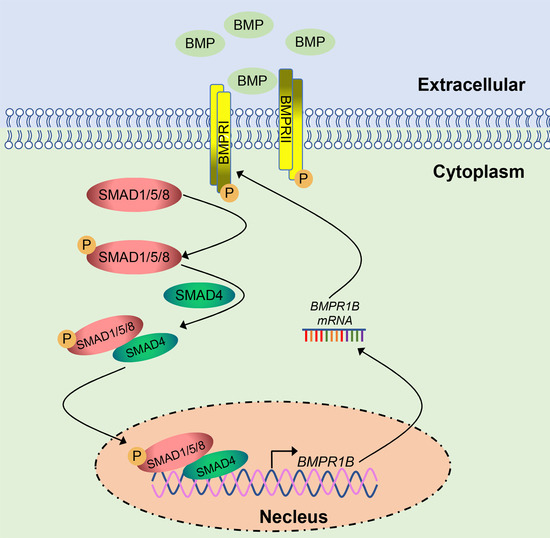
Smad4 Feedback Enhances BMPR1B Transcription in Ovine Granulosa Cells
Abstract
:1. Introduction
2. Results
2.1. Identification of the Transcription Start Sites of the Ovine BMPR1B Gene
2.2. Identification of PII Promoter Region of the Ovine BMPR1B Gene
2.3. Characterization of the PII Core Promoter Region of the Ovine BMPR1B Gene
2.4. Smad4 Enhances PII Promoter Activity of the Ovine BMPR1B Gene
2.5. Smad4 Direct Binds to SBE Motif of the Ovine BMPR1B Promoter Region
2.6. Smad4 Positive Feedback Regulates BMPR1B Expression in the Ovine Granulosa Cells
2.7. Smad4 Rescues BMPR1B-siRNA-Induced Cell Apoptosis in the Ovine Granulosa Cells
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. Cell Culture
4.3. Nucleic Acid Extraction and qRT-PCR
4.4. 5′ Rapid Amplification of cDNA Ends (RACE)
4.5. Bioinformatics Analysis
4.6. Plasmid Construction
4.7. Luciferase Activity Assay
4.8. Western Blotting
4.9. Apoptosis Analysis
4.10. Chromatin Immunoprecipitation
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chen, W.; Ten, D.P. Immunoregulation by members of the TGFβ superfamily. Nat. Rev. Immunol. 2016, 16, 723–740. [Google Scholar] [CrossRef] [PubMed]
- Ongaro, L.; Schang, G.; Ho, C.C.; Zhou, X.; Bernard, D.J. TGFβ superfamily regulation of follicle-stimulating hormone synthesis by gonadotrope cells: Is there a role for bone morphogenetic proteins? Endocrinology 2019, 60, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Sartori, R.; Gregorevic, P.; Sandri, M. TGFβ and BMP signaling in skeletal muscle: Potential significance for muscle-related disease. Trends Endocrinol. Metab. 2014, 25, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Du, X.; Zhou, J.; Pan, Z.; Liu, H.; Li, Q. MicroRNA-26b functions as a proapoptotic factor in porcine follicular Granulosa cells by targeting Sma-and Mad-related protein 4. Biol. Reprod. 2014, 91, 146. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, S.; Nahm, M.; Kopke, D.; Kim, J.; Cho, E.; Lee, M.J.; Lee, M.; Kim, S.H.; Broadie, K.; et al. BMP-dependent synaptic development requires Abi-Abl-Rac signaling of BMP receptor macropinocytosis. Nat. Commun. 2019, 10, 684. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, E.; Han, Y.; Chen, L.; Xie, Z. Differential expression of mRNAs encoding BMP/Smad pathway molecules in antral follicles of high- and low-fecundity Hu sheep. Anim. Reprod. Sci. 2010, 120, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Li, W.T.; Zhang, M.M.; Li, Q.G.; Tang, H.; Zhang, L.F.; Wang, K.J.; Zhu, M.Z.; Lu, Y.F.; Bao, H.G.; Zhang, Y.M.; et al. Whole-genome resequencing reveals candidate mutations for pig prolificacy. Proc. Biol. Sci. 2017, 284, 20172437. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Ye, J.; Han, X.; Qiao, R.; Li, X.; Lv, G.; Wang, K. Whole-genome sequencing identifies potential candidate genes for reproductive traits in pigs. Genomics 2019. [Google Scholar] [CrossRef]
