CrossRef] [Green Version]
- Conner, J.A.; Podio, M.; Ozias-Akins, P. Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes. Plant Rep. 2017, 30, 41–52. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.-M.; van Lammeren, A.A.M.; Miki, B.L.A.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [Green Version]
- Khanday, I.; Skinner, D.; Yang, B.; Mercier, R.; Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 2019, 565, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Worthington, M.; Heffelfinger, C.; Bernal, D.; Quintero, C.; Zapata, Y.P.; Perez, J.G.; De Vega, J.; Miles, J.; Dellaporta, S.; Tohme, J. A parthenogenesis gene candidate and evidence for segmental allopolyploidy in apomictic Brachiaria decumbens. Genetics 2016, 203, 1117–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, J.B.; Alexander, P.J.; Allphin, L.; Al-Shehbaz, I.A.; Rushworth, C.; Bailey, C.D.; Windham, M.D. Does hybridization drive the transition to asexuality in diploid Boechera? Evolution 2012, 66, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Colombo, L.; Battaglia, R.; Kater, M.M. Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 2008, 13, 444–450. [Google Scholar] [CrossRef]
- Wijnker, E.; Schnittger, A. Control of the meiotic cell division program in plants. Plant Reprod. 2013, 26, 143–158. [Google Scholar] [CrossRef] [Green Version]
- Drews, G.N.; Koltunow, A.M. The female gametophyte. Arab. B. 2011, 9, e0155. [Google Scholar] [CrossRef]
- Schmidt, A.; Schmid, M.W.; Grossniklaus, U. Plant germline formation: Common concepts and developmental flexibility in sexual and asexual reproduction. Development 2015, 142, 229–241. [Google Scholar] [CrossRef] [Green Version]
- Grossniklaus, U.; Schneitz, K. The molecular and genetic basis of ovule and megagametophyte development. Semin. Cell Dev. Biol. 1998, 9, 227–238. [Google Scholar] [CrossRef]
- Ohad, N.; Yadegari, R.; Margossian, L.; Hannon, M.; Michaeli, D.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. Mutations in FIE, a WD Polycomb group gene, allow endosperm development without fertilization. Plant Cell 1999, 11, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Bilodeau, P.; Dennis, E.S.; Peacock, W.J.; Chaudhury, A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 2000, 97, 10637–10642. [Google Scholar] [CrossRef] [Green Version]
- Köhler, C.; Hennig, L.; Spillane, C.; Pien, S.; Gruissem, W.; Grossniklaus, U. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 2003, 17, 1540–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Aguilar, M.; Michaud, C.; Leblanc, O.; Grimanelli, D. Inactivation of a DNA methylation pathway in Maize reproductive organs results in apomixis-like phenotypes. Plant Cell 2010, 22, 3249–3267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Bramsiepe, J.; Van Durme, M.; Komaki, S.; Prusicki, M.A.; Maruyama, D.; Forner, J.; Medzihradszky, A.; Wijnker, E.; Harashima, H.; et al. RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science 2017, 356, eaaf6532. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Wang, S.; Venglat, P.; Zhao, L.; Cheng, Y.; Ye, S.; Qin, Y.; Datla, R.; Zhou, Y.; Wang, H. Arabidopsis ICK/KRP cyclin-dependent kinase inhibitors function to ensure the formation of one megaspore mother cell and one functional megaspore per ovule. PLOS Genet. 2018, 14, e1007230. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Zhao, L.; Zhao, Y.; Li, S.; Won, S.; Cai, H.; Wang, L.; Li, Z.; Chen, P.; Qin, Y.; et al. The THO complex non-cell-autonomously represses female germline specification through the TAS3-ARF3 module. Curr. Biol. 2017, 27, 1597–1609.e2. [Google Scholar] [CrossRef] [Green Version]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Cai, H.; Su, Z.; Wang, L.; Huang, X.; Zhang, M.; Chen, P.; Dai, X.; Zhao, H.; Palanivelu, R.; et al. KLU suppresses megasporocyte cell fate through SWR1-mediated activation of WRKY28 expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E526–E535. [Google Scholar] [CrossRef] [Green Version]
