Self-Service Data Science in Healthcare with Automated Machine Learning
Abstract
:1. Introduction
- Which AutoML method performs best on a benchmark test, given medical datasets?
- What are the requirements of healthcare professionals for starting to use AutoML in their daily practice?
- How does the selected AutoML method suit healthcare professionals in their knowledge discovery process?
2. Overview of AutoML Methods
2.1. Healthcare
2.2. Fixed Pipelines
2.3. Neural Networks
2.4. Evolutionary Methods
2.5. Distributed Methods
2.6. Overview of Methods
3. Research Method
4. Results
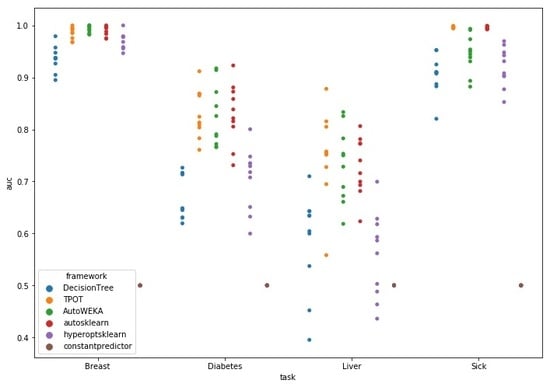
4.1. Benchmark Test
4.2. AutoML Requirements Evaluation
4.3. Artefact Evaluation
5. Discussion
5.1. Lessons Learned
5.1.1. Suitability of AutoML Methods for Researcher-Physicians
5.1.2. Bias in Medical Analytics Publications
5.2. Validity
5.2.1. Descriptive Validity
5.2.2. Interpretive Validity
5.2.3. External Validity
6. Conclusions
7. Future Research
7.1. AutoML Model Uncertainty
7.2. AutoML Use Cases
7.3. AutoML Interpretability
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Tool | Library/Package | Optimization | Pre-Processor | Post-Processor | Extra Feature(s) | Analysis Capabilities | Code Link |
|---|---|---|---|---|---|---|---|
| Auto-Weka 2.0 | WEKA | Tree-based hierarchical BO | Yes | No | Binary classification Multi-label classification Regression | https://github.com/automl/autoweka | |
| Auto-Sklearn | scikit-learn | Tree-based BO | Yes | Yes | Meta-learner | Binary classification Multi-label classification Regression | https://github.com/automl/auto-sklearn |
| Hyperopt-Sklearn | scikit-learn | Tree-based BO | Yes | No | Binary classification Multi-label classification | https://github.com/hyperopt/hyperopt-sklearn | |
| TPOT | scikit-learn DEAP | Tree-based GP | Yes | No | Binary classification Multi-label classification Regression | https://github.com/EpistasisLab/tpot | |
| Layered TPOT | scikit-learn | Tree-based GP | Yes | No | Binary classification Multi-label classification Regression | https://github.com/PG-TUe/tpot/tree/layered | |
| Auto-Net 1.0 | Lasagne | Feed-forward NN on Stochastic Gradient Descent | Yes | No | Binary classification Multi-label classification Regression | No implementation found | |
| Auto-Net 2.0 | PyTorch | BO and Hyperband (BOHB) | Yes | No | Binary classification Multi-label classification Regression | No implementation found | |
| FLASH | scikit-learn | BO with expected improvement | Yes | No | Pipeline caching | Binary classification | https://github.com/yuyuz/FLASH |
| RECIPE | scikit-learn | Grammar-based GP | Yes | No | Binary classification | https://github.com/RecipeML/Recipe | |
| Auto-Prognosis | scikit-learn | BO and GP | Yes | Yes | Meta-learner, Explainer | Binary classification Survival analysis Temporal analysis | https://github.com/ahmedmalaa/AutoPrognosis |
| ML-Plan | WEKA scikit-learn | HTN and EA | Yes | No | Binary classification Multi-label classification | https://github.com/fmohr/ML-Plan | |
| Auto- stacker | scikit-learn XGBoost | Hierarchical stacking and EA | No | No | Binary classification Multi-label classification | No implementation found | |
| Alpha3DM | PyTorch | NN and Monte Carlo Tree Search | Yes | Yes | Binary classification Multi-label classification Regression | No implementation found | |
| PoSH AUTO-sklearn | scikit-learn | BO with successive halving | Yes | Yes | Meta-learner | Binary classification Multi-label classification Regression | No implementation found |
