Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems
Abstract
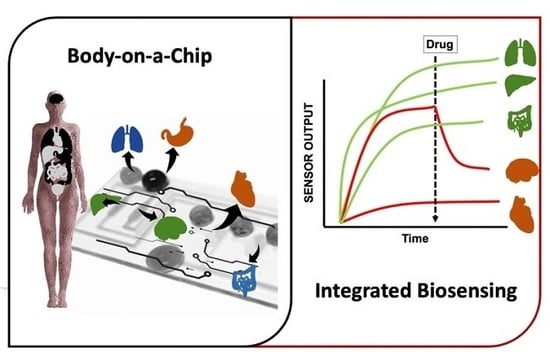
:1. Introduction
2. Biosensors for Organs-on-a-Chip Systems with Single Tissue Models
2.1. Biosensors for Analysis of Organ & Cancer Tissue Metabolism
2.2. Monitoring in Endothelial & Epithelial Barrier-on-a-Chip Models
2.3. Cardiac and Skeletal Muscle-on-a-Chip Systems with Integrated Biosensors
2.4. In Situ Analysis of Human Microfluidic Nervous Systems and Blood Brain Barrier Models
3. Bio Multi Organ and Human-on-a-Chip Systems
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mak, I.W.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114. [Google Scholar] [PubMed]
- Liu, K.D.; Humphreys, B.D.; Endre, Z.H. The ten barriers for translation of animal data on AKI to the clinical setting. Intensive Care Med. 2017, 43, 898–900. [Google Scholar] [CrossRef] [PubMed]
- Leenaars, C.H.; Kouwenaar, C.; Stafleu, F.R.; Bleich, A.; Ritskes-Hoitinga, M.; De Vries, R.B.; Meijboom, F.L. Animal to human translation: A systematic scoping review of reported concordance rates. J. Transl. Med. 2019, 17, 223. [Google Scholar] [CrossRef]
- Rothbauer, M.; Rosser, J.M.; Zirath, H.; Ertl, P. Tomorrow today: Organ-on-a-chip advances towards clinically relevant pharmaceutical and medical in vitro models. Curr. Opin. Biotechnol. 2019, 55, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef]
- van Midwoud, P.M.; Verpoorte, E.; Groothuis, G.M. Microfluidic devices for in vitro studies on liver drug metabolism and toxicity. Integr. Biol. 2011, 3, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Choi, J.-H.; Kim, H.J. Human Gut-on-a-Chip Technology: Will this Revolutionize Our Understanding of IBD and Future Treatments; Taylor & Francis: London, UK, 2016. [Google Scholar]
- Weber, E.J.; Chapron, A.; Chapron, B.D.; Voellinger, J.L.; Lidberg, K.A.; Yeung, C.K.; Wang, Z.; Yamaura, Y.; Hailey, D.W.; Neumann, T. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int. 2016, 90, 627–637. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.I.; Abaci, H.E.; Shuler, M.L. Microfluidic blood–brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol. Bioeng. 2017, 114, 184–194. [Google Scholar] [CrossRef]
- Günther, A.; Yasotharan, S.; Vagaon, A.; Lochovsky, C.; Pinto, S.; Yang, J.; Lau, C.; Voigtlaender-Bolz, J.; Bolz, S.-S. A microfluidic platform for probing small artery structure and function. Lab Chip 2010, 10, 2341–2349. [Google Scholar] [CrossRef]
- Bachmann, B.; Spitz, S.; Rothbauer, M.; Jordan, C.; Purtscher, M.; Zirath, H.; Schuller, P.; Eilenberger, C.; Ali, S.F.; Mühleder, S. Engineering of 3D pre-vascular networks within fibrin hydrogel constructs by microfluidic control over reciprocal cell signaling. Biomicrofluidics 2018, 12, 042216. [Google Scholar] [CrossRef]
- Jastrzebska, E.; Tomecka, E.; Jesion, I. Heart-on-a-chip based on stem cell biology. Biosens. Bioelectron. 2016, 75, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Marsano, A.; Conficconi, C.; Lemme, M.; Occhetta, P.; Gaudiello, E.; Votta, E.; Cerino, G.; Redaelli, A.; Rasponi, M. Beating heart on a chip: A novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016, 16, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.; Aung, A.; Varghese, S. Skeletal muscle-on-a-chip: An in vitro model to evaluate tissue formation and injury. Lab Chip 2017, 17, 3447–3461. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Romero, R.; Han, Y.M.; Kim, H.C.; Kim, C.J.; Hong, J.-S.; Huh, D. Placenta-on-a-chip: A novel platform to study the biology of the human placenta. J. Matern.-Fetal Neonatal Med. 2016, 29, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.N.; Lancaster, K.Z.; Hogue, I.B.; Meng, F.; Kong, Y.L.; Enquist, L.W.; McAlpine, M.C. 3D printed nervous system on a chip. Lab Chip 2016, 16, 1393–1400. [Google Scholar] [CrossRef] [PubMed]