- Nonomura, K.-I.; Miyoshi, K.; Eiguchi, M.; Suzuki, T.; Miyao, A.; Hirochika, H.; Kurata, N. The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 2003, 15, 1728–1739. [Google Scholar] [CrossRef] [Green Version]
- Nonomura, K.-I.; Morohoshi, A.; Nakano, M.; Eiguchi, M.; Miyao, A.; Hirochika, H.; Kurata, N. A germ cell-specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 2007, 19, 2583–2594. [Google Scholar] [CrossRef] [Green Version]
- Olmedo-Monfil, V.; Durán-Figueroa, N.; Arteaga-Vázquez, M.; Demesa-Arévalo, E.; Autran, D.; Grimanelli, D.; Slotkin, R.K.; Martienssen, R.A.; Vielle-Calzada, J.-P. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 2010, 464, 628–632. [Google Scholar] [CrossRef]
- Singh, M.; Goel, S.; Meeley, R.B.; Dantec, C.; Parrinello, H.; Michaud, C.; Leblanc, O.; Grimanelli, D. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 2011, 23, 443–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leblanc, O.; Dueñas, M.; Hernández, M.; Bello, S.; Garcia, V.; Berthaud, J.; Savidan, Y. Chromosome doubling in Tripsacum: The production of artificial, sexual tetraploid plants. Plant Breed. 1995, 114, 226–230. [Google Scholar] [CrossRef]
- Rodríguez-Leal, D.; León-Martínez, G.; Abad-Vivero, U.; Vielle-Calzada, J.P. Natural variation in epigenetic pathways affects the specification of female gamete precursors in Arabidopsis. Plant Cell 2015, 27, 1034–1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baroux, C.; Raissig, M.T.; Grossniklaus, U. Epigenetic regulation and reprogramming during gamete formation in plants. Curr. Opin. Genet. Dev. 2011, 21, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Grimanelli, D. Epigenetic regulation of reproductive development and the emergence of apomixis in angiosperms. Curr. Opin. Plant Biol. 2012, 15, 57–62. [Google Scholar] [CrossRef]
- Rodriguez-Leal, D.; Vielle-Calzada, J.P. Regulation of apomixis: Learning from sexual experience. Curr. Opin. Plant Biol. 2012, 15, 549–555. [Google Scholar] [CrossRef]
- Mercier, R. SWITCH1 (SWI1): A novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev. 2001, 15, 1859–1871. [Google Scholar] [CrossRef] [Green Version]
- Siddiqi, I.; Ganesh, G.; Grossniklaus, U.; Subbiah, V. The dyad gene is required for progression through female meiosis in Arabidopsis. Development 2000, 127, 197–207. [Google Scholar]
- Ravi, M.; Marimuthu, M.P.A.; Siddiqi, I. Gamete formation without meiosis in Arabidopsis. Nature 2008, 451, 1121–1124. [Google Scholar] [CrossRef]
- d’Erfurth, I.; Jolivet, S.; Froger, N.; Catrice, O.; Novatchkova, M.; Mercier, R. Turning meiosis into mitosis. PLoS Biol. 2009, 7, e1000124. [Google Scholar] [CrossRef] [Green Version]
- d’Erfurth, I.; Cromer, L.; Jolivet, S.; Girard, C.; Horlow, C.; Sun, Y.; To, J.P.C.; Berchowitz, L.E.; Copenhaver, G.P.; Mercier, R. The CYCLIN-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet. 2010, 6, e1000989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazaki, S.; Ito, M. Calcium signals for egg activation in mammals. J. Pharmacol. Sci. 2006, 100, 545–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uranga, J.A.; Pedersen, R.A.; Arechaga, J. Parthenogenetic activation of mouse oocytes using calcium ionophores and protein kinase C stimulators. Int. J. Dev. Biol. 1996, 40, 515–519. [Google Scholar] [PubMed]
- Antoine, A.-F.; Faure, J.-E.; Dumas, C.; Feijó, J.A. Differential contribution of cytoplasmic Ca2+ and Ca2+ influx to gamete fusion and egg activation in maize. Nat. Cell Biol. 2001, 3, 1120–1123. [Google Scholar] [CrossRef] [PubMed]
- Guitton, A.-E.; Berger, F. Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr. Biol. 2005, 15, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.C.M.; Luo, M.; Berger, F.; Koltunow, A.M.G. Polycomb group gene function in sexual and asexual seed development in angiosperms. Sex. Plant Reprod. 2010, 23, 123–133. [Google Scholar] [CrossRef]
