Variations in the Morphology, Mechanics and Adhesion of Persister and Resister E. coli Cells in Response to Ampicillin: AFM Study
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of Ampicillin Concentration and Exposure Time on Bacterial Viability
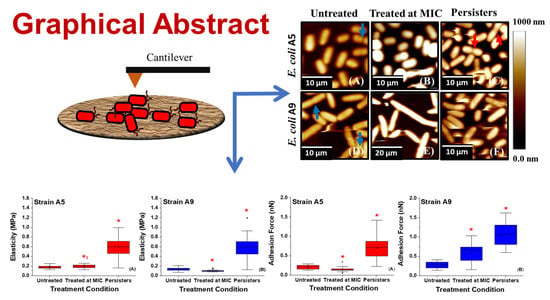
2.2. Effect of Ampicillin on Bacterial Morphology and Dimensions as Probed by AFM and SEM
2.3. Effect of Ampicillin Concentration and Exposure Time on Cellular Surface Area (SA), Volume (V), and Surface Area to Volume Ratio (SA/V)
2.4. Effect of Ampicillin Concentration and Exposure Time on Bacterial Surface Roughness
2.5. Effect of Ampicillin Concentration and Exposure Time on Adhesion Forces Measured between the Biopolymers of MDR E. coli Cells and a Model Surface of Si3N4
2.6. Variations in the Thicknesses and Grafting Densities of Bacterial Surface Biopolymer Brushes in Response to Ampicillin at Different Concentrations and Times
2.7. Variations in the Elasticities of Bacterial Cells in Response to Ampicillin at Different Concentrations and Times
2.8. Possible Mechanisms Employed by MDR E. coli Resistant and Persistent Cells to Resist Ampicillin
2.8.1. Mechanism 1: Dormancy
2.8.2. Mechanism 2: Increase in Cellular Elasticity
2.9. Implications of Our Findings on the Design of Effective Antibiotics against Persistent E. coli Cells
3. Materials and Methods
3.1. Cells and Chemicals
3.2. Choice of MDR E. coli as the Bacterial Model
3.3. Choice of the Model Antibiotic-Ampicillin
3.4. Bacterial Growth Conditions and Isolation of Persister Cells
3.5. AFM Experiments
3.6. Scanning Electron Microscopy (SEM) Imaging
3.7. Bacterial Morphology Analysis
3.8. Analysis of Bacterial Surface Roughness
3.9. Analysis of Adhesion Forces
3.10. Estimating the Length and the Grafting Density of a Bacterial Surface Biopolymer Brush Using the Steric Model
3.11. Estimation of the Young’s Modulus of the Bacterial Cell
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mohammad, H.; Mayhoub, A.S.; Cushman, M.; Seleem, M.N. Anti-biofilm activity and synergism of novel thiazole compounds with glycopeptide antibiotics against multidrug-resistant Staphylococci. J. Antibiot. (Tokyo) 2015, 68, 259–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solomon, S.L.; Oliver, K.B. Antibiotic resistance threats in the United States: Stepping back from the brink. Am. Fam. Physician 2014, 89, 938–941. [Google Scholar] [PubMed]
- Aeschlimann, J.R. The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other Gram-negative bacteria. Pharmacotherapy 2003, 23, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Babic, M.; Hujer, A.M.; Bonomo, R.A. What’s new in antibiotic resistance? Focus on beta-lactamases. Drug Resist. Update 2006, 9, 142–156. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.; Mishra, S.; Jena, P.; Jacob, B.; Sarkar, B.; Sonawane, A. An investigation on the antibacterial, cytotoxic, and antibiofilm efficacy of starch-stabilized silver nanoparticles. Nanomedicine 2012, 8, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Read, A.F.; Woods, R.J. Antibiotic resistance management. Evol. Med. Public Health 2014, 2014, 147. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef]
- Shah, D.; Zhang, Z.; Khodursky, A.; Kaldalu, N.; Kurg, K.; Lewis, K. Persisters: A distinct physiological state of E. coli. BMC Microbiol. 2006, 6, 53. [Google Scholar]
- Bigger, J.W. Treatment of infections with penicilin by intreatment sterilization. Lancet 1944, 244, 497–500. [Google Scholar] [CrossRef]
- Hobby, G.L.; Meyer, K.; Chaffee, E. Observations on the mehanism of action of penicillin. Exp. Biol. Med. 1942, 50, 281–285. [Google Scholar] [CrossRef]
- Gefen, O.; Balaban, N.Q. The importance of being persistent: Heterogeneity of bacterial populations under antibiotic stress: Review article. FEMS Microbiol. Rev. 2009, 33, 704–717. [Google Scholar] [CrossRef] [PubMed]