- Arık, Y.B.; van der Helm, M.W.; Odijk, M.; Segerink, L.I.; Passier, R.; van den Berg, A.; van der Meer, A.D. Barriers-on-chips: Measurement of barrier function of tissues in organs-on-chips. Biomicrofluidics 2018, 12, 042218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Chen, Q.; Liu, W.; He, Z.; Lin, J.-M. Recent advances in microfluidic 3D cellular scaffolds for drug assays. TrAC Trends Anal. Chem. 2017, 87, 19–31. [Google Scholar] [CrossRef]
- Kaushik, G.; Leijten, J.; Khademhosseini, A. Concise review: Organ engineering: Design, technology, and integration. Stem Cells 2017, 35, 51–60. [Google Scholar] [CrossRef]
- Wall, M.; Butler, D.; El Haj, A.; Bodle, J.C.; Loboa, E.G.; Banes, A.J. Key developments that impacted the field of mechanobiology and mechanotransduction. J. Orthop. Res. 2018, 36, 605–619. [Google Scholar] [CrossRef]
- Wang, X.; Phan, D.T.; Sobrino, A.; George, S.C.; Hughes, C.C.; Lee, A.P. Engineering anastomosis between living capillary networks and endothelial cell-lined microfluidic channels. Lab Chip 2016, 16, 282–290. [Google Scholar] [CrossRef]
- Ertl, P.; Sticker, D.; Charwat, V.; Kasper, C.; Lepperdinger, G. Lab-on-a-chip technologies for stem cell analysis. Trends Biotechnol. 2014, 32, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Sticker, D.; Lechner, S.; Jungreuthmayer, C.; Zanghellini, J.; Ertl, P. Microfluidic migration and wound healing assay based on mechanically induced injuries of defined and highly reproducible areas. Anal. Chem. 2017, 89, 2326–2333. [Google Scholar] [CrossRef]
- Shen, C.; Meng, Q.; Zhang, G. Increased curvature of hollow fiber membranes could up-regulate differential functions of renal tubular cell layers. Biotechnol. Bioeng. 2013, 110, 2173–2183. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, V.; Rbaibi, Y.; Pastor-Soler, N.M.; Carattino, M.D.; Weisz, O.A. Shear stress-dependent regulation of apical endocytosis in renal proximal tubule cells mediated by primary cilia. Proc. Natl. Acad. Sci. USA 2014, 111, 8506–8511. [Google Scholar] [CrossRef] [Green Version]
- Ergir, E.E.; Bachmann, B.; Redl, H.R.; Forte, G.; Ertl, P. Small force, big impact: Next generation organ-on-a-chip systems incorporating biomechanical cues. Front. Physiol. 2018, 9, 1417. [Google Scholar] [CrossRef]
- Rothbauer, M.; Praisler, I.; Docter, D.; Stauber, R.; Ertl, P. Microfluidic impedimetric cell regeneration assay to monitor the enhanced cytotoxic effect of nanomaterial perfusion. Biosensors 2015, 5, 736–749. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Zirath, H.; Rothbauer, M.; Spitz, S.; Bachmann, B.; Jordan, C.; Müller, B.; Ehgartner, J.; Priglinger, E.; Mühleder, S.; Redl, H. Every breath you take: Non-invasive real-time oxygen biosensing in two-and 3D microfluidic cell models. Front. Physiol. 2018, 9, 815. [Google Scholar] [CrossRef]
- Novak, R.; Ranu, N.; Mathies, R.A. Rapid fabrication of nickel molds for prototyping embossed plastic microfluidic devices. Lab Chip 2013, 13, 1468–1471. [Google Scholar] [CrossRef] [Green Version]
- Tsao, C.-W. Polymer microfluidics: Simple, low-cost fabrication process bridging academic lab research to commercialized production. Micromachines 2016, 7, 225. [Google Scholar] [CrossRef]
- Lafleur, J.P.; Joensson, A.; Senkbeil, S.; Kutter, J.P. Recent advances in lab-on-a-chip for biosensing applications. Biosens. Bioelectron. 2016, 76, 213–233. [Google Scholar] [CrossRef] [PubMed]