- Zuo, J.; Niu, Q.-W.; Frugis, G.; Chua, N.-H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002, 30, 349–359. [Google Scholar] [CrossRef]
- Lotan, T.; Ohto, M.; Yee, K.M.; West, M.A.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811. [Google Scholar] [CrossRef] [Green Version]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef]
- Gilles, L.M.; Khaled, A.; Laffaire, J.; Chaignon, S.; Gendrot, G.; Laplaige, J.; Bergès, H.; Beydon, G.; Bayle, V.; Barret, P.; et al. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 2017, 36, 707–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Li, X.; Meng, D.; Zhong, Y.; Chen, C.; Dong, X.; Xu, X.; Chen, B.; Li, W.; Li, L.; et al. A 4-bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant 2017, 10, 520–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, L.; Zhang, Y.; Liu, C.; Liu, Y.; Wang, Y.; Liang, D.; Liu, J.; Sahoo, G.; Kelliher, T. OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 2018, 4, 530–533. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhong, Y.; Qi, X.; Chen, M.; Liu, Z.; Chen, C.; Tian, X.; Li, J.; Jiao, Y.; Wang, D.; et al. Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat. Plant Biotechnol. J. 2020, 18, 316–318. [Google Scholar] [CrossRef] [Green Version]
- Matzk, F.; Meyer, H.-M.; Bäumlein, H.; Balzer, H.-J.; Schubert, I. A novel approach to the analysis of the initiation of embryo development in gramineae. Sex. Plant Rep. 1995, 8, 266–272. [Google Scholar] [CrossRef]
- Horst, N.A.; Katz, A.; Pereman, I.; Decker, E.L.; Ohad, N.; Reski, R. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nat. Plants 2016, 2, 15209. [Google Scholar] [CrossRef]
- Ikeda, T.; Tanaka, W.; Toriba, T.; Suzuki, C.; Maeno, A.; Tsuda, K.; Shiroishi, T.; Kurata, T.; Sakamoto, T.; Murai, M.; et al. BELL 1-like homeobox genes regulate inflorescence architecture and meristem maintenance in rice. Plant J. 2019, 98, 465–478. [Google Scholar] [CrossRef]
- Brambilla, V.; Battaglia, R.; Colombo, M.; Masiero, S.; Bencivenga, S.; Kater, M.M.; Colombo, L. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 2007, 19, 2544–2556. [Google Scholar] [CrossRef] [Green Version]
- Rövekamp, M.; Bowman, J.L.; Grossniklaus, U. Marchantia MpRKD regulates the gametophyte-sporophyte transition by keeping egg cells quiescent in the absence of fertilization. Curr. Biol. 2016, 26, 1782–1789. [Google Scholar] [CrossRef] [Green Version]
- Köszegi, D.; Johnston, A.J.; Rutten, T.; Czihal, A.; Altschmied, L.; Kumlehn, J.; Wüst, S.E.J.; Kirioukhova, O.; Gheyselinck, J.; Grossniklaus, U.; et al. Members of the RKD transcription factor family induce an egg cell-like gene expression program. Plant J. 2011, 67, 280–291. [Google Scholar] [CrossRef] [Green Version]
- León-Martínez, G.; Vielle-Calzada, J.P. Apomixis in flowering plants: Developmental and evolutionary considerations. Curr. Top. Dev. Biol. 2019, 131, 565–604. [Google Scholar] [PubMed]
- Hands, P.; Rabiger, D.S.; Koltunow, A. Mechanisms of endosperm initiation. Plant Rep. 2016, 29, 215–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossniklaus, U.; Vielle-Calzada, J.; Hoeppner, M.; Gagliano, W. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 1998, 280, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, A.M.; Ming, L.; Miller, C.; Craig, S.; Dennis, E.S.; Peacock, W.J. Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1997, 94, 4223–4228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, Y.B.; Pirrotta, V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007, 8, 9–22. [Google Scholar] [CrossRef]
- Makarevich, G.; Villar, C.B.R.; Erilova, A.; Kohler, C. Mechanism of PHERES1 imprinting in Arabidopsis. J. Cell Sci. 2008, 121, 906–912. [Google Scholar] [CrossRef] [Green Version]