- Schneider, J.F.; Nonneman, D.J.; Wiedmann, R.T.; Vallet, J.L.; Rohrer, G.A. Genomewide association and identification of candidate genes for ovulation rate in swine. J. Anim. Sci. 2014, 92, 3792–3803. [Google Scholar] [CrossRef]
- Abdoli, R.; Zamani, P.; Mirhoseini, S.Z.; Ghavi Hossein-Zadeh, N.; Nadri, S. A review on prolificacy genes in sheep. Reprod. Domest. Anim. 2016, 51, 631–637. [Google Scholar] [CrossRef]
- Du, X.; Pan, Z.; Li, Q.; Liu, H.; Li, Q. SMAD4 feedback regulates the canonical TGF-β signaling pathway to control granulosa cell apoptosis. Cell Death Dis. 2018, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Reheman, A.; Xu, Y.; Li, Q. miR-125b contributes to ovarian granulosa cell apoptosis through targeting BMPR1B, a major gene for sheep prolificacy. Reprod. Sci. 2019, 26, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Mulsant, P.; Lecerf, F.; Fabre, S.; Schibler, L.; Monget, P.; Lanneluc, I.; Pisselet, C.; Riquet, J.; Monniaux, D.; Callebaut, I.; et al. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc. Natl. Acad. Sci. USA 2001, 98, 5104–5109. [Google Scholar] [CrossRef] [PubMed]
- Regan, S.L.; McFarlane, J.R.; O’Shea, T.; Andronicos, N.; Arfuso, F.; Dharmarajan, A.; Almahbobi, G. Flow cytometric analysis of FSHR, BMRR1B, LHR and apoptosis in granulosa cells and ovulation rate in merino sheep. Reproduction 2015, 150, 151–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Du, X.; Pan, Z.; Zhang, L.; Li, Q. The transcription factor SMAD4 and miR-10b contribute to E2 release and cell apoptosis in ovarian granulosa cells by targeting CYP19A1. Mol. Cell. Endocrinol. 2018, 476, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Hua, G.H.; Yang, L.G. A review of research progress of FecB gene in Chinese breeds of sheep. Anim. Reprod. Sci. 2009, 116, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Pan, B.; Yang, L.; Wang, B.; Li, J. Beta defensin 3 enhances ovarian granulosa cell proliferation and migration via ERK1/2 pathway in vitro. Biol. Reprod. 2018. [Google Scholar] [CrossRef] [PubMed]
- Borroni, R.; Liu, Z.; Simpson, E.R.; Hinshelwood, M.M. A putative binding site for Sp1 is involved in transcriptional regulation of CYP17 gene expression in bovine ovary. Endocrinology 1997, 138, 2011–2020. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.C.; Chiu, Y.N.; Hu, M.C.; Guo, I.C.; Chung, B.C. Regulation of steroid production: Analysis of Cyp11a1 promoter. Mol. Cell. Endocrinol. 2011, 336, 80–84. [Google Scholar] [CrossRef]
- Anjali, G.; Kaur, S.; Lakra, R.; Taneja, J.; Kalsey, G.S.; Nagendra, A.; Shrivastav, T.G.; Devi, M.G.; Malhotra, N.; Kriplani, A.; et al. FSH stimulates IRS-2 expression in human granulosa cells through cAMP/SP1, an inoperative FSH action in PCOS patients. Cell. Signal. 2015, 27, 2452–2466. [Google Scholar] [CrossRef]
- Yeh, H.Y.; Sun, D.; Peng, Y.C.; Wu, Y.L. Regulation of the regulator of G protein signaling 2 expression and cellular localization by PKA and PKC pathways in mouse granulosa cells. Biochem. Biophys. Res. Commun. 2018, 503, 950–955. [Google Scholar] [CrossRef] [PubMed]
