ParaBTM: A Parallel Processing Framework for Biomedical Text Mining on Supercomputers
Abstract
:1. Introduction
2. Results and Discussion
- <PMID>|t|<Title of the paper>
- <PMID>|a|<The rest of the paper>
3. Materials and Methods
3.1. Data Sources and Storage
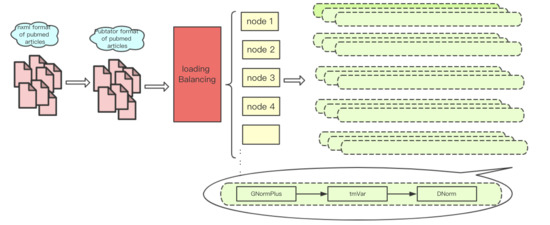
3.2. Parallel Processing
3.2.1. MPI-Based Multi-Node Computation
3.2.2. Load Balancing Strategy
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rebholz-Schuhmann, D.; Oellrich, A.; Hoehndorf, R. Text-mining solutions for biomedical research: Enabling integrative biology. Nat. Rev. Genet. 2012, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-H.; Harris, B.R.; Kao, H.-Y.; Lu, Z. tmVar: A text mining approach for extracting sequence variants in biomedical literature. Bioinformatics 2013, 29, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Rocktäschel, T.; Hakenberg, J.; Lichtblau, Y.; Leser, U. SETH detects and normalizes genetic variants in text. Bioinformatics 2016, 32, 2883–2885. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-H.; Kao, H.-Y. Cross-species gene normalization by species inference. BMC Bioinform. 2011, 12 (Suppl. 8), S5. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Shen, H.B. OUGENE: A disease associated over-expressed and under-expressed gene database. Sci. Bull. 2016, 61, 752–754. [Google Scholar] [CrossRef]
- Leaman, R.; Lu, Z. TaggerOne: Joint named entity recognition and normalization with semi-Markov Models. Bioinformatics 2016, 32, 2839–2846. [Google Scholar] [CrossRef] [PubMed]
- Leaman, R.; Islamaj Dogan, R.; Lu, Z. DNorm: Disease name normalization with pairwise learning to rank. Bioinformatics 2013, 29, 2909–2917. [Google Scholar] [CrossRef] [PubMed]
- Quan, C.; Wang, M.; Ren, F. An unsupervised text mining method for relation extraction from biomedical literature. PLoS ONE 2014, 9, e102039. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhang, M.; Xie, Y.; Wang, F.; Chen, M.; Zhu, K.Q.; Wei, J. DTMiner: Identification of potential disease targets through biomedical literature mining. Bioinformatics 2016, 32, 3619–3626. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, Q. A knowledge-driven conditional approach to extract pharmacogenomics specific drug-gene relationships from free text. J. Biomed. Inform. 2012, 45, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Percha, B.; Garten, Y.; Altman, R.B. Discovery and explanation of drug-drug interactions via text mining. Pac. Symp. Biocomput. 2012, 410–421. [Google Scholar] [CrossRef]
- Segura-Bedmar, I.; Martínez, P.; de Pablo-Sánchez, C. Using a shallow linguistic kernel for drug-drug interaction extraction. J. Biomed. Inform. 2011, 44, 789–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo, À.; Piñero, J.; Queralt-Rosinach, N.; Rautschka, M.; Furlong, L.I. Extraction of relations between genes and diseases from text and large-scale data analysis: Implications for translational research. BMC Bioinform. 2015, 16, 55. [Google Scholar] [CrossRef] [PubMed]
- Pletscher-Frankild, S.; Pallejà, A.; Tsafou, K.; Binder, J.X.; Jensen, L.J. DISEASES: Text mining and data integration of disease–gene associations. Methods 2015, 74, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Gerner, M.; Sarafraz, F.; Nenadic, G.; Bergman, C.M. BioContext: An integrated text mining system for large-scale extraction and contextualisation of biomolecular events. Bioinformatics 2012, 28, 2154–2161. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-H.; Kao, H.-Y.; Lu, Z. GNormPlus: An Integrative Approach for Tagging Genes, Gene Families, and Protein Domains. BioMed Res. Int. 2015, 2015, 918710–918717. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Xiao, L.; Yang, C.; Lu, Y. MilkyWay-2 supercomputer: System and application. Front. Comput. Sci. 2014, 8, 345–356. [Google Scholar] [CrossRef]
