Salinity Response in Chloroplasts: Insights from Gene Characterization
Abstract
:1. Introduction
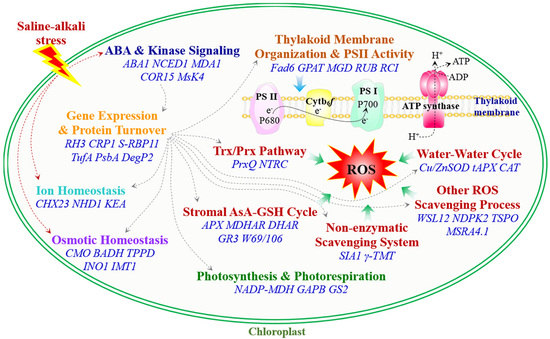
2. Salinity-Induced Diverse ROS Scavenging Pathways in Chloroplasts
2.1. The Water-Water Cycle Detoxifies O2− and H2O2
2.2. Stromal Ascorbate (AsA)-Glutathione (GSH) Cycle
2.3. Thioredoxin/Peroxiredoxin (Trx/Prx) and Glutathione Peroxidase (GPX) Pathway
2.4. Non-Enzymatic OH• and 1O2 Scavenging System
2.5. Other Genes Involved in Chloroplast ROS Scavenging
3. Thylakoid Membrane Organization and Photosynthesis
4. Osmotic and Ion Homeostasis
5. ABA and Kinase Signaling Pathways
6. Chloroplast Gene Expression and Protein Turnover
7. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| 3-PGA | Glycerate-3-phosphate |
| ABA | Abscisic acid |
| ABC1K | Activity of bc1 complex-like kinase |
| ABI4 | Abscisic acid insensitive 4 |
| APX/tAPX | Ascorbate peroxidase/Thylakoid ascorbate peroxidase |
| AsA | Ascorbate |
| BADH | Betaine aldehyde dehydrogenase |
| CAM | Crassulacean acid metabolism |
| CAT | Catalase |
| CMO | Choline monooxygenase |
| CO2 | Carbon dioxide |
| COR | Cold-responsive protein |
| Cu/Zn SOD | Copper/zinc superoxide dismutase |
| DegP2 | Prokaryotic trypsin-type Deg/Htr serine protease |
| DGDG | Digalactodiacylglycerol |
| DHA | Dehydroascorbate |
| DHAR | Dehydroascorbate reductase |
| EF-Tu | Translation elongation factor |
| ETC | Electron transport chain |
| GAPB | Glyceraldehyde 3-phosphate dehydrogenase beta subunit |
| GPAT | Glycerol-3-phosphate acyltransferase |
| GPX | Glutathione peroxidase |
| GR | Glutathione reductase |
| GS2 | Glutamine synthetase |
| GSH | Reduced glutathione |
| GSSG | Oxidized glutathione |
| H2O2 | Hydrogen peroxide |
| HKT1 | High-affinity K+ transporter 1 |
| KEA | K+-efflux antiporter |
| MDHAR | Monodehydroascorbate reductase |
| Met | Methionine |
| MGD | Monogalactosyldiacylglycerol synthase |
| MGDG | Monogalactosyldiacylglycerol |
| MSR | Methionine sulfoxide reductase |
| NADP-MDH | NADP+-dependent malate dehydrogenase |
| NDPK2 | Nucleoside diphosphate kinase 2 |
| NTRC | NADPH thioredoxin reductase |
| 1O2 | Singlet oxygen |
| O2 | Oxygen |
| O2− | Superoxide anion |
| C | Oxaloacetate |
| OH• | Hydroxyl radical |
| PS II | Photosystem II |
| γ-TMT | γ-tocopherol methyltransferase |
| RBP | RNA binding protein |
| RH | DEAD-box RNA helicase |
| ROS | Reactive oxygen species |
| RRM | RNA-recognition motif |
| RUB | Rubredoxin |
| Rubisco | Ribulose-1,5-bisphosphate carboxylase/oxygenase |
| TPP | Trehalose-6-phosphate phosphatase |
| Trx/Prx | Thioredoxin/Peroxiredoxin |
| TSPO | 18 kDa translocator protein |
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Chapter twenty-four-mechanisms of high salinity tolerance in plants. Methods Enzymol. 2007, 428, 419–438. [Google Scholar] [PubMed]
- Van Wijk, K.J. Proteomics of the chloroplast: Experimentation and prediction. Trends Plant Sci. 2000, 5, 420–425. [Google Scholar] [CrossRef]
- Baginsky, S.; Gruissem, W. Chloroplast proteomics: Potentials and challenges. J. Exp. Bot. 2004, 55, 1213–1220. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, F.; Salamini, F.; Leister, D. A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 2000, 5, 141–142. [Google Scholar] [CrossRef]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Leister, D. Chloroplast research in the genomic age. Trends Genet. 2003, 19, 47–56. [Google Scholar] [CrossRef]