- Sultana, S.T.; Call, D.R.; Beyenal, H. Eradication of Pseudomonas aeruginosa biofilms and persister cells using an electrochemical scaffold and enhanced antibiotic susceptibility. NPJ Biofilms Microbiomes 2016, 2, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amato, S.M.; Brynildsen, M.P. Nutrient transitions are a source of persisters in Escherichia coli biofilms. PLoS ONE 2014, 9, e93110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.; Conery, A.L.; Rajamuthiah, R.; Fuchs, B.B.; Ausubel, F.M.; Mylonakis, E. Identification of an antimicrobial agent effective against methicillin-resistant Staphylococcus aureus persisters using a fluorescence-based screening strategy. PLoS ONE 2015, 10, e0127640. [Google Scholar] [CrossRef]
- Kwan, B.W.; Valenta, J.A.; Benedik, M.J.; Wood, T.K. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 2013, 57, 1468–1473. [Google Scholar] [CrossRef] [Green Version]
- Lewis, K. Persister Cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Megaw, J.; Gilmore, B.F. Archaeal persisters: Persister cell formation as a stress response in Haloferax volcanii. Front. Microbiol. 2017, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Pu, Y.; Zhao, Z.; Li, Y.; Zou, J.; Ma, Q.; Zhao, Y.; Ke, Y.; Zhu, Y.; Chen, H.; Baker, M.A.B.; et al. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol. Cell 2016, 62, 284–294. [Google Scholar] [CrossRef] [Green Version]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, N.; Wood, T.L.; Martínez-Vázquez, M.; García-Contreras, R.; Wood, T.K. DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol. Bioeng. 2016, 113, 1984–1992. [Google Scholar] [CrossRef]
- Kwan, B.W.; Chowdhury, N.; Wood, T.K. Combatting bacterial infections by killing persister cells with mitomycin C. Environ. Microbiol. 2015, 17, 4406–4414. [Google Scholar] [CrossRef] [PubMed]
- Amato, S.M.; Orman, M.A.; Brynildsen, M.P. Metabolic control of persister formation in Escherichia coli. Mol. Cell 2013, 50, 475–487. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.; Joshi-Datar, A.; Lepine, F.; Bauerle, E.; Olakanmi, O.; Beer, K.; McKay, G.; Siehnel, R.; Schafhauser, J.; Wang, Y.; et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 2011, 80, 54–68. [Google Scholar] [CrossRef] [Green Version]
- Williamson, K.S.; Richards, L.A.; Perez-Osorio, A.C.; Pitts, B.; Mclmmerney, K.; Stewart, P.S.; Franklin, M.J. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 2012, 194, 2062–2073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 2008, 68, 223–240. [Google Scholar] [CrossRef]
- Moyed, H.S.; Bertrand, K.P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983, 155, 768–775. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, M.A.; Piro, K.M.; Xu, W.; Hansen, S.; Lewis, K.; Brennan, R.G. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 2009, 80, 396–401. [Google Scholar] [CrossRef] [Green Version]
- Korch, S.B.; Henderson, T.A.; Hill, T.M. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 2003, 50, 1199–1213. [Google Scholar] [CrossRef]
- Korch, S.B.; Hill, T.M. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: Effects on macromolecular synthesis and persister formation. J. Bacteriol. 2006, 188, 3826–3836. [Google Scholar] [CrossRef] [Green Version]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Uzoechi, S.C.; Abu-Lail, N.I. The effects of beta-Lactam antibiotics on surface modifications of multidrug-resistant Escherichia coli: A multiscale approach. Microsc. Microanal. 2019, 25, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Uzoechi, S.C.; Abu-lail, N.I. Changes in cellular elasticities and conformational properties of bacterial surface biopolymers of multidrug-resistant Escherichia coli (MDR-E. coli) strains in response to ampicillin. Cell Surf. 2019, 5, 100019. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Shah, D.; Spoering, A.; Kaldalu, N.; Lewis, K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. Bacteriology 2004, 186, 8172–8180. [Google Scholar] [CrossRef] [Green Version]