| Auto-Keras | scikit-learn | BO guided network morphism | Yes | No | Binary classification Multi-label classification | https://autokeras.com/ | |
| ATM | scikit-learn | Conditional Parameter Tree | Yes | No | Meta-learner | Binary classificationMulti-label classification | https://github.com/HDI-Project/ATM |
Appendix B
| # | User Story | Frequency | Category |
|---|---|---|---|
| 1 | As a researcher-physician, I want to know how a prediction mechanism works, so that I can trust it more easily. | 5 | MX |
| 2 | As a researcher-physician, I want to be able to perform ML without having to code, so that I do not have to spend time learning how to program. | 5 | MC |
| 3 | As a researcher-physician, I want to use logistic regression, so that I can follow the medical guidelines for research. | 4 | MC |
| 4 | As a researcher-physician, I want to see which variables are included and excluded in the model, so that I can assess variable importance. | 4 | MX |
| 5 | As a researcher-physician, I want to see the difference between models with different variables included so that I can assess variable importance. | 4 | MX |
| 6 | As a researcher-physician, I want to transfer my model into a calculation tool, so that it can be used in clinical practice. | 4 | MU |
| 7 | As a researcher-physician, I want to have results within a day, so that I do not have to wait. | 4 | MC, MU |
| 8 | As a researcher-physician, I want to know the statistical power of a created model, so that I know if I can use it. | 4 | MX |
| 9 | As a researcher-physician, I want that the AutoML method explains its decisions, so that I can check its reasoning. | 3 | MX |
| 10 | As a researcher-physician, I want to have a graphical user interface, so that the chance of making errors is less than while coding. | 2 | UI |
| 11 | As a researcher-physician, I want to see the importance of each variable. So that I can check the reasoning of the computer. | 2 | MX |
| 12 | As a researcher-physician, I want to use code, so that I can trace back the decisions that I have made. | 2 | UI, MX |
| 13 | As a researcher-physician, I want to know what happens with missing data, so that I can evaluate the model correctly. | 2 | MC, MX |
| 14 | As a researcher-physician, I want to get suggestions for variables to include by the computer, so that I can improve my models. | 2 | MC |
| 15 | As a researcher-physician, I want to see the amount of variance that is explained by my model, so that I can assess the model quality. | 2 | MX |
Appendix C



Appendix D
- Artefact A is preferred over Artefact B to upload a dataset.
- Artefact A is preferred over Artefact B to create a subset.
- Artefact B is preferred over Artefact A to find the way in the different steps.
- Artefact B is preferred over Artefact A for the explanations of the workflow.
- Artefact B is preferred over Artefact A for progress reporting on model construction.
- Artefact B is preferred over Artefact A for model construction.
- Artefact A is preferred over Artefact B for comparing results of model creation.
- Artefact A is preferred over Artefact B for the explanation of missing data handling.
- Artefact B is preferred over Artefact A for reading the produced model.
- Users consider accuracy to be a good measure of model performance.
- Users want to know the statistical power of the created model.
- Users want to know the importance of each variable in the created model.