- Sasanuma, H.; Tsuda, M.; Morimoto, S.; Saha, LK.; Rahman, M.M.; Kiyooka, Y.; Fujiike, H.; Cherniack, AD.; Itou, J.; Callen, M.E.; et al. BRCA1 ensures genome integrity by eliminating estrogen-induced pathological topoisomerase II-DNA complexes. Proc. Natl. Acad. Sci. USA 2018, 115, E10642–E10651. [Google Scholar] [CrossRef] [PubMed]
- Daum, H.; Peretz, T.; Laufer, N. BRCA mutations and reproduction. Fertil. Steril. 2018, 109, 33–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mylona, A.; Theillet, F.X.; Foster, C.; Cheng, T.M.; Miralles, F.; Bates, P.A.; Selenko, P.; Treisman, R. Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science 2016, 354, 233–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, M.M.; Tai, C.J.; Kang, S.K.; Nathwani, P.S.; Pang, S.F.; Leung, P.C. Direct action of melatonin in human granulosa-luteal cells. J. Clin. Endocrinol. Metab. 2001, 86, 4789–4797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, F.; Feng, X.; Yang, H.; Zhu, A.; Pang, J.; Han, L.; Zhang, T.; Yao, X.; Wang, F. Genome-wide analysis of DNA methylation profiles on sheep ovaries associated with prolificacy using whole-genome Bisulfite sequencing. BMC Genom. 2017, 18, 759. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Toke, N.H.; Luo, S.; Vasoya, R.P.; Fullem, R.L.; Parthasarathy, A.; Perekatt, A.O.; Verzi, M. A reinforcing HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nat. Genet. 2019, 51, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Monsivais, D.; Clementi, C.; Peng, J.; Titus, M.M.; Barrish, J.P.; Creighton, C.J.; Lydon, J.P.; DeMayo, F.J.; Matzuk, M.M. Uterine ALK3 is essential during the window of implantation. Proc. Natl. Acad. Sci. USA 2016, 113, E387–E395. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Du, X.; Liu, L.; Cao, Q.; Pan, Z.; Li, Q. miR-1306 mediates the feedback regulation of TGF-β/SMAD signaling pathway in granulosa cells. Cells 2019, 8, 298. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Zhang, L.; Li, X.; Pan, Z.; Liu, H.; Li, Q. TGF-β signaling controls FSHR signaling-reduced ovarian granulosa cell apoptosis through theSMAD4/miR-143 axis. Cell Death. Dis. 2016, 7, e2476. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Xiong, X.; Chen, Y.G. Feedback regulation of TGF-beta signaling. Acta Biochim. Biophys. Sin. Shanghai 2018, 50, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Kucuksayan, H.; Akgun, S.; Ozes, O.N.; Alikanoglu, A.S.; Yildiz, M.; Dal, E.; Akca, H. TGF-beta-SMAD-miR-520e axis regulates NSCLC metastasis through a TGFBR2-mediated negative feedback loop. Carcinogenesis 2018. [Google Scholar] [CrossRef] [PubMed]
- Nakano, N.; Maeyama, K.; Sakata, N.; Itoh, F.; Akatsu, R.; Nakata, M.; Katsu, Y.; Ikeno, S.; Togawa, Y.; Vo Nguyen, T.T.; et al. C18 ORF1, a novel negative regulator of transforming growth factor-β signaling. J. Biol. Chem. 2014, 289, 12680–12692. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Itoh, S.; Goto, T.; Ohnishi, E.; Inamitsu, M.; Itoh, F.; Satoh, K.; Wiercinska, E.; Yang, W.; Shi, L.; et al. TMEPAI, a transmembrane TGF-beta-inducible protein, sequesters Smad proteins from active participation in TGF-beta signaling. Mol. Cell. 2010, 37, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, H.; Li, Y.; Liu, S.; Chen, J.; Ying, H. miR-24 and miR-122 negatively regulate the transforming growth factor-beta/Smad signaling pathway in skeletal muscle fibrosis. Mol. Ther. Nucleic Acids 2018, 11, 528–537. [Google Scholar] [CrossRef]