- Gabardo, C.; Soleymani, L. Deposition, patterning, and utility of conductive materials for the rapid prototyping of chemical and bioanalytical devices. Analyst 2016, 141, 3511–3525. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothbauer, M.; Wartmann, D.; Charwat, V.; Ertl, P. Recent advances and future applications of microfluidic live-cell microarrays. Biotechnol. Adv. 2015, 33, 948–961. [Google Scholar] [CrossRef] [PubMed]
- Gruber, P.; Marques, M.P.; Szita, N.; Mayr, T. Integration and application of optical chemical sensors in microbioreactors. Lab Chip 2017, 17, 2693–2712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pires, N.; Dong, T.; Hanke, U.; Hoivik, N. Recent developments in optical detection technologies in lab-on-a-chip devices for biosensing applications. Sensors 2014, 14, 15458–15479. [Google Scholar] [CrossRef] [PubMed]
- Kieninger, J.; Weltin, A.; Flamm, H.; Urban, G.A. Microsensor systems for cell metabolism–from 2D culture to organ-on-chip. Lab Chip 2018, 18, 1274–1291. [Google Scholar] [CrossRef]
- Caballero, D.; Kaushik, S.; Correlo, V.; Oliveira, J.M.; Reis, R.; Kundu, S. Organ-on-chip models of cancer metastasis for future personalized medicine: From chip to the patient. Biomaterials 2017, 149, 98–115. [Google Scholar] [CrossRef]
- Perrier, R.; Pirog, A.; Jaffredo, M.; Gaitan, J.; Catargi, B.; Renaud, S.; Raoux, M.; Lang, J. Bioelectronic organ-based sensor for microfluidic real-time analysis of the demand in insulin. Biosens. Bioelectron. 2018, 117, 253–259. [Google Scholar] [CrossRef]
- Bavli, D.; Prill, S.; Ezra, E.; Levy, G.; Cohen, M.; Vinken, M.; Vanfleteren, J.; Jaeger, M.; Nahmias, Y. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 2016, 113, E2231–E2240. [Google Scholar] [CrossRef]
- Misun, P.M.; Rothe, J.; Schmid, Y.R.F.; Hierlemann, A.; Frey, O. Multi-analyte biosensor interface for real-time monitoring of 3D microtissue spheroids in hanging-drop networks. Microsyst. Nanoeng. 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakolish, C.M.; Esch, M.B.; Hickman, J.J.; Shuler, M.L.; Mahler, G.J. Modeling barrier tissues in vitro: Methods, achievements, and challenges. EBioMedicine 2016, 5, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.I.; Oleaga, C.; Long, C.J.; Esch, M.B.; McAleer, C.W.; Miller, P.G.; Hickman, J.J.; Shuler, M.L. Self-contained, low-cost Body-on-a-Chip systems for drug development. Exp. Biol. Med. 2017, 242, 1701–1713. [Google Scholar] [CrossRef] [PubMed]
- Mermoud, Y.; Felder, M.; Stucki, J.; Stucki, A.; Guenat, O. Microimpedance tomography system to monitor cell activity and membrane movements in a breathing lung-on-chip. Sens. Actuators B Chem. 2017, 255, 3647–3653. [Google Scholar] [CrossRef]
- Henry, O.Y.F.; Villenave, R.; Cronce, M.J.; Leineweber, W.D.; Benz, M.A.; Ingber, D.E. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip 2017, 17, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, Q.; Ting, F.C. In vitro micro-physiological immune-competent model of the human skin. Lab Chip 2016, 16, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef] [PubMed]
- Pitsalidis, C.; Ferro, M.P.; Iandolo, D.; Tzounis, L.; Inal, S.; Owens, R.M. Transistor in a tube: A route to 3D bioelectronics. Sci. Adv. 2018, 4, eaat4253. [Google Scholar] [CrossRef]
- Machuca, M.A.O.; Garibay, X.G.F.; Castaño, A.G.; De Chiara, F.; Albors, A.H.; Trias, J.B.; Azcon, J.R. Muscle-on-a-chip with on-site multiplexed biosensing system for in situ-monitoring of secreted IL-6 and TNF-α. Lab Chip 2019, 19, 2568–2580. [Google Scholar] [CrossRef]
- Huebsch, N.; Charrez, B.; Siemons, B.; Boggess, S.C.; Wall, S.; Charwat, V.; Jæger, K.H.; Montiel, F.T.L.; Jeffreys, N