References
- Wang, X.; Noor-E-Alam, M.; Islam, M.; Hasan, M.; Germack, H. A Systematic Review on Healthcare Analytics: Application and Theoretical Perspective of Data Mining. Healthcare 2018, 6, 54. [Google Scholar]
- Chawla, N.V.; Davis, D.A. Bringing big data to personalized healthcare: A patient-centered framework. J. Gen. Intern. Med. 2013, 28, 660–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghupathi, W.; Raghupathi, V. Big data analytics in healthcare: Promise and potential. Heal. Inf. Sci. Syst. 2014, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Spruit, M.; Lytras, M. Applied data science in patient-centric healthcare: Adaptive analytic systems for empowering physicians and patients. Telemat. Inform. 2018, 35, 643–653. [Google Scholar] [CrossRef]
- Spruit, M.; Vries de, N. Self-Service Data Science for Adverse Event Prediction with AutoCrisp. In Proceedings of the Springer Proceedings in Complexity, Research & Innovation Forum, Athens, Greece, 15–17 April 2020. [Google Scholar]
- Spruit, M.; Jagesar, R. Power to the People!—Meta-Algorithmic Modelling in Applied Data Science. In Proceedings of the 8th International Conference on Knowledge Discovery and Information Retrieval, Porto, Portugal, 9–11 November 2016; Volume 1, pp. 400–406. [Google Scholar]
- Spruit, M.R.; Dedding, T.; Vijlbrief, D. Self-Service Data Science for Healthcare Professionals: A Data Preparation Approach. In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020)—Volume 5: HEALTHINF, Valletta, Malta, 24–26 February 2020; pp. 724–734. [Google Scholar]
- Thornton, C.; Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Auto-WEKA: Combined Selection and Hyperparameter Optimization of Classification Algorithms. In Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 22–27 August 2013. [Google Scholar]
- Hutter, F.; Kotthoff, L.; Vanschoren, J. Automatic Machine Learning: Methods, Systems, Challenges; Springer: New York, NY, USA, 2019. [Google Scholar]
- Zhang, Y.; Bahadori, M.T.; Su, H.; Sun, J. FLASH: Fast Bayesian Optimization for Data Analytic Pipelines. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1–21. [Google Scholar]
- Alaa, A.M.; van der Schaar, M. AutoPrognosis: Automated Clinical Prognostic Modeling via Bayesian Optimization with Structured Kernel Learning. In Proceedings of the International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; pp. 139–148. [Google Scholar]
- Kotthoff, L.; Thornton, C.; Hoos, H.H.; Hutter, F.; Leyton-Brown, K. Auto-WEKA 2.0: Automatic model selection and hyperparameter optimization in WEKA. J. Phys. B At. Mol. Opt. Phys. 2017, 18, 1–5. [Google Scholar]
- Komer, B.; Bergstra, J.; Eliasmith, C. Hyperopt-Sklearn: Automatic HyperparameterConfiguration for Scikit-Learn. ICML Work. AutoML 2014, 2825–2830. [Google Scholar] [CrossRef] [Green Version]
- Feurer, M.; Klein, A.; Eggensperger, K.; Springenberg, J.T.; Blum, M.; Hutter, F. Efficient and Robust Automated Machine Learning. In Proceedings of the 28th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; MIT Press: Cambridge, MA, USA, 2015; pp. 2755–2763. [Google Scholar]
- Feurer, M.; Eggensperger, K.; Falkner, S.; Lindauer, M.; Hutter, F. Practical Automated Machine Learning for the AutoML Challenge 2018. In Proceedings of the ICML 2018 AutoML Workshop, Stockholm, Sweden, 10–15 July 2018. [Google Scholar]
- Mohr, F.; Wever, M.; Hüllermeier, E. ML-Plan: Automated machine learning via hierarchical planning. Mach. Learn. 2018, 107, 1495–1515. [Google Scholar] [CrossRef] [Green Version]