- Sun, Q.; Mao, S.; Li, H.; Zen, K.; Zhang, C.Y.; Li, L. Role of miR-17 family in the negative feedback loop of bone morphogenetic protein signaling in neuron. PLoS ONE 2013, 8, e83067. [Google Scholar] [CrossRef] [PubMed]
- Akizu, N.; Estarás, C.; Guerrero, L.; Martí, E.; Martínez-Balbás, M.A. H3K27me3 regulates BMP activity in developing spinal cord. Development 2010, 137, 2915–2925. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Du, S.Y.; Ding, M.; Dou, X.; Zhang, F.F.; Wu, Z.Y.; Qian, S.W.; Zhang, W.; Tang, Q.Q.; Xu, C.J. The BMP4-Smad signaling pathway regulates hyperandrogenism development in a female mouse model. J. Biol. Chem. 2017, 292, 11740–11750. [Google Scholar] [CrossRef] [Green Version]
- Passos, M.J.; Vasconcelos, G.L.; Silva, A.W.; Brito, I.R.; Saraiva, M.V.; Magalhães, D.M.; Costa, J.J.; Donato, M.A.; Ribeiro, R.P.; Cunha, E.V.; et al. Accelerated growth of bovine preantral follicles in vitro after stimulation with both FSH and BMP-15 is accompanied by ultrastructural changes and increased atresia. Theriogenology 2013, 79, 1269–1277. [Google Scholar] [CrossRef]
- Monniaux, D. Driving folliculogenesis by the oocyte-somatic cell dialog: Lessons from genetic models. Theriogenology 2016, 86, 41–53. [Google Scholar] [CrossRef]
- Yao, Y.; Niu, J.; Sizhu, S.; Li, B.; Chen, Y.; Li, R.; Yangzong, Q.; Li, Q.; Xu, Y. microRNA-125b regulates apoptosis by targeting bone morphogenetic protein receptor 1B in yak granulosa cells. DNA Cell Biol. 2018, 37, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Kayamori, T.; Murayama, C.; Miyamoto, A. Bone morphogenetic protein (BMP)-4 and BMP-7 suppress granulosa cell apoptosis via different pathways: BMP-4 via PI3K/PDK-1/Akt and BMP-7 via PI3K/PDK-1/PKC. Biochem. Biophys. Res. Commun. 2012, 417, 869–873. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Castrillon, D.H.; Zhou, W.; Richards, J.S. FOXO1/3 depletion in granulosa cells alters follicle growth, death and regulation of pituitary FSH. Mol. Endocrinol. 2013, 27, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Persani, L.; Rossetti, R.; Di Pasquale, E.; Cacciatore, C.; Fabre, S. The fundamental role of bone morphogenetic protein 15 in ovarian function and its involvement in female fertility disorders. Hum. Reprod. Update 2014, 20, 869–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, X.; Jing, X.; Wu, X.; Bi, X.; Liu, J.; Long, Z.; Zhang, X.; Zhang, D.; Jia, H.; Su, D.; et al. Abnormal expression levels of BMP15/Smad1 are associated with granulosa cell apoptosis in patients with polycystic ovary syndrome. Mol. Med. Rep. 2017, 16, 8231–8236. [Google Scholar] [CrossRef] [PubMed]
