Comprehensive Exonic Sequencing of Known Ataxia Genes in Episodic Ataxia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cohort Summary
2.2. Whole Exome Sequencing Pipeline
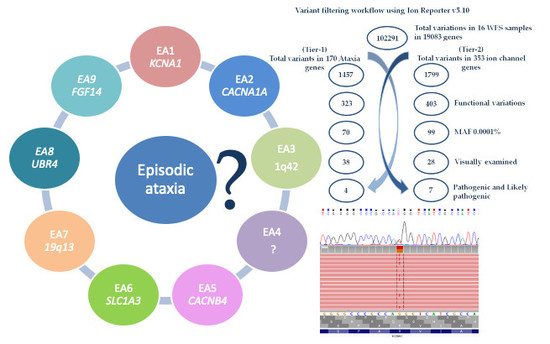
2.3. Variant Filtering Workflow
2.4. Sanger Sequencing (SS) Method
3. Results
3.1. Genetic Analysis
3.2. Ataxia Gene-Panel Analysis (Tier-1)
3.3. Ion Channel Analysis (Tier-2)
4. Discussion
4.1. Ataxia Gene-Panel Variants (Tier-1)
4.2. Ion Channel Gene-Panel Variants (Tier-2)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| WES | Whole Exome Sequencing |
| EAs | Episodic Ataxias |
| ACMG | American College of Medical Genetics and Genomics standards and guidelines |
| PCR | Polymerase Chain Reaction |
| BDT | BigDye terminator |
| HGVS | Human Genome Variation Society |
| MAF | Minor Allele frequency |
| VOUS | Variants with unknown significance |
References
- Jen, J.C.; Wan, J. Episodic ataxias. Handbook Clin. Neurol. 2018, 155, 205–215. [Google Scholar]
- Humbertclaude, V.; Remerand, G.; Hadjadj, J.; Rejou, F.; Coubes, C.; Pinson, L.; Meyer, P.; Roubertie, A. FGF14-related episodic ataxia: Delineating the phenotype of Episodic Ataxia type 9. Ann. Clin. Transl. Neurol. 2020, 7, 565–572. [Google Scholar]
- Browne, D.L.; Gancher, S.; Nutt, J.G.; Brunt, E.R.P.; Smith, E.A.; Kramer, P.; Litt, M. Episodic ataxia/myokymia syndrome is associated with point mutations in the human potassium channel gene, KCNA1. Nat. Genet. 1994, 8, 136–140. [Google Scholar] [CrossRef]
- Strupp, M.; Zwergal, A.; Brandt, T. Episodic ataxia type 2. Neurotherapeutics 2007, 4, 267–273. [Google Scholar] [CrossRef] [Green Version]
- Ophoff, R.A.; Terwindt, G.M.; Vergouwe, M.N.; Van Eijk, R.; Oefner, P.J.; Hoffman, S.M.; Lamerdin, J.E.; Mohrenweiser, H.W.; E Bulman, D.; Ferrari, M.; et al. Familial hemiplegic migraine and episodic ataxia Type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 1996, 87, 543–552. [Google Scholar] [CrossRef] [Green Version]
- Maksemous, N.; Roy, B.; Smith, R.A.; Griffiths, L.R. Next-generation sequencing identifies novel CACNA1A gene mutations in episodic ataxia type 2. Mol. Genet. Genom. Med. 2016, 4, 211–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maksemous, N.; Smith, R.A.; Sutherland, H.G.; Sampaio, H.; Griffiths, L.R. Whole-exome sequencing implicates SCN2A in episodic ataxia, but multiple ion channel variants may contribute to phenotypic complexity. Int. J. Mol. Sci. 2018, 19, 3113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, T.D. Hereditary Ataxia Overview. In GeneReviews(R); Adam, M.P., Ardinger, H.H., Eds.; GeneReviews® [Internet]; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Conroy, J.; Mcgettigan, P.; Murphy, R.; Webb, D.; Murphy, S.; McCoy, B.; Albertyn, C.; McCreary, D.; McDonagh, C.; Walsh, O.; et al. A novel locus for episodic ataxia: UBR4 the likely candidate. Eur. J. Hum. Genet. 2014, 22, 505–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damji, K.F.; Allingham, R.R.; Pollock, S.C.; Small, K.; Lewis, K.E.; Stajich, J.M.; Yamaoka, L.H.; Vance, J.M.; Pericak-Vance, M.A. Periodic vestibulocerebellar ataxia, an autosomal dominant ataxia with defective smooth pursuit, is genetically distinct from other autosomal dominant ataxias. Arch. Neurol. 