- Bergstra, J.S.; Bardenet, R.; Bengio, Y.; Kégl, B. Algorithms for hyper-parameter optimization. In Proceedings of the Advances in Neural Information Processing Systems, Granada, Spain, 12–14 December 2011; pp. 2546–2554. [Google Scholar]
- Bergstra, J.; Yamins, D.; Cox, D.D. Hyperopt: A Python Library for Optimizing the Hyperparameters of Machine Learning Algorithms. Comput. Sci. Discov. 2013, 1–8. [Google Scholar] [CrossRef]
- Vanschoren, J.; van Rijn, J.N.; Bischl, B.; Torgo, L. OpenML: Networked science in machine learning. ACM SIGKDD Explor. Newsl. 2014. [Google Scholar] [CrossRef] [Green Version]
- Nau, D.; Au, T.C.; Ilghami, O.; Kuter, U.; Murdock, J.W.; Wu, D.; Yaman, F. SHOP2: An HTN planning system. J. Artif. Intell. Res. 2003, 20, 379–404. [Google Scholar] [CrossRef]
- Mendoza, H.; Klein, A.; Feurer, M.; Springenberg, J.T.; Hutter, F. Towards Automatically-Tuned Neural Networks. Proc. Work. Autom. Mach. Learn. 2016, 64, 58–65. [Google Scholar]
- Drori Krishnamurthy, Y.; Rampin, R.; Lourenço, R.; Ono, J.P.; Cho, K.; Silva, C.T.; Freire, J. AlphaD3M: Machine Learning Pipeline Synthesis. In Proceedings of the ICML 2018 AutoML Workshop, Stockholm, Sweden, 10–15 July 2018. [Google Scholar]
- Jin, H.; Song, Q.; Hu, X. Auto-Keras: Efficient Neural Architecture Search with Network Morphism. arXiv 2018, arXiv:1806.10282. [Google Scholar]
- Olson, R.S.; Moore, J.H. TPOT: A tree-based pipeline optimization tool for automating machine learning. In Work. Automated Machine Learning; Springer: New York, NY, USA, 2006; pp. 66–74. [Google Scholar]
- Gijsbers, P.; Vanschoren, J.; Olson, R.S. Layered TPOT: Speeding up tree-based pipeline optimization. In Proceedings of the European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases, Skopje, Macedonia, 18–22 September 2017; pp. 49–68. [Google Scholar]
- de Sá, A.G.C.S.; Pinto, W.J.G.S.; Oliveira, L.O.V.B.; Pappa, G.L. RECIPE: A grammar-based framework for automatically evolving classification pipelines. Lect. Notes Comput. Sci. (Incl. Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinform.) 2017, 10196 LNCS, 246–261. [Google Scholar]
- Chen, B.; Wu, H.; Mo, W.; Chattopadhyay, I.; Lipson, H. Autostacker: A Compositional Evolutionary Learning System. In Proceedings of the Genetic and Evolutionary Computation Conference, Kyoto, Japan, 15–19 July 2018; pp. 402–409. [Google Scholar]
- Hornby, G.S. ALPS: The Age-Layered Population Structure for Reducing the Problem of Premature Convergence. In Proceedings of the 8th Annual Conference on Genetic and Evolutionary Computation, Seattle, DC, USA, 8–12 July 2006. [Google Scholar]
- Swearingen, T.; Drevo, W.; Cyphers, B.; Cuesta-Infante, A.; Ross, A.; Veeramachaneni, K. ATM: A distributed, collaborative, scalable system for automated machine learning. In Proceedings of the 2017 IEEE International Conference on Big Data, Big Data 2017, Boston, MA, USA, 11–14 December 2017; pp. 151–162. [Google Scholar]
- Hevner, A.R.; Ram, S.; March, S.; Park, J. Design Science in Information Systems. MIS Q. 2004, 28, 75–105. [Google Scholar]
- Gijsbers, P.; Ledell, E.; Thomas, J.; Poirier, S.; Bischl, B.; Vanschoren, J. An Open Source AutoML Benchmark. In Proceedings of the ICML Workshop on AutoML, Long Beach, CA, USA, 14 June 2019; pp. 1–8. [Google Scholar]
- Bischl, B.; Casalicchio, G.; Feurer, M.; Hutter, F.; Lang, M.; Mantovani, R.G.; van Rijn, J.N.; Vanschoren, J. OpenML Benchmarking Suites and the OpenML100. arxiv 2017, arXiv:1708.03731. [Google Scholar]
- Mangasariona, O.L.; Wolberg, W.H. Cancer Diagnosis via Linear Programming. SIAM News 1990, 23, 1–18. [Google Scholar]
- Dua, D.; Graff, C. UCI Machine Learning Repository. 2019. Available online: https://archive.ics.uci.edu/ml/index.php (accessed on 28 April 2019).