- Grossniklaus, U.; Paro, R. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb. Perspect. Biol. 2014, 6, a019331. [Google Scholar] [CrossRef] [Green Version]
- Vinkenoog, R.; Spielman, M.; Adams, S.; Fischer, R.L.; Dickinson, H.G.; Scott, R.J. Hypomethylation promotes autonomous endosperm development and rescues postfertilization lethality in fie mutants. Plant Cell 2000, 12, 2271–2282. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, J.C.M.; Tucker, M.R.; Johnson, S.D.; Hrmova, M.; Koltunow, A.M.G. Sexual and apomictic seed formation in Hieracium requires the plant Polycomb-group gene FERTILIZATION INDEPENDENT ENDOSPERM. Plant Cell 2008, 20, 2372–2386. [Google Scholar] [CrossRef] [Green Version]
- Vogel, B. New Plant Breeding Techniques; Commissioned by the Swiss Federal Office for the Environment [BAFU/FOEN]: Bern, Switzerland, 2016. [Google Scholar]
- Pillot, M.; Baroux, C.; Vazquez, M.A.; Autran, D.; Leblanc, O.; Vielle-Calzada, J.P.; Grossniklaus, U.; Grimanelli, D. Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell 2010, 22, 307–320. [Google Scholar] [CrossRef] [Green Version]
- She, W.; Grimanelli, D.; Rutowicz, K.; Whitehead, M.W.J.; Puzio, M.; Kotliński, M.; Jerzmanowski, A.; Baroux, C. Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development 2013, 140, 4008–4019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossniklaus, U. From sexuality to apomixis: Molecular and genetic approaches. In The flowering of Apomixis: From Mechanisms to Genetic Engineering; Savidan, Y., Carman, J.G., Dresselhaus, T., Eds.; CIMMYT, IRD, European Commission OC VI (FAIR): Mexico City, Mexico, 2001; pp. 168–211. ISBN 970-648-074-9. [Google Scholar]
- Koltunow, A.M. Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 1993, 5, 1425–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimanelli, D.; Leblanc, O.; Perotti, E.; Grossniklaus, U. Developmental genetics of gametophytic apomixis. Trends Genet. 2001, 17, 597–604. [Google Scholar] [CrossRef]
- Richards, A.J. Apomixis in flowering plants: An overview. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2003, 358, 1085–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carman, J.G.; Jamison, M.; Elliott, E.; Dwivedi, K.K.; Naumova, T.N. Apospory appears to accelerate onset of meiosis and sexual embryo sac formation in Sorghum ovules. BMC Plant Biol. 2011, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- Albertini, E.; Barcaccia, G.; Carman, J.G.; Pupilli, F. Did apomixis evolve from sex or was it the other way around? J. Exp. Bot. 2019, 70, 2951–2964. [Google Scholar] [CrossRef]
- Wilson, Z.A.; Yang, C. Plant gametogenesis: Conservation and contrasts in development. Reproduction 2004, 128, 483–492. [Google Scholar] [CrossRef]
- Hisanaga, T.; Yamaoka, S.; Kawashima, T.; Higo, A.; Nakajima, K.; Araki, T.; Kohchi, T.; Berger, F. Building new insights in plant gametogenesis from an evolutionary perspective. Nat. Plants 2019, 5, 663–669. [Google Scholar] [CrossRef]
- Mogie, M. The Evolution of Asexual Reproduction in Plants; Chapman & Hall: London, UK, 1992; ISBN 13: 0-412-44220-5. [Google Scholar]
- Savidan, Y. Apomixis: Genetics and Breeding. In Plant Breeding Reviews; John Wiley & Sons, Inc.: Oxford, UK, 2000; pp. 13–86. [Google Scholar]
- Hojsgaard, D. Transient activation of apomixis in sexual neotriploids may retain genomically altered states and enhance polyploid establishment. Front. Plant Sci. 2018, 9, 230. [Google Scholar] [CrossRef] [Green Version]
- Barke, B.H.; Daubert, M.; Hörandl, E. Establishment of apomixis in diploid F2 hybrids and inheritance of apospory from F1 to F2 hybrids of the Ranunculus auricomus complex. Front. Plant Sci. 2018, 9, 1111. [Google Scholar] [CrossRef]
- Sartor, M.E.; Quarin, C.L.; Espinoza, F. Mode of reproduction of colchicine-induced Paspalum plicatulum tetraploids. Crop Sci. 2009, 49, 1270–1276. [Google Scholar] [CrossRef]