- Peltier, J.B.; Emanuelsson, O.; Kalume, D.E.; Ytterberg, J.; Friso, G.; Rudella, A.; Liberles, D.A.; Söderberg, L.; Roepstorff, P.; von Heijne, G.; et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 2002, 14, 211–236. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liang, W.; Xing, J.; Tan, F.; Chen, Y.; Huang, L.; Cheng, C.L.; Chen, W. Dynamics of chloroplast proteome in salt-stressed mangrove Kandelia candel (L.) Druce. J. Proteome Res. 2013, 12, 5124–5136. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Feng, J.; Jiang, P.; Chen, X.; Bao, H.; Nie, L.; Jiang, D.; Lv, S.; Kuang, T.; Li, Y. Coordination of carbon fixation and nitrogen metabolism in Salicornia europaea under salinity: Comparative proteomic analysis on chloroplast proteins. Proteomics 2011, 11, 4346–4367. [Google Scholar] [CrossRef] [PubMed]
- Zörb, C.; Herbst, R.; Forreiter, C.; Schubert, S. Short-term effects of salt exposure on the maize chloroplast protein pattern. Proteomics 2009, 9, 4209–4220. [Google Scholar] [CrossRef] [PubMed]
- Do Amaral, M.N.; Arge, L.W.P.; Benitez, L.C.; Danielowski, R.; da Silveira Silveira, S.F.; da Rosa Farias, D.; de Oliveira, A.C.; da Maia, L.C.; Braga, E.J.B. Comparative transcriptomics of rice plants under cold, iron, and salt stresses. Funct. Integr. Genom. 2016, 16, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Edreva, A. Generation and scavenging of reactive oxygen species in chloroplasts: A submolecular approach. Agr. Ecosyst. Environ. 2005, 106, 119–133. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, J.; Li, X.; Xu, J.; Wang, L. Identification and characterization of a PutCu/Zn-SOD gene from Puccinellia tenuiflora (Turcz.) Scribn. et Merr. Plant Growth Regul. 2016, 79, 55–64. [Google Scholar] [CrossRef]
- Badawi, G.H.; Yamauchi, Y.; Shimada, E.; Sasaki, R.; Kawano, N.; Tanaka, K.; Tanaka, K. Enhanced tolerance to salt stress and water deficit by overexpressing superoxide dismutase in tobacco (Nicotiana tabacum) chloroplasts. Plant Sci. 2004, 166, 919–928. [Google Scholar] [CrossRef]
- Jing, X.; Hou, P.; Lu, Y.; Deng, S.; Li, N.; Zhao, R.; Sun, J.; Wang, Y.; Han, Y.; Lang, T.; et al. Overexpression of copper/zinc superoxide dismutase from mangrove Kandelia candel in tobacco enhances salinity tolerance by the reduction of reactive oxygen species in chloroplast. Front. Plant Sci. 2015, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Tseng, M.J.; Liu, C.W.; Yiu, J.C. Enhanced tolerance to sulfur dioxide and salt stress of transgenic Chinese cabbage plants expressing both superoxide dismutase and catalase in chloroplasts. Plant Physiol. Biochem. 2007, 45, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Wu, J.; Li, Y.; Nan, Z.; Guo, X.; Wang, Y.; Zhang, A.; Wang, Z.; Xia, G.; Tian, Y. Synergistic effects of GhSOD1 and GhCAT1 overexpression in cotton chloroplasts on enhancing tolerance to methyl viologen and salt stresses. PLoS ONE 2013, 8, e54002. [Google Scholar] [CrossRef] [PubMed]
- Badawi, G.H.; Kawano, N.; Yamauchi, Y.; Shimada, E.; Sasaki, R.; Kubo, A.; Tanaka, K. Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiol. Plant. 2004, 121, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Sheptovitsky, Y.G.; Brudvig, G.W. Isolation and characterization of spinach photosystem II membrane-associated catalase and polyphenol oxidase. Biochemistry 1996, 35, 16255–16263. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, K.; George, S.; Venkataraman, G.; Parida, A. A salt-inducible chloroplastic monodehydroascorbate reductase from halophyte Avicennia marina confers salt stress tolerance on transgenic plants. Biochimie 2010, 92, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Le Martret, B.; Poage, M.; Shiel, K.; Nugent, G.D.; Dix, P.J. Tobacco chloroplast transformants expressing genes encoding dehydroascorbate reductase, glutathione reductase, and glutathione-S-transferase, exhibit altered anti-oxidant metabolism and improved abiotic stress tolerance. Plant Biotechnol. J. 2011, 9, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.M.; Lin, W.R.; Kao, Y.T.; Hsu, Y.T.; Yeh, C.H.; Hong, C.Y.; Kao, C.H. Identification and characterization of a novel chloroplast/mitochondria co-localized glutathione reductase 3 involved in salt stress response in rice. Plant Mol. Biol. 2013, 83, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.M.; Lin, W.R.; Kao, C.H.; Hong, C.Y. Gene knockout of glutathione reductase 3 results in increased sensitivity to salt stress in rice. Plant Mol. Boil. 2015, 87, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.W.; Chen, S.H.; Guo, X.L.; Zhang, H.; Zhao, Y.X. Overexpression of a chloroplast-located peroxiredoxin Q gene, SsPrxQ, increases the salt and low-temperature tolerance of Arabidopsis. J. Integr. Plant Biol. 2006, 48, 1244–1249. [Google Scholar] [CrossRef]
- Serrato, A.J.; Pérez-Ruiz, J.M.; Spínola, M.C.; Cejudo, F.J. A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 43821–43827. [Google Scholar] [CrossRef] [PubMed]
- Zhai, C.Z.; Zhao, L.; Yin, L.J.; Chen, M.; Wang, Q.Y.; Li, L.C.; Xu, Z.S.; Ma, Y.Z. Two wheat glutathione peroxidase genes whose products are located in chloroplasts improve salt and H2O2 tolerances in Arabidopsis. PLoS ONE 2013, 8, e73989. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, Q.; Li, T.; Du, J.; Yang, S.; Yang, C. AtSIA1, an ABC1-like kinase, regulates salt response in Arabidopsis. Biologia 2012, 67, 1107–1111. [Google Scholar] [CrossRef]
- Jin, S.; Daniell, H. Expression of γ-tocopherol methyltransferase in chloroplasts results in massive proliferation of the inner envelope membrane and decreases susceptibility to salt and metal-induced oxidative stresses by reducing reactive oxygen species. Plant Biotechnol. J. 2014, 12, 1274–1285. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Hu, S.; Wu, L.; Ge, C.; Cui, Y.; Chen, P.; Wang, X.; Xu, J.; Ren, D.; Dong, G.; et al. White stripe leaf 12 (WSL12), encoding a nucleoside diphosphate kinase 2 (OsNDPK2), regulates chloroplast development and abiotic stress response in rice (Oryza sativa L.). Mol. Breed. 2016, 36, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Lim, S.; Yang, K.S.; Kim, C.Y.; Kwon, S.Y.; Lee, H.S.; Wang, X.; Zhou, Z.; Ma, D.; Yun, D.J.; et al. Expression of Arabidopsis NDPK2 increases antioxidant enzyme activities and enhances tolerance to multiple environmental stresses in transgenic sweetpotato plants. Mol. Breed. 2009, 24, 233–244. [Google Scholar] [CrossRef]
- Balsemão-Pires, E.; Jaillais, Y.; Olson, B.J.; Andrade, L.R.; Umen, J.G.; Chory, J.; Sachetto-Martins, G. The Arabidopsis translocator protein (AtTSPO) is regulated at multiple levels in response to salt stress and perturbations in tetrapyrrole metabolism. BMC Plant Biol. 2011, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wu, Y.; Wang, Y.; Chen, Y.; Chu, C. OsMSRA4.1 and OsMSRB1.1, two rice plastidial methionine sulfoxide reductases, are involved in abiotic stress responses. Planta 2009, 230, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.T.; Zhu, J.Q.; Zhu, Q.; Liu, H.; Gao, X.S.; Zhang, H.X. Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2009, 390, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.L.; Li, F.; Su, N.; Sun, X.L.; Zhao, S.J.; Meng, Q.W. The increase in unsaturation of fatty acids of phosphatidylglycerol in thylakoid membrane enhanced salt tolerance in tomato. Photosynthetica 2010, 48, 400–408. [Google Scholar] [CrossRef]