- Tamayo, J.; Humphris, A.D.L.; Owen, R.J.; Miles, M.J. High-Q dynamic force microscopy in liquid and its application to living cells. Biophys. J. 2001, 81, 526–537. [Google Scholar] [CrossRef] [Green Version]
- Beniac, D.R.; Siemens, C.G.; Wright, C.J.; Booth, T.F.A. Filtration based technique for simultaneous SEM and TEM sample preparation for the rapid detection of pathogens. Viruses 2014, 6, 3358–3371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golding, C.G.; Lamboo, L.L.; Beniac, D.R.; Booth, T.F. The scanning electron microscope in microbiology and diagnosis of infectious disease. Sci. Rep. 2016, 6, 26516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyström, T. Stationary-phase physiology. Annu. Rev. Microbiol. 2004, 58, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Justice, S.S.; Hunstad, D.A.; Cegelski, L.; Hultgren, S.J. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 2008, 6, 162–168. [Google Scholar] [CrossRef]
- Si, F.; Li, D.; Cox, S.E.; Sauls, J.T.; Azizi, O.; Sou, C.; Schwartz, A.B.; Erickstad, M.J.; Jun, Y.; Li, X.; et al. Invariance of initiation mass and predictability of cell size in Escherichia coli. Curr. Biol. 2017, 27, 1278–1287. [Google Scholar] [CrossRef]
- Yoon, M.Y.; Lee, K.M.; Park, Y.; Yoon, S.S. Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS ONE 2011, 6, e16105. [Google Scholar] [CrossRef] [Green Version]
- Young, K.D. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 2006, 70, 660–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, K.D. Bacterial shape: Two-dimensional questions and possibilities. Annu. Rev. Microbiol. 2010, 64, 223–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2012, 10, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Carballido-Lopez, R. The actin-like MreB proteins in Bacillus subtilis a new turn. Front. Biosci. 2012, 4, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaritsky, A.; Woldringh, C.L.; Helmstetter, C.E.; Grover, N.B. Dimensional rearrangement of Escherichia coli B/r cells during a nutritional shift-down. J. Gen. Microbiol. 1993, 139, 2711–2714. [Google Scholar] [CrossRef] [Green Version]
- Spoering, A.M.Y.L.; Lewis, K.I.M. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 2001, 183, 6746–6751. [Google Scholar] [CrossRef] [Green Version]
- Alves, C.S.; Melo, M.N.; Franquelim, H.G.; Ferre, R.; Planas, M.; Feliu, L.; Bardají, E.; Kowalczyk, W.; Andreu, D.; Santos, N.C.; et al. Escherichia coli cell surface perturbation and disruption induced by antimicrobial peptides BP100 and pepR. J. Biol. Chem. 2010, 285, 27536–27544. [Google Scholar] [CrossRef] [Green Version]
- Su, H.N.; Chen, Z.H.; Song, X.Y.; Chen, X.L.; Shi, M.; Zhou, B.C.; Zhao, X.; Zhang, Y.Z. Antimicrobial peptide trichokonin VI-induced alterations in the morphological and nanomechanical properties of Bacillus subtilis. PLoS ONE 2012, 7, e45818. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, D.; Strzelecki, J.; Pawlak, K.; Dahm, H.; Balter, A. Short communication effect of ampicillin on adhesive properties of bacteria examined by atomic force microscopy. Micron 2018, 112, 84–90. [Google Scholar] [CrossRef]
- Park, B.J.; Abu-Lail, N.I. Atomic force microscopy investigations of heterogeneities in the adhesion energies measured between pathogenic and non-pathogenic Listeria species and silicon nitride as they correlate to virulence and adherence. Biofouling 2011, 27, 543–559. [Google Scholar] [CrossRef] [Green Version]