- Mansouri-Attia, N.; James, R.; Ligon, A.; Li, X.; Pangas, S.A. Soy promotes juvenile granulosa cell tumor development in mice and in the human granulosa cell tumor-derived COV434 cell line. Biol. Reprod. 2014, 91, 100. [Google Scholar] [CrossRef] [PubMed]
- Nie, M.; Yu, S.; Peng, S.; Fang, Y.; Wang, H.; Yang, X. miR-23a and miR-27a promote human granulosa cell apoptosis by targeting SMAD5. Biol. Reprod. 2015, 93, 98. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, J.; Xu, B.; Chrusciel, M.; Gao, J.; Bazert, M.; Stelmaszewska, J.; Xu, Y.; Zhang, H.; Pawelczyk, L.; et al. Functional characterization of microRNA-27a-3p expression in human polycystic ovary syndrome. Endocrinology 2018, 159, 297–309. [Google Scholar] [CrossRef]







| Primer | Gene | Primer Sequence (5′-3′) | Size (bp) | Tm (°C) | Usage |
|---|---|---|---|---|---|
| P1 | BMPR1B | GATTACGCCAAGCTTTTCGCCACGCCACTTTCCCATCC | RACE | ||
| P2 | BMPR1B | F: GAGCTCAGCTGGGTCAGGCTCCTTTC R: CTCGAGAGACTCCTCTGCTGCCACTC | 200 | 58 | Deletion construction |
| P3 | BMPR1B | F: GAGCTCACGATTCCCAAAGAATTACC R: CTCGAGAGACTCCTCTGCTGCCACTC | 403 | 56 | Deletion construction |
| P4 | BMPR1B | F: GAGCTCTTCCCAGCATGAATCAGAGTC R: CTCGAGAGACTCCTCTGCTGCCACTC | 921 | 57 | Deletion construction |
| P5 | BMPR1B | F: GAGCTCGGAACTGAGGATGTTGGATTG R: CTCGAGAGACTCCTCTGCTGCCACTC | 1322 | 56 | Deletion construction |
| P6 | BMPR1B | F: AGCTCCCATGTCAGCACAGTATAGGTGTGTGCCTCTCCGG R: ACTGTGCTGACATGGGAGCT | 298 | 56 | Promoter construction |
| P7 | BMPR1B | F: GTATAGGTGTGTGTCTCTCCGGAATGCCTCCACAGCTGCCCA R: CCGGAGAGACACACACCTATAC | 298 | 56 | Mutation construction |
| P8 | BMPR1B | F: TGCTGGGCTCTTTAGTGG R: CTGGACAATGGTGGTGGC | 237 | 60 | qPCR for variant I |
| P9 | BMPR1B | F: GGCAAGAAACAGGAGGCT R: CCACAGGCATCCCAGAGT | 241 | 60 | qPCR for variant II |
| P10 | BMPR1B | F: CATAGACAACAGCCCACCAG R: GACCACAGGCATCCCAGA | 264 | 60 | qPCR for variant III |
| P11 | BMPR1B | F: AGCACTCAAGGCAAACCA R: GGCCATGATGTAAGACTGAAAG | 243 | 60 | qPCR |
| P12 | GAPDH | F: TGGAATGACATCTCGGTCTGGTA R: CACCATGGCTCAGAAGCACAC | 247 | 58 | qPCR |
| P13 | BMPR1B | F: CCCAAAGAATTACCATTCATGGC R: GACATTCCGGAGAGACACACACCTA | 118 | 63 | ChIP-PCR |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdurahman, A.; Du, X.; Yao, Y.; Sulaiman, Y.; Aniwashi, J.; Li, Q. Smad4 Feedback Enhances BMPR1B Transcription in Ovine Granulosa Cells. Int. J. Mol. Sci. 2019, 20, 2732. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20112732
Abdurahman A, Du X, Yao Y, Sulaiman Y, Aniwashi J, Li Q. Smad4 Feedback Enhances BMPR1B Transcription in Ovine Granulosa Cells. International Journal of Molecular Sciences. 2019; 20(11):2732. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20112732
Chicago/Turabian StyleAbdurahman, Anwar, Xing Du, Yilong Yao, Yiming Sulaiman, Jueken Aniwashi, and Qifa Li. 2019. "Smad4 Feedback Enhances BMPR1B Transcription in Ovine Granulosa Cells" International Journal of Molecular Sciences 20, no. 11: 2732. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20112732