- Wang, S.; Uddin, M.I.; Tanaka, K.; Yin, L.; Shi, Z.; Qi, Y.; Mano, J.; Matsui, K.; Shimomura, N.; Sakaki, T.; et al. Maintenance of chloroplast structure and function by overexpression of the OsMGD gene leads to enhanced salt tolerance in tobacco. Plant Physiol. 2014, 165, 1144–1155. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, P.; Takano, T.; Liu, S. A chloroplast-localized rubredoxin family protein gene from Puccinellia tenuiflora (PutRUB) increases NaCl and NaHCO3 tolerance by decreasing H2O2 accumulation. Int. J. Mol. Sci. 2016, 17, 804. [Google Scholar] [CrossRef] [PubMed]
- Khurana, N.; Chauhan, H.; Khurana, P. Characterization of a chloroplast localized wheat membrane protein (TaRCI) and its role in heat, drought and salinity stress tolerance in Arabidopsis thaliana. Plant Gene 2015, 4, 45–54. [Google Scholar] [CrossRef]
- Cushman, J.C. Molecular cloning and expression of chloroplast NADP-malate dehydrogenase during Crassulacean acid metabolism induction by salt stress. Photosynth. Res. 1993, 35, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Guo, A.; Jin, X.; Yang, Q.; Wang, D.; Sun, Y.; Huang, Q.; Wang, L.; Peng, C.; Wang, X. The β subunit of glyceraldehyde 3-phosphate dehydrogenase is an important factor for maintaining photosynthesis and plant development under salt stress-based on an integrative analysis of the structural, physiological and proteomic changes in chloroplasts in Thellungiella halophila. Plant Sci. 2015, 236, 223–238. [Google Scholar] [PubMed]
- Hoshida, H.; Tanaka, Y.; Hibino, T.; Hayashi, Y.; Tanaka, A.; Takabe, T.; Takabe, T. Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol. Biol. 2000, 43, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Zhao, R.; Li, F.; Tang, W.; Han, L. Simultaneous expression of Spinacia oleracea chloroplast choline monooxygenase (CMO) and betaine aldehyde dehydrogenase (BADH) genes contribute to dwarfism in transgenic Lolium perenne. Plant Mol. Biol. Report. 2011, 29, 379–388. [Google Scholar] [CrossRef]
- Zhang, J.; Tan, W.; Yang, X.H.; Zhang, H.X. Plastid-expressed choline monooxygenase gene improves salt and drought tolerance through accumulation of glycine betaine in tobacco. Plant Cell Rep. 2008, 27, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Dhingra, A.; Daniell, H. Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confers enhanced salt tolerance. Plant Physiol. 2004, 136, 2843–2854. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liang, Z.; Wen, X.; Lu, C. Genetic engineering of the biosynthesis of glycinebetaine leads to increased tolerance of photosynthesis to salt stress in transgenic tobacco plants. Plant Mol. Biol. 2008, 66, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Broyart, C.; Rabanal, F.A.; Jonak, C. The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxid. Redox Signal. 2014, 21, 1289–1304. [Google Scholar] [CrossRef] [PubMed]
- Patra, B.; Ray, S.; Richter, A.; Majumder, A.L. Enhanced salt tolerance of transgenic tobacco plants by co-expression of PcINO1 and McIMT1 is accompanied by increased level of myo-inositol and methylated inositol. Protoplasma 2010, 245, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Song, C.P.; Guo, Y.; Qiu, Q.; Lambert, G.; Galbraith, D.W.; Jagendorf, A.; Zhu, J.K. A probable Na+(K+)/H+ exchanger on the chloroplast envelope functions in pH homeostasis and chloroplast development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10211–10216. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Kunz, H.H.; Schroeder, J.I.; Kemp, G.; Young, H.S.; Neuhaus, H.E. Decreased capacity for sodium export out of Arabidopsis chloroplasts impairs salt tolerance, photosynthesis and plant performance. Plant J. 2014, 78, 646–658. [Google Scholar] [CrossRef] [PubMed]