- Park, B.J.; Haines, T.; Abu-Lail, N.I. A correlation between the virulence and the adhesion of Listeria monocytogenes to silicon nitride: An atomic force microscopy study. Colloid Surf. B 2009, 73, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Berne, C.; Ducret, A.; Hardy, G.G.; Brun, Y.V. Adhesins involved in attachment to abiotic surfaces by Gram-negative bacteria. Microbiol. Spectr. 2015, 3, 1–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Garrett, T.R.; Bhakoo, M.; Zhang, Z. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Vu, B.; Chen, M.; Crawford, R.J.; Ivanova, E.P. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 2009, 14, 2535–2554. [Google Scholar] [CrossRef]
- Abu-Lail, N.I.; Camesano, T.A. Role of ionic strength on the relationship of biopolymer conformation, DLVO contributions, and steric interactions to bioadhesion of Pseudomonas putida KT2442. Biomacromolecules 2003, 4, 1000–1012. [Google Scholar] [CrossRef]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J.; Cruz, S. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 2015, 3, 1–30. [Google Scholar] [CrossRef] [Green Version]
- Sans-Serramitjana, E.; Fusté, E.; Martínez-Garriga, B.; Merlos, A.; Pastor, M.; Pedraz, J.L.; Esquisabel, A.; Bachiller, D.; Vinuesa, T.; Viñas, M. Killing effect of nanoencapsulated colistin sulfate on Pseudomonas aeruginosa from cystic fibrosis patients. J. Cyst. Fibros. 2016, 15, 611–618. [Google Scholar] [CrossRef] [Green Version]
- Falagas, M.E.; Kasiakou, S.K. Colistin: The revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin. Infect. Dis. 2005, 40, 1331–1341. [Google Scholar] [CrossRef] [Green Version]
- Jacoby, G.A.; Medeiros, A.A. More extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 1991, 35, 1697–1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomson, J.M.; Bonomo, R.A. The threat of antibiotic resistance in Gram-negative pathogenic bacteria: β-lactams in peril! Curr. Opin. Microbiol. 2005, 8, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Eastman, T.; Zhu, D.A. dhesion forces between surface-modified AFM tips and a mica surface. Langmuir 1996, 12, 2859–2862. [Google Scholar] [CrossRef]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renner, L.D.; Weibel, D.B. Physiochemical regulation of biofilm formation. MRS Bull. 2011, 36, 347–355. [Google Scholar] [CrossRef] [Green Version]
- Camesano, T.A.; Abu-Lail, N.I. Heterogeneity in bacterial surface polysaccharides, probed on a single-molecule basis. Biomacromolecules 2002, 3, 661–667. [Google Scholar] [CrossRef]
- Van der Mei, H.C.; Busscher, H.J. Bacterial cell surface heterogeneity: A pathogen’s disguise. PLoS Pathog. 2012, 8, e1002821. [Google Scholar] [CrossRef] [Green Version]
- Zobell, C.E. The effect of solid surfaces upon bacterial activity. J. Bacteriol. 1943, 46, 39–56. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, P.; Das, J.; Foley, I. Biofilm Susceptibility to antimicrobials. Adv. Dent. Res. 1997, 11, 160–167. [Google Scholar] [CrossRef]
- Stewart, P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef]
- Ramadan, M.A.; Tawfik, A.F.; Shibl, A.M.; Gemmell, C.G. Post-antibiotic effect of azithromycin and erythromycin on streptococcal susceptibility to phagocytosis. J. Med. Microbiol. 1995, 42, 362–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajkarimi, M.; Harrison, S.H.; Hung, A.M.; Graves, J.L. Mechanobiology of antimicrobial resistant Escherichia coli and Listeria innocua. PLoS ONE 2016, 11, e0149769. [Google Scholar] [CrossRef] [PubMed]
- Vidya, K.; Mallya, P.; Rao, P. Inhibition of bacterial adhesion by subinhibitory concentrations of antibiotics. Indian J. Med. Microbiol. 2005, 23, 102–105. [Google Scholar] [PubMed]
- Ghuysen, J.-M. Molecular structures of penicillin-binding proteins and P-lactamases. Trends Microbiol. 1994, 2, 372–380. [Google Scholar] [CrossRef]