1996, 53, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Németh, A.H.; Kwasniewska, A.C.; Lise, S.; Schnekenberg, R.P.; Becker, E.B.E.; Bera, K.D.; Shanks, M.E.; Gregory, L.; Buck, D.; Cader, M.Z.; et al. Next generation sequencing for molecular diagnosis of neurological disorders using ataxias as a model. Brain A J. Neurol. 2013, 136, 3106–3118. [Google Scholar] [CrossRef]
- Dunn, P.J.; Maher, B.H.; Albury, C.L.; Stuart, S.; Sutherland, H.G.; Maksemous, N.; Benton, M.C.; Smith, R.A.; Haupt, L.M.; Griffiths, L.R. Tiered analysis of whole-exome sequencing for epilepsy diagnosis. Mol. Genet. Genom. MGG 2020, 295, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, O.; Sutherland, H.G.; Maksemous, N.; Smith, R.A.; Haupt, L.M.; Griffiths, L.R. Exploring neuronal vulnerability to head trauma using a whole exome approach. J. Neurotrauma 2020. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Maksemous, N.; Smith, R.; Menon, S.; Davies, G.; Griffiths, L.R. Two novel mutations and a previously unreported intronic polymorphism in the NOTCH3 gene. Mutat. Res. 2012, 732, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.T.; Silver, K.; Young, R.; Debrosse, S.D.; Ebel, R.S.; Swoboda, K.J.; Acsadi, G. Fever-induced paroxysmal weakness and encephalopathy, a new phenotype of ATP1A3 mutation. Pediatr. Neurol. 2017, 73, 101–105. [Google Scholar] [CrossRef]
- Motley, A.M.; Brites, P.; Gerez, L.; Hogenhout, E.; Haasjes, J.; Benne, R.; Tabak, H.F.; Wanders, R.J.A.; Waterham, H.R. Mutational spectrum in the PEX7 gene and functional analysis of mutant alleles in 78 patients with rhizomelic chondrodysplasia punctata type 1. Am. J. Hum. Genet. 2002, 70, 612–624. [Google Scholar] [CrossRef] [Green Version]
- Van den Brink, D.M.; Brites, P.; Haasjes, J.; Wierzbicki, A.S.; Mitchell, J.; Lambert-Hamill, M.; de Belleroche, J.; Jansen, G.A.; Waterham, H.R.; Wanders, R.J.A. Identification of PEX7 as the second gene involved in Refsum disease. Am. J. Hum. Genet. 2003, 72, 471–477. [Google Scholar] [CrossRef] [Green Version]
- Tomlinson, S.E.; Rajakulendran, S.; Tan, S.V.; Graves, T.D.; Bamiou, D.-E.; Labrum, R.W.; Burke, D.; Sue, C.M.; Giunti, P.; Schorge, S.; et al. Clinical, genetic, neurophysiological and functional study of new mutations in episodic ataxia type 1. J. Neurol. Neurosurg. Psychiatry 2013, 84, 1107–1112. [Google Scholar] [CrossRef] [Green Version]
- Wallace, R.H.; Wang, D.W.; Singh, R.; Scheffer, I.E.; George, A.L.; Phillips, H.A.; Saar, K.; Reis, A.; Johnson, E.W.; Sutherland, G.R.; et al. Febrile seizures and generalized epilepsy associated with a mutation in the Na + -channel beta1 subunit gene SCN1B. Nat. Genet. 1998, 19, 366–3670. [Google Scholar] [CrossRef]
- Schwarz, N.; Bast, T.; Gaily, E.; Golla, G.; Gorman, K.; Griffiths, L.; Hahn, A.; Hukin, J.; King, M.; Korff, C.; et al. Clinical and genetic spectrum of SCN2A-associated episodic ataxia. Eur. J. Paediatr. Neurol. EJPN Off. J. Eur. Paediatr. Neurol. Soc. 2019, 23, 438–447. [Google Scholar] [CrossRef] [Green Version]