- Kunz, H.H.; Gierth, M.; Herdean, A.; Satoh-Cruz, M.; Kramer, D.M.; Spetea, C.; Schroeder, J.I. Plastidial transporters KEA1, -2, and-3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 7480–7485. [Google Scholar] [CrossRef] [PubMed]
- Li, P.C.; Huang, J.G.; Yu, S.W.; Li, Y.Y.; Sun, P.; Wu, C.A.; Zheng, C.C. Arabidopsis YL1/BPG2 is involved in seedling shoot response to salt stress through ABI4. Sci. Rep. 2016, 6, 30163. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Lee, H.; Ishitani, M.; Zhu, J.K. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 2002, 277, 8588–8596. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Robles, P.; Micol, J.L.; Quesada, V. Arabidopsis MDA1, a nuclear-encoded protein, functions in chloroplast development and abiotic stress responses. PLoS ONE 2012, 7, e42924. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Hou, L.; Li, W.C.; Cheng, J.F.; Fu, Y.Q. COR15B expression is affected by chloroplast functionality and its role in response to salt stress in Arabidopsis thaliana. Biol. Plant. 2014, 58, 667–675. [Google Scholar] [CrossRef]
- Kempa, S.; Rozhon, W.; Samaj, J.; Erban, A.; Baluška, F.; Becker, T.; Haselmayer, J.; Schleiff, E.; Kopka, J.; Hirt, H.; et al. A plastid-localized glycogen synthase kinase 3 modulates stress tolerance and carbohydrate metabolism. Plant J. 2007, 49, 1076–1090. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Xu, T.; Lee, K.; Lee, K.H.; Kang, H. A chloroplast-localized DEAD-box RNA helicase AtRH3 is essential for intron splicing and plays an important role in the growth and stress response in Arabidopsis thaliana. Plant Physiol. Biochem. 2014, 82, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Sy, N.D.; Lee, H.J.; Kwak, K.J.; Gu, L.; Kim, J.I.; Kang, H. Functional characterization of a chloroplast-targeted RNA-binding protein CRP1 in Arabidopsis thaliana under abiotic stress conditions. J. Plant Biol. 2014, 57, 349–356. [Google Scholar] [CrossRef]
- Lee, S.Y.; Seok, H.Y.; Tarte, V.N.; Woo, D.H.; Le, D.H.; Lee, E.H.; Moon, Y.H. The Arabidopsis chloroplast protein S-RBP11 is involved in oxidative and salt stress responses. Plant Cell Rep. 2014, 33, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Mishra, R.N.; Agarwal, P.K.; Goswami, M.; Nair, S.; Sopory, S.K.; Reddy, M.K. A pea chloroplast translation elongation factor that is regulated by abiotic factors. Biochem. Biophys. Res. Commun. 2004, 320, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Allakhverdiev, S.I.; Nishiyama, Y.; Miyairi, S.; Yamamoto, H.; Inagaki, N.; Kanesaki, Y.; Murata, N. Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbA genes in Synechocystis. Plant Physiol. 2002, 130, 1443–1453. [Google Scholar] [CrossRef] [PubMed]
- Haußühl, K.; Andersson, B.; Adamska, I. A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 2001, 20, 713–722. [Google Scholar] [PubMed]
- Sevilla, F.; Camejo, D.; Ortiz-Espín, A.; Calderón, A.; Lázaro, J.J.; Jiménez, A. The thioredoxin/peroxiredoxin/sulfiredoxin system: Current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J. Exp. Bot. 2015, 66, 2945–2955. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Martinis, J.; Glauser, G.; Valimareanu, S.; Kessler, F. A chloroplast ABC1-like kinase regulates vitamin E metabolism in Arabidopsis. Plant Physiol. 2013, 162, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.A.; Moon, H.; Kim, G.; Lim, C.J.; Hong, J.C.; Lim, C.O.; Yun, D.J. NDP kinase 2 regulates expression of antioxidant genes in Arabidopsis. Proc. Jpn. Acad. Ser. B 2003, 79, 86–91. [Google Scholar] [CrossRef]