- Vollmer, W.; Seligman, S.J. Architecture of peptidoglycan: More data and more models. Trends Microbiol. 2010, 18, 59–66. [Google Scholar] [CrossRef]
- Formosa, C.; Grare, M.; Jauvert, E.; Coutable, A.; Regnouf-de-Vains, J.B.; Mourer, M.; Duval, R.E.; Dague, E. Nanoscale analysis of the effects of antibiotics and CX1 on a Pseudomonas aeruginosa multidrug-resistant strain. Sci. Rep. 2012, 2, 575. [Google Scholar] [CrossRef] [Green Version]
- Perry, C.C.; Weatherly, M.; Beale, T.; Randriamahefa, A. Atomic force microscopy study of the antimicrobial activity of aqueous garlic versus ampicillin against Escherichia coli and Staphylococcus aureus. J. Sci. Food Agric. 2009, 89, 958–964. [Google Scholar] [CrossRef]
- Cerf, A.; Cau, J.C.; Vieu, C.; Dague, E. Nanomechanical properties of dead or alive single-patterned bacteria. Langmuir 2009, 25, 5731–5736. [Google Scholar] [CrossRef]
- Longo, G.; Rio, L.M.; Trampuz, A.; Dietler, G.; Bizzini, A.; Kasas, S. Antibiotic-induced modifications of the stiffness of bacterial membranes. J. Microbiol. Methods 2013, 93, 80–84. [Google Scholar] [CrossRef]
- Braga, P.C.; Ricci, D. Atomic force microscopy: Application to investigation of Escherichia coli morphology before and after exposure to cefodizime. Antimicrob. Agents Chemother. 1998, 42, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Scheffers, D.J.; Pinho, M.G. Bacterial cell wall synthesis: New insights from localization Studies. Microbiol. Mol. Biol. Rev. 2005, 69, 585–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikaiido, H.; Pages, J.M. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 2012, 36, 340–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luby, E.; Ibekwe, A.M.; Zilles, J.; Pruden, A. Molecular methods for assessment of antibiotic resistance in agricultural ecosystems: Prospects and challenges. J. Environ. Qual. 2016, 45, 441–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mokrozub, V.V.; Lazarenko, L.M.; Sichel, L.M.; Babenko, L.P.; Lytvyn, P.M.; Demchenko, O.M.; Melnichenko, Y.O.; Boyko, N.V.; Biavati, B.; DiGioia, D.; et al. The role of beneficial bacteria wall elasticity in regulating innate immune response. EPMA J. 2015, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Clinical Lab Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI supplement M100; Clincial and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Filho, R.P.; Polli, M.C.; Filho, S.B.; Garcia, M.; Ferreira, E.I. Prodrugs available on the Brazilian pharmaceutical market and their corresponding bioactivation pathways. Brazilian J. Pharm. Sci. 2010, 46, 393–420. [Google Scholar] [CrossRef] [Green Version]
- Marques, C.N.H. Isolation of persister cells from biofilm and planktonic populations of Pseudomonas aeruginosa. Bio-Protocol 2015, 5, 1590. [Google Scholar] [CrossRef]
- Cañas-Duarte, S.J.; Restrepo, S.; Pedraza, J.M. Novel protocol for persister cells isolation. PLoS ONE 2014, 9, e88660. [Google Scholar] [CrossRef]
- Grant, L.M.; Ducker, W.A. Effect of substrate hydrophobicity on surface−aggregate geometry: Zwitterionic and nonionic surfactants. J. Phys. Chem. B 1997, 101, 5337–5345. [Google Scholar] [CrossRef]
- Hutter, J.L.; Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. [Google Scholar] [CrossRef] [Green Version]
- Camesano, T.A.; Natan, M.J.; Logan, b.E. Observation of changes in bacterial cell morphology using tapping mode atomic force microscopy. Langmuir 2000, 16, 4563–4572. [Google Scholar] [CrossRef]