- Varshney, R.K.; Shi, C.; Thudi, M.; Mariac, C.; Wallace, J.; Qi, P.; Zhang, H.; Zhao, Y.; Wang, X.; Rathore, A.; et al. Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat. Biotechnol. 2017, 35, 969–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carballo, J.; Santos, B.A.C.M.; Zappacosta, D.; Garbus, I.; Selva, J.P.; Gallo, C.A.; Díaz, A.; Albertini, E.; Caccamo, M.; Echenique, V. A high-quality genome of Eragrostis curvula grass provides insights into Poaceae evolution and supports new strategies to enhance forage quality. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, J.T.; Willis, J.H.; Mitchell-Olds, T. Evolutionary genetics of plant adaptation. Trends Genet. 2011, 27, 258–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.R.; Wang, B.; Mojica, J.P.; Mandáková, T.; Prasad, K.V.S.K.; Goicoechea, J.L.; Perera, N.; Hellsten, U.; Hundley, H.N.; Johnson, J.; et al. Young inversion with multiple linked QTLs under selection in a hybrid zone. Nat. Ecol. Evol. 2017, 1, 0119. [Google Scholar] [CrossRef]
- Kliver, S.; Rayko, M.; Komissarov, A.; Bakin, E.; Zhernakova, D.; Prasad, K.; Rushworth, C.; Baskar, R.; Smetanin, D.; Schmutz, J.; et al. Assembly of the Boechera retrofracta genome and evolutionary analysis of apomixis-associated genes. Genes (Basel) 2018, 9, 185. [Google Scholar] [CrossRef] [Green Version]
- Sharbel, T.F.; Rigault, P. Comparative Genomics of Apomictic Plants: Advancing Novel Tools for Niche Breeding. Available online: https://www.researchgate.net/project/Comparative-Genomics-of-Apomictic-Plants-Advancing-Novel-Tools-for-Niche-Breeding (accessed on 11 March 2020).
- Brukhin, V.; Grossniklaus, U. Sequencing, Assembly, and Annotation of the Highly Heterozygous Genome of the Apomictic Plant Boechera divaricarpa. Available online: https://www.researchgate.net/project/Sequencing-Assembly-and-Annotation-of-the-Highly-Heterozygous-Genome-of-the-Apomictic-Plant-Boechera-divaricarpa (accessed on 11 March 2020).
- Muir, P.; Li, S.; Lou, S.; Wang, D.; Spakowicz, D.J.; Salichos, L.; Zhang, J.; Weinstock, G.M.; Isaacs, F.; Rozowsky, J.; et al. The real cost of sequencing: Scaling computation to keep pace with data generation. Genome Biol. 2016, 17, 53. [Google Scholar] [CrossRef] [Green Version]
- Jiao, W.-B.; Schneeberger, K. The impact of third generation genomic technologies on plant genome assembly. Curr. Opin. Plant Biol. 2017, 36, 64–70. [Google Scholar] [CrossRef]
- Kyriakidou, M.; Tai, H.H.; Anglin, N.L.; Ellis, D.; Strömvik, M.V. Current strategies of polyploid plant genome sequence assembly. Front. Plant Sci. 2018, 9, 1660. [Google Scholar] [CrossRef]
- Kadota, M.; Nishimura, O.; Miura, H.; Tanaka, K.; Hiratani, I.; Kuraku, S. Multifaceted Hi-C benchmarking: What makes a difference in chromosome-scale genome scaffolding? Gigascience 2020, 9, 1–15. [Google Scholar] [CrossRef]
- Blackburn, A.N.; Blondell, L.; Kos, M.Z.; Blackburn, N.B.; Peralta, J.M.; Stevens, P.T.; Lehman, D.M.; Blangero, J.; Göring, H.H.H. Genotype phasing in pedigrees using whole-genome sequence data. Eur. J. Hum. Genet. 2020. [Google Scholar] [CrossRef] [PubMed]
- Castel, S.E.; Mohammadi, P.; Chung, W.K.; Shen, Y.; Lappalainen, T. Rare variant phasing and haplotypic expression from RNA sequencing with phASER. Nat. Commun. 2016, 7, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hojsgaard, D. Apomixis Technology: Separating the Wheat from the Chaff. Genes 2020, 11, 411. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11040411
Hojsgaard D. Apomixis Technology: Separating the Wheat from the Chaff. Genes. 2020; 11(4):411. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11040411
Chicago/Turabian StyleHojsgaard, Diego. 2020. "Apomixis Technology: Separating the Wheat from the Chaff" Genes 11, no. 4: 411. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11040411