- Upchurch, R.G. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 2008, 30, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Loll, B.; Kern, J.; Saenger, W.; Zouni, A.; Biesiadka, J. Lipids in photosystem II: Interactions with protein and cofactors. Biochim. Biophys. Acta 2007, 1767, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.G. Membrane lipids: It’s only a phase. Curr. Biol. 2000, 10, R377–R380. [Google Scholar] [CrossRef]
- Jordan, P.; Fromme, P.; Witt, H.T.; Klukas, O.; Saenger, W.; Krauß, N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 2001, 411, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Mizusawa, N.; Wada, H. The role of lipids in photosystem II. Biochim. Biophys. Acta 2012, 1817, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Shimojima, M.; Ohta, H. Critical regulation of galactolipid synthesis controls membrane differentiation and remodeling in distinct plant organs and following environmental changes. Prog. Lipid Res. 2011, 50, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, D.M. Microbial detoxification of superoxide: The non-heme iron reductive paradigm for combating oxidative stress. Acc. Chem. Res. 2004, 37, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Calderon, R.H.; García-Cerdán, J.G.; Malnoë, A.; Cook, R.; Russell, J.J.; Gaw, C.; Dent, R.M.; de Vitry, C.; Niyogi, K.K. A conserved rubredoxin is necessary for photosystem II accumulation in diverse oxygenic photoautotrophs. J. Biol. Chem. 2013, 288, 26688–26696. [Google Scholar] [CrossRef] [PubMed]
- Majumder, A.L.; Sengupta, S.; Goswami, L. Osmolyte regulation in abiotic stress. In Abiotic Stress Adaptation in Plants; Pareek, A., Sopory, S.K., Bohnert, H.J., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 349–370. [Google Scholar]
- Rhodes, D.; Hanson, A.D. Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 357–384. [Google Scholar] [CrossRef]
- Papageorgiou, G.C.; Murata, N. The unusually strong stabilizing effects of glycine betaine on the structure and function of the oxygen-evolving photosystem II complex. Photosynth. Res. 1995, 44, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H.; Murata, N. Glycinebetaine: An effective protectant against abiotic stress in plants. Trends Plant Sci. 2008, 13, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Wang, W.; Liang, C.; Wang, X.; Wang, G.; Wang, W. Overaccumulation of glycine betaine makes the function of the thylakoid membrane better in wheat under salt stress. Crop J. 2017, 5, 73–82. [Google Scholar] [CrossRef]
- Berthomieu, P.; Conéjéro, G.; Nublat, A.; Brackenbury, W.J.; Lambert, C.; Savio, C.; Uozumi, N.; Oiki, S.; Yamada, K.; Cellier, F. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003, 22, 2004–2014. [Google Scholar] [CrossRef] [PubMed]
- Shkolnik-Inbar, D.; Adler, G.; Bar-Zvi, D. ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J. 2013, 73, 993–1005. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; Kim, D.H.; Hwang, I. ABA homeostasis and signaling involving multiple subcellular compartments and multiple receptors. Plant Cell Rep. 2013, 32, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Park, S.J.; Kang, H. Regulation of RNA metabolism in plant development and stress responses. J. Plant Biol. 2013, 56, 123–129. [Google Scholar] [CrossRef]