- Park, B.; Abu-Lail, N.I. Variations in the nanomechanical properties of virulent and avirulent Listeria monocytogenes. Soft Matter 2010, 6, 3898–3909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, L.C.; Mergulhão, F.J. SEM analysis of surface impact on biofilm antibiotic treatment SEM analysis of surface impact on biofilm antibiotic treatment. Scanning 2017, 2017, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishino, T.; Nakazawa, S. 1033–1042 Bacteriological study on effects of beta lactam group antibiotics in high concentrations. Antimicrob. Agents Chemother. 1976, 9, 1033–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorn, R.M.S.; Greenman, J. A novel in vitro flat-bed perfusion biofilm model for determining the potential antimicrobial efficacy of topical wound treatments. J. Appl. Microbiol. 2009, 107, 2070–2079. [Google Scholar] [CrossRef] [PubMed]
- Young, K.D. Bacterial morphology: Why have different shapes? Curr. Opin. Microbiol. 2007, 10, 596–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girasole, M.; Pompeo, G.; Cricenti, A.; Congiu-Castellano, A.; Andreola, F.; Serafino, A.; Frazer, B.H.; Boumis, G.; Amiconi, G. Roughness of the plasma membrane as an independent morphological parameter to study RBCs: A quantitative atomic force microscopy investigation. Biochim. Biophys. Acta-Biomembr. 2007, 1768, 1268–1276. [Google Scholar] [CrossRef] [Green Version]
- Park, B.J.; Abu-Lail, N.I. The role of the pH conditions of growth on the bioadhesion of individual and lawns of pathogenic Listeria monocytogenes cells. J. Colloid Interface Sci. 2011, 358, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Abu-Lail, N.I. The Effect of Biopolymer Properties on Bacterial Adhesion: An Atomic Force Microscopy (AFM) Study; WPI: Worcester, MA, USA, 2003. [Google Scholar]
- Alexander, S. Adsorption of chain molecules with a polar head a scaling description. J. Phys. Chem. B 1977, 38, 983–987. [Google Scholar] [CrossRef]
- Butt, H.-J.; Kappl, M.; Mueller, H.; Raiteri, R.; Meyer, W.; Rühe, J. Steric forces measured with the atomic force microscope at various temperatures. Langmuir 1999, 15, 2559–2565. [Google Scholar] [CrossRef]
- Abu-Lail, N.I.; Camesano, T.A. The effect of solvent polarity on the molecular surface properties and adhesion of Escherichia coli. Colloid Surf. B 2006, 51, 62–70. [Google Scholar] [CrossRef]
- Eskhan, A.O.; Abu-Lail, N.I. Cellular and molecular investigations of the adhesion and mechanics of Listeria monocytogenes lineages’ I and II environmental and epidemic strains. J. Colloid Interface Sci. 2013, 394, 554–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramezanian, S.; Uzoechi, S.C.; Muhunthan, B.; Abu-Lail, N.I. Role of ionic strength in the thicknesses of the biopolymer fringes, spring constants, and Young’s moduli of Pseudomonas putida. J. Vac. Sci. Technol. B Nanotechnol. Microelectron. 2018, 36, 021801. [Google Scholar] [CrossRef]
- Vadillo-Rodriguez, V.; Schooling, S.R.; Dutcher, J.R. In situ characterization of differences in the viscoelastic response of individual Gram-negative and Gram-positive bacterial cells. J. Bacteriol. 2009, 191, 5518–5525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y. Opening the box of a boxplot. Am. Stat. 1988, 42, 257–262. [Google Scholar]












© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
C. Uzoechi, S.; I. Abu-Lail, N. Variations in the Morphology, Mechanics and Adhesion of Persister and Resister E. coli Cells in Response to Ampicillin: AFM Study. Antibiotics 2020, 9, 235. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9050235
C. Uzoechi S, I. Abu-Lail N. Variations in the Morphology, Mechanics and Adhesion of Persister and Resister E. coli Cells in Response to Ampicillin: AFM Study. Antibiotics. 2020; 9(5):235. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9050235
Chicago/Turabian StyleC. Uzoechi, Samuel, and Nehal I. Abu-Lail. 2020. "Variations in the Morphology, Mechanics and Adhesion of Persister and Resister E. coli Cells in Response to Ampicillin: AFM Study" Antibiotics 9, no. 5: 235. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9050235