- Brashear, A.; Mink, J.W.; Hill, D.F.; Boggs, N.; McCall, W.V.; Stacy, M.A.; Snively, B.; Light, L.S.; Sweadner, K.J.; Ozelius, L.J.; et al. ATP1A3 mutations in infants: A new rapid-onset dystonia-Parkinsonism phenotype characterized by motor delay and ataxia. Dev. Med. Child Neurol. 2012, 54, 1065–1067. [Google Scholar] [CrossRef] [PubMed]
- Herndon, J.H., Jr.; Steinberg, D.; Uhlendorf, B.W. Refsum’s disease: Defective oxidation of phytanic acid in tissue cultures derived from homozygotes and heterozygotes. N. Eng. J. Med. 1969, 281, 1034–1438. [Google Scholar] [CrossRef]
- Chen, T.T.; Klassen, T.L.; Goldman, A.M.; Marini, C.; Guerrini, R.; Noebels, J.L. Novel brain expression of ClC-1 chloride channels and enrichment of CLCN1 variants in epilepsy. Neurology 2013, 80, 1078–1085. [Google Scholar] [CrossRef] [PubMed]
- Dupré, N.; Chrestian, N.; Bouchard, J.-P.; Rossignol, E.; Brunet, D.; Sternberg, D.; Brais, B.; Mathieu, J.; Puymirat, J. Clinical, electrophysiologic, and genetic study of non-dystrophic myotonia in French-Canadians. Neuromuscul. Disord. NMD 2009, 19, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Matthews, E.; Fialho, D.; Tan, S.V.; Venance, S.L.; Cannon, S.C.; Sternberg, D.; Fontaine, B.; Amato, A.A.; Barohn, R.J.; Griggs, R.; et al. The non-dystrophic myotonias: Molecular pathogenesis, diagnosis and treatment. Brain A J. Neurol. 2009, 133, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Helbig, K.L.; Lauerer, R.; Bahr, J.C.; Souza, I.A.; Myers, C.T.; Uysal, B.; Schwarz, N.; Gandini, M.A.; Huang, S.; Keren, B.; et al. De novo pathogenic variants in CACNA1E cause developmental and epileptic encephalopathy with contractures, macrocephaly, and dyskinesias. Am. J. Hum. Genet. 2018, 103, 666–678. [Google Scholar] [CrossRef] [Green Version]
- Heyne, H.; Singh, T.; Stamberger, H.; Jamra, R.A.; Caglayan, H.; Craiu, D.C.; De Jonghe, P.; Guerrini, R.; Helbig, K.L.; Koeleman, B.P.C.; et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet. 2018, 50, 1048–1053. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Westenbroek, R.E.; Xu, X.; Edwards, C.A.; Sorenson, D.R.; Chen, Y.; McEwen, D.P.; O’Malley, H.A.; Bharucha, V.; Meadows, L.S.; et al. Mice lacking sodium channel beta1 subunits display defects in neuronal excitability, sodium channel expression, and nodal architecture. J. Neurosci. Off. J. Soc. Neurosci. 2004, 24, 4030–4042. [Google Scholar] [CrossRef] [Green Version]
- Catterall, W.A. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron 2000, 26, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.A.; Pappas, C.; Dahle, E.J.; Claes, L.R.F.; Pruess, T.H.; De Jonghe, P.; Thompson, J.; Dixon, M.; Gurnett, C.A.; Peiffer, A.; et al. A Role of SCN9A in human epilepsies, as a cause of febrile seizures and as a potential modifier of dravet syndrome. PLoS Genet. 2009, 5, e1000649. [Google Scholar] [CrossRef]

| ID | Age at Test Request (Years) | Gender | Age at Onset (Years) | Familial History | Clinical Information | Acetazolamide Response |
|---|---|---|---|---|---|---|