- Rajkowitsch, L.; Chen, D.; Stampfl, S.; Semrad, K.; Waldsich, C.; Mayer, O.; Jantsch, M.F.; Konrat, R.; Bläsi, U.; Schroeder, R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007, 4, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Caldas, T.D.; El Yaagoubi, A.; Richarme, G. Chaperone properties of bacterial elongation factor EF-Tu. J. Biol. Chem. 1998, 273, 11478–11482. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Badger, M.R. Photoprotection in plants: A new light on photosystem II damage. Trends Plant Sci. 2011, 16, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Nixon, P.J.; Barker, M.; Boehm, M.; de Vries, R.; Komenda, J. FtsH-mediated repair of the photosystem II complex in response to light stress. J. Exp. Bot. 2005, 56, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Nie, L.; Jiang, P.; Feng, J.; Lv, S.; Chen, X.; Bao, H.; Guo, J.; Tai, F.; Wang, J.; et al. Transcriptome analysis of Salicornia europaea under saline conditions revealed the adaptive primary metabolic pathways as early events to facilitate salt adaptation. PLoS ONE 2013, 8, e80595. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Plant Species | Encoding Protein | Salt Treatment Condition | Ref. |
|---|---|---|---|---|
| Stress and defense (22) | ||||
| Cu/Zn SOD | Alkaligrass (Puccinellia tenuiflora) | Copper/zinc superoxide dismutase | NaCl (0, 50, 100 mM; 21 days); NaHCO3 (0, 3, 5 mM; 21 days) | [16] |
| Cu/Zn SOD | Rice (Oryza sativa) | Copper/zinc superoxide dismutase | NaCl (300 mM; 2, 4, 6, 8, 10 days) | [17] |
| Cu/Zn SOD | Mangrove (Kandelia candel) | Copper/zinc superoxide dismutase | NaCl (100, 300 mM; 8, 24 h; 1, 2, 3 weeks) | [18] |
| Cu/Zn SOD | Maize (Zea mays) | Copper/zinc superoxide dismutase | NaCl (0, 50, 100, 150, 200 mM; 10 days; 4 weeks) | [19] |
| Cu/Zn SOD | Cotton (Gossypium hirsutum) | Copper/zinc superoxide dismutase | NaCl (50, 100, 150, 200 mM; 1, 2, 3, 4 weeks) | [20] |
| APX | Arabidopsis thaliana | Ascorbate peroxidase | NaCl (300 mM; 8 days) | [21] |
| CAT | Maize (Z. mays) | Catalase | NaCl (0, 50, 100, 150, 200 mM; 10 days; 4 weeks) | [19] |
| CAT | Cotton (G. hirsutum) | Catalase | NaCl (50, 100, 150, 200 mM; 1, 2, 3, 4 weeks) | [20] |
| MDHAR | Mangrove (Avicennia marina) | Monodehydroascorbate reductase | NaCl (200 mM; 4 days) | [23] |
| DHAR | Rice (O. sativa) | Dehydroascorbate reductase | NaCl (100, 150, 200 mM; 12, 14 days) | [24] |
| GR3 | Rice (O. sativa) | Glutathione reductase | NaCl (200 mM; 0, 4, 8, 12, 16, 20, 24 days) | [25] |
| GR3 | Rice (O. sativa) | Glutathione reductase | NaCl (100 mM; 7, 15 days) | [26] |
| PrxQ | Suaeda salsa | Peroxiredoxin Q | NaCl (0, 100, 150 mM; 3 weeks) | [27] |
| NTRC | Rice (O. sativa) | NADPH thioredoxin reductase | NaCl (170 mM; 1, 3 days) | [28] |
| W69 | Wheat (Triticum aestivum) | Glutathione peroxidase | NaCl (150 mM; 7 days) | [29] |
| W106 | Wheat (T. aestivum) | Glutathione peroxidase | NaCl (150 mM; 7 days) | [29] |
| SIA1 | Arabidopsis | ABC1-like kinase | NaCl (200 mM; 3 days) | [30] |
| γ-TMT | Arabidopsis | γ-tocopherol methyltransferase | NaCl (0, 200, 300, 400 mM; 12, 24, 48 h; 4 weeks) | [31] |
| WSL12 | Rice (O. sativa) | Nucleoside diphosphate kinase | NaCl (100, 150, 200 mM; 0, 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 5.5, 6, 6.5 days) | [32] |
| NDPK2 | Arabidopsis | Nucleoside diphosphate kinase 2 | NaCl (200 mM; 14 days) | [33] |
| TSPO | Arabidopsis | 18 kDa translocator protein | NaCl (150 mM; 1, 3, 6, 12, 24 h) | [34] |
| MSRA4.1 | Rice (O. sativa) | Methionine sulfoxide reductase | NaCl (100 mM; 2 days) | [35] |
| Thylakoid membrane organization and PSII activity (5) | ||||
| Fad6 | Arabidopsis | ω-6 desaturase | NaCl (0, 75, 100, 125 mM; 8 days); (300 mM; 0, 1, 3, 6, 12, 24 h) | [36] |
| GPAT | Tomato (Lycopersicon esculentum) | Glycerol-3-phosphate acyltransferase | NaCl (200 mM; 1, 3, 5, 7 days); (150 mM; 30 days) | [37] |
| MGD | Rice (O. sativa) | Monogalactosyl-diacylglycerol synthase | NaCl (0, 200 mM; 10 days) | [38] |
| RUB | Alkaligrass (P. tenuiflora) | Rubredoxin family protein | NaCl (100, 125 mM; 10 days); NaHCO3 (1.5, 3 mM; 10 days) | [39] |