- Quinlan, J.R. Simplifying decision trees. International journal of man-machine studies. Int. J. Man Mach. Stud. 1987, 27, 221–234. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.; Dieste, O.; Hickey, A.; Juristo, N.; Moreno, A.M. Effectiveness of Requirements Elicitation Techniques: Empirical Results Derived from a Systematic Review. In Proceedings of the 14th IEEE International Requirements Engineering Conference, Minneapolis/St. Paul, MN, USA, 11–15 September 2006; pp. 179–188. [Google Scholar]
- Al-Busaidi, Z.Q. Qualitative research and its uses in health care. Sultan Qaboos Univ. Med. J. 2008, 8, 9–11. [Google Scholar] [PubMed]
- Pope, C.; van Royen, P.; Baker, R. Qualitative methods in research on healthcare quality. Qual. Saf. Health Care 2002, 11, 52–148. [Google Scholar] [CrossRef] [Green Version]
- Castillo-Montoya, M. Preparing for interview research: The interview protocol refinement framework. Qual. Rep. 2016, 21, 811–831. [Google Scholar]
- Venable, J.; Pries-Heje, J.; Baskerville, R. FEDS: A Framework for Evaluation in Design Science Research. Eur. J. Inf. Syst. 2016, 25, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Offermann, P.; Levina, O.; Schönherr, M.; Bub, U. Outline of a Design Science Research Project. In Proceedings of the 4th International Conference Design Science Research in Information Systems and Technologies, Philadelphia, PA, USA, 6–8 May 2009. [Google Scholar]
- Cohen, M. User Stories Applied: For Agile Software Development; Addison-Wesley Professional: Boston, MA, USA, 2004. [Google Scholar]
- Janssen, J.H.N. The Right to Explanation: Means for ‘White-Boxing’ the Black-Box? Master’s Thesis, Tilburg University, Tilburg, The Netherlands, 2019. [Google Scholar]
- Chapman, P.; Clinton, J.; Kerber, R.; Khabaza, T.; Reinartz, T.; Shearer, C.; Wirth, R. The CRISP-DM user guide. In Proceedings of the 4th CRISP-DM SIG Workshop, Brussels, Belgium, 18 March 1999; Volume 1999. [Google Scholar]
- Christodoulou, E.; Ma, J.; Collins, G.S.; Steyerberg, E.W.; Verbakel, J.Y.; van Calster, B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol. 2019, 110, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Gareth, J.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning with Applications in R; Springer: New York, NY, USA, 2013. [Google Scholar]
- Kotsiantis, S.B. Supervised Machine Learning: A Review of Classification Techniques. Informatica 2007, 31, 249–268. [Google Scholar]
- Vollmer, S.; Mateen, B.; Bohner, G.; Király, F.; Ghani, R.; Jonsson, P.; Cumbers, S.; Jonas, A.; McAllister, K.; Myles, P.; et al. ine learning and AI research for Patient Benefit: 20 Critical Questions on Transparency, Replicability, Ethics and Effectiveness. arXiv 2018, arXiv:1812.10404. [Google Scholar]
- Kaplan, B.; Maxwell, J. Qualitative Research Methods for Evaluating Computer Information Systems. In Evaluating the Organizational Impact of Healthcare Information Systems; Springer: New York, NY, USA, 2005. [Google Scholar]
- Burke Johnson, R. Examining the Validity Structure of Qualitative Research. Education 1997, 118, 282–292. [Google Scholar]
- Kiegelmann, M. The Role of the Researcher in Qualitative Psychology; Ingeborg Huber Verlag: Tübingen, Germany, 2002. [Google Scholar]