- Wu, C.; Schwartz, J.M.; Brabant, G.; Nenadic, G. Molecular profiling of thyroid cancer subtypes using large-scale text mining. BMC Med. Genom. 2014, 7 (Suppl. 3), S3. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Kaur, G. A Review of Load Balancing Strategies for Distributed Systems. IJCA 2015, 121, 45–47. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |










| Items | Content |
|---|---|
| Manufacturer | NUDT |
| Cores | 3,120,000 |
| Memory | 1,024,000 GB |
| CPU | Intel Xeon E5-2692v2 12 C 2.2 GHz |
| Interconnect | TH Express-2 |
| Linpack Performance(Rmax) | 33,862.7 TFlop/s |
| Theoretical Peak(Rpeak) | 54,902.4 TFlop/s |
| HPCG [TFlop/s] | 580.109 |
| Operating System | Kylin Linux |
| MPI | MPICH2 with a customized GLEX channel |
| Number of Papers | Time(s) | ||||
|---|---|---|---|---|---|
| Distribution | GNormPlus | tmVar | Dnorm | Total | |
| 10 | 2.11 | 1742.16 | 416.71 | 58.29 | 2219.27 |
| 20 | 4.2 | 2764.13 | 444.86 | 65.14 | 3278.33 |
| 40 | 7.76 | 4865.14 | 488.08 | 83.19 | 5444.17 |
| 80 | 17.17 | 7864.37 | 744.86 | 177.45 | 8803.85 |
| Sizes of Papers (M) | Time (s) | ||||
|---|---|---|---|---|---|
| Distribution | GNormPlus | tmVar | Dnorm | Total | |
| 1 | 1.93 | 1511.35 | 239.74 | 59.51 | 1812.53 |
| 2 | 4.80 | 2696.64 | 319.31 | 62.94 | 3083.69 |
| 4 | 16.24 | 4944.35 | 633.57 | 86.24 | 5680.40 |
| 8 | 21.77 | 8604.43 | 985.63 | 130.33 | 9742.16 |
| (a) Maximum times on different numbers of parallel processes with different strategies. | |||
| Number of Processes | Maximum Time among All Processes (s) | ||
| Random | Round-Robin | Short-Board | |
| 2 | 8676.60 | 8651.42 | 8466.89 |
| 4 | 4466.81 | 5211.96 | 4596.09 |
| 8 | 2792.29 | 2706.41 | 2386.45 |
| 16 | 1737.94 | 1457.01 | 1475.07 |
| 32 | 932.85 | 942.10 | 930.25 |
| 64 | 609.06 | 579.8 | 553.42 |
| (b) Average times on different numbers of parallel processes with different strategies. | |||
| Number of Processes | Average Time of All Processes (s) | ||
| Random | Round-Robin | Short-Board | |
| 2 | 8513.10 | 8374.08 | 8392.46 |
| 4 | 4001.38 | 4535.42 | 4393.07 |
| 8 | 2287.19 | 2354.14 | 2242.42 |
| 16 | 1265.46 | 1174.12 | 1270.33 |
| 32 | 624.73 | 666.91 | 640.63 |
| 64 | 379.08 | 376.06 | 366.84 |
| (c) Load balancing efficiencies on different numbers of parallel processes with different strategies. | |||
| Number of Processes | Efficiency (Average/Maximum) | ||
| Random | Round-Robin | Short-Board | |
| 2 | 0.98 | 0.97 | 0.99 |
| 4 | 0.90 | 0.87 | 0.96 |
| 8 | 0.82 | 0.87 | 0.94 |
| 16 | 0.73 | 0.81 | 0.86 |
| 32 | 0.67 | 0.71 | 0.69 |
| 64 | 0.62 | 0.65 | 0.66 |
| Number of Processes | Time (s) | ||||
|---|---|---|---|---|---|
| Distribution | GNormPlus | tmVar2.0 | Dnorm | Total | |
| 1 | 18,934.18 | 5,874,482.04 | 654,145.38 | 82,455.3 | 6,630,016.9 |
| 128 | 3643 | 23,733 | 16,214 | 233 | 43,823 |
| Speed-up (x) | 5.20 | 247.52 | 40.34 | 353.89 | 151.29 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, Y.; Wu, C.; Yang, X.; Wang, W.; Zhu, E.; Yin, J. ParaBTM: A Parallel Processing Framework for Biomedical Text Mining on Supercomputers. Molecules 2018, 23, 1028. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23051028
Xing Y, Wu C, Yang X, Wang W, Zhu E, Yin J. ParaBTM: A Parallel Processing Framework for Biomedical Text Mining on Supercomputers. Molecules. 2018; 23(5):1028. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23051028
Chicago/Turabian StyleXing, Yuting, Chengkun Wu, Xi Yang, Wei Wang, En Zhu, and Jianping Yin. 2018. "ParaBTM: A Parallel Processing Framework for Biomedical Text Mining on Supercomputers" Molecules 23, no. 5: 1028. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23051028