| 1 | 2.5 | F | 2 | NO | Episodes of ataxia | - |
| 2 | 15 | M | - | YES | Episodes of ataxia associated with fever and gait ataxia and past pointing. Attacks triggered by a mild head trauma | Positive |
| 4 | 37 | F | - | - | Episodes of ataxia, possible hemiplegic migraine | - |
| 6 | 6 | M | 1 | YES | Episodes of ataxia associated with fever; gait ataxia; 4–5 attacks/year that can last for several days | Negative |
| 7 | 60 | M | - | - | Episodes of ataxia | - |
| 10 | 37 | F | - | - | Daily attack | - |
| 11 | 26 | M | - | - | Episodes of ataxia | - |
| 12 | 76 | F | 61 | YES | Attacks of vertigo, +/− headache every few months | Partial |
| 14 | 27 | F | - | - | Severe ataxia, nausea, vomiting, nystagmus; Four attacks/year | Positive |
| 16 | 42 | F | - | YES | Episodic ataxia | - |
| 19 | 38 | F | - | - | Vertigo, fluctuating ataxia, abnormal nerve excitability | - |
| 20 | 80 | F | - | - | Episodes of ataxia | - |
| 21 | 80 | M | 60 | - | Late onset of episodic ataxia | - |
| 23 | 24 | M | - | - | Unsteadiness, muscle myokemia, exercise induced | Partial |
| 25 | 59 | M | - | YES | Ataxia, no headache. | - |
| 31 | 56 | F | - | NO | Progressive ataxia, vertigo | - |
| ID | Locus (hg19) | Ref | Genes | Transcript | Amino Acid Change | Coding | GnomAD Frequency | ACMG Rules | Verdict |
|---|---|---|---|---|---|---|---|---|---|
| 2 | chr2:166231195 | G | SCN2A | NM_001040143.1 | p.Val1325Phe | c.3973G > T | - | PM1,PM2,PP2,PP3 | Likely Pathogenic |
| 6 | chr19:42474691 | C | ATP1A3 | NM_152296.4 | p.Arg756His | c.2267G > A | - | PM1,PM2,PM5,PP2,PP3,PP5 | Pathogenic |
| 14 | chr7:143048918 | G | CLCN1 | NM_000083.2 | p.Gly945fs | c.2831dup | 0.000016 | PVS1,PM2,PP3 | Pathogenic |
| 19 | chr6:137143923 | C | PEX7 | NM_000288.3 | p.Tyr40Ter | c.120C > G | 0.0000907 | PVS1, PS3, PM2, PP3. | Pathogenic |
| 21 | chr1:181689430 | A | CACNA1E | NM_001205293.1 | p.Ile614Val | c.1840A > G | 0.00000804 | PM1,PM2,PP2,PP3 | Likely Pathogenic |
| 23 | chr12:5021044 | G | KCNA1 | NM_000217.2 | p.Arg167Met | c.500G > T | - | PM1,PM2,PP2,PP3 | Likely Pathogenic |
| 25 | chr19:35524558 | C | SCN1B | NM_199037.4 | p.Cys121Trp | c.363C > G | 0.0000141 | PM1,PM2,PP3,PP5 | Likely Pathogenic |
| 31 | chr2:167094721 | A | SCN9A | NM_002977.3 | p.Tyr1217Ter | c.3651T > G | - | PVS1,PM2,PP3 | Pathogenic |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maksemous, N.; Sutherland, H.G.; Smith, R.A.; Haupt, L.M.; Griffiths, L.R. Comprehensive Exonic Sequencing of Known Ataxia Genes in Episodic Ataxia. Biomedicines 2020, 8, 134. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines8050134
Maksemous N, Sutherland HG, Smith RA, Haupt LM, Griffiths LR. Comprehensive Exonic Sequencing of Known Ataxia Genes in Episodic Ataxia. Biomedicines. 2020; 8(5):134. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines8050134
Chicago/Turabian StyleMaksemous, Neven, Heidi G. Sutherland, Robert A. Smith, Larisa M. Haupt, and Lyn R. Griffiths. 2020. "Comprehensive Exonic Sequencing of Known Ataxia Genes in Episodic Ataxia" Biomedicines 8, no. 5: 134. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines8050134