- Polit, D.F.; Beck, C.T. Generalization in quantitative and qualitative research: Myths and strategies. Int. J. Nurs. Stud. 2010, 47, 1451–1458. [Google Scholar] [CrossRef] [Green Version]
- Michael, W.D.; Dustin, T.; Edward, C.; Jonas, K.; Jeremy, N.; Ghassen, J.; Katherine, H.; Andrew, M.D. Analyzing the Role of Model Uncertainty for Electronic Health Records. arXiv 2019, arXiv:1906.03842. [Google Scholar]
- Sung, N.; Crowley, J.W.F.; Genel, M.; Salber, P.; Sandy, L.; Sherwood, L.M.; Johnson, S.; Catanese, V.; Tilson, H.; Getz, K.; et al. Central Challenges Facing the National Clinical Research Enterprise. JAMA J. Am. Med. Assoc. 2003, 289, 1278–1287. [Google Scholar] [CrossRef]
- Molnar, C. Interpretable Machine Learning; Leanpub: Munich, Germany, 2019. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. Model-Agnostic Interpretability of Machine Learning. In Proceedings of the ICML Workshop on Human Interpretability in Machine Learning, New York, NY, USA, 23 June 2016. [Google Scholar]
- Shapley, L.S. A value for n-person games. In Contributions to the Theory of Games; Princeton University Press: Princeton, NJ, USA, 1953; pp. 307–317. [Google Scholar]
- Bartoletti, I. AI in Healthcare: Ethical and Privacy Challenges. In Artificial Intelligence in Medicine; Elsevier: Amsterdam, The Netherlands, 2019; pp. 7–10. [Google Scholar]




| Breast | Diabetes | Liver | Sick | |
|---|---|---|---|---|
| H-statistic | 11.36 | 18.64 | 17.93 | 27.87 |
| P-value | 0.995 ** | 0.324 ** | 0.455 ** | 0.386 ** |
| Breast | Diabetes | Liver | Sick | |||||
|---|---|---|---|---|---|---|---|---|
| p | U | p | U | p | U | p | U | |
| DecisionTree | 0.164 ** | 2.0 | 0.908 ** | 0.0 | 0.895 ** | 8.0 | 0.913 ** | 0.0 |
| TPOT | 0.236 | 40.0 | ||||||
| AutoWEKA | 0.395 | 46.0 | 0.455 | 48.0 | 0.263 | 40.0 | 0.913 ** | 0.0 |
| Autosklearn | 0.425 | 47.0 | 0.213 | 39.0 | 0.5 | 49.5 | ||
| Hyperoptsklearn | 0.8 * | 17.5 | 0.164 ** | 2.0 | 0.657 ** | 7.0 | 0.913 ** | 0.0 |
| Category/Method | Auto Sklearn | Auto-WEKA | TPOT | Hyperopt-Sklearn |
|---|---|---|---|---|
| User interaction (UI) | n/a | n/a | n/a | n/a |
| Model construction (MC) | 3 | 3 | 4 | 2 |
| Model explanation (MX) | 0 | 0 | 0 | 0 |
| Model usage (MU) | 1 | 1 | 1 | 1 |
| Total matches | 4 | 4 | 5 | 3 |
| Category | Preference | Score |
|---|---|---|
| User interaction | ||
| Upload dataset | GUI | 4/5 |
| Create a subset | Code | 3/5 |
| Workflow | Code | 4/5 |
| Workflow explanation | Code | 4/5 |
| Model construction | ||
| Progress reporting | Code | 4/5 |
| Model construction | Code | 5/5 |
| Model explanation | ||
| Compare results | GUI | 4/5 |
| Explanation missing data | Code | 4/5 |
| Readability | GUI | 4/5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ooms, R.; Spruit, M. Self-Service Data Science in Healthcare with Automated Machine Learning. Appl. Sci. 2020, 10, 2992. https://0-doi-org.brum.beds.ac.uk/10.3390/app10092992
Ooms R, Spruit M. Self-Service Data Science in Healthcare with Automated Machine Learning. Applied Sciences. 2020; 10(9):2992. https://0-doi-org.brum.beds.ac.uk/10.3390/app10092992
Chicago/Turabian StyleOoms, Richard, and Marco Spruit. 2020. "Self-Service Data Science in Healthcare with Automated Machine Learning" Applied Sciences 10, no. 9: 2992. https://0-doi-org.brum.beds.ac.uk/10.3390/app10092992