| RCI | Wheat (T. aestivum) | Rare cold inducible protein | NaCl (150 mM; 2 weeks) | [40] |
| Photosynthesis and photorespiration (3) | ||||
| NADP-MDH | Mesembryanthemum crystallinum | NADP+-dependent malate dehydrogenase | NaCl (400 mM; 1, 6, 12, 30, 72, 126 h) | [41] |
| GAPB | Thellungiella halophila | Glyceraldehyde 3 phosphate dehydrogenase β subunit | NaCl (200 mM; 2 weeks) | [42] |
| GS2 | Rice (O. sativa) | Glutamine synthetase | NaCl (150 mM, 12 days) | [43] |
| Osmotic and ion homoestasis (12) | ||||
| CMO | Spinach (Spinacia oleracea) | Choline monooxygenase | NaCl (50, 100 mM; 0, 3, 6, 9, 12, 15 weeks) | [44] |
| CMO | Beet (Beta vulgaris) | Choline monooxygenase | NaCl (0, 100, 150 mM; 36 days) | [45] |
| BADH | Spinach (S. oleracea) | Betaine aldehyde dehydrogenase | NaCl (50, 100 mM; 0, 3, 6, 9, 12, 15 weeks) | [44] |
| BADH | Tobacco (Nicotiana tabacum) | Betaine aldehyde dehydrogenase | NaCl (100, 200, 300, 400, 500 mM; 1 month) | [46] |
| BADH | Spinach (S. oleracea) | Betaine aldehyde dehydrogenase | NaCl (0, 75, 150 mM; 3 weeks) | [47] |
| TPPD | Arabidopsis | Trehalose-6-phosphate phosphatase | NaCl (200 mM; 0, 1, 3, 8 h) | [48] |
| INO1 | Porteresia coarctata | l-myo-inositol 1-phosphate synthase | NaCl (100, 200, 300, 400 mM; 96 h) | [49] |
| IMT1 | M. crystallinum | Inositol methyl transferase | NaCl (100, 200, 300, 400 mM; 96 h) | [49] |
| CHX23 | Arabidopsis | Na+(K+)/H+ exchanger | NaCl (75 mM; 12 days) | [50] |
| NHD1 | Arabidopsis | Sodium hydrogen antiporter | NaCl (150 mM; 72 h) | [51] |
| KEA | Arabidopsis | K+/H+ antiporter | NaCl (75 mM) | [52] |
| YL1 | Arabidopsis | YqeH-type GTPase | NaCl (0, 150 mM; 2 days) | [53] |
| ABA and kinase signaling (5) | ||||
| ABA1 | Arabidopsis | Zeaxanthin epoxidase | NaCl (300 mM; 3 h) | [54] |
| NCED1 | Cowpea (Vigna unguiculata) | 9-cis-epoxycarotenoid dioxygenase | NaCl (250 mM; 0, 1, 2, 5, 10, 24 h) | [55] |
| MDA1 | Arabidopsis | Transcription termination factor | NaCl (100, 150, 200 mM; 4, 9, 10, 14 days) | [56] |
| COR15 | Arabidopsis | 15 kDa protein | NaCl (150 mM; 3 days) | [57] |
| MsK4 | Alfalfa (Medicago sativa) | Glycogen synthase kinase 3 like kinase | NaCl (100 mM; 4 weeks) | [58] |
| Gene expression and protein turnover (6) | ||||
| RH3 | Arabidopsis | DEAD-box RNA helicase | NaCl (100 mM; 0, 4, 12, 24 h) | [59] |
| CRP1 | Arabidopsis | Chloroplast-targeted RNA-binding protein 1 | NaCl (150 mM, 7 days) | [60] |
| S-RBP11 | Arabidopsis | RNA-binding group protein | NaCl (0, 130, 140, 150, 160 mM; 14 days); (300 mM; 0, 1, 2, 4, 8 h) | [61] |
| TufA | Pea (Pisum sativum) | Chloroplast translation elongation factor | NaCl (100, 200, 500 mM; 4, 6, 24 h) | [62] |
| PsbA | Synechocystis sp. PCC 6803 | Photosystem II D1 protein | NaCl (20 mM, 500 mM, 1000 mM; 0, 1, 2, 3, 4 h) | [63] |
| DegP2 | Arabidopsis | Prokaryotic trypsin-type Deg/Htr serine protease | NaCl (400 mM; 2 h) | [64] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suo, J.; Zhao, Q.; David, L.; Chen, S.; Dai, S. Salinity Response in Chloroplasts: Insights from Gene Characterization. Int. J. Mol. Sci. 2017, 18, 1011. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18051011
Suo J, Zhao Q, David L, Chen S, Dai S. Salinity Response in Chloroplasts: Insights from Gene Characterization. International Journal of Molecular Sciences. 2017; 18(5):1011. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18051011
Chicago/Turabian StyleSuo, Jinwei, Qi Zhao, Lisa David, Sixue Chen, and Shaojun Dai. 2017. "Salinity Response in Chloroplasts: Insights from Gene Characterization" International Journal of Molecular Sciences 18, no. 5: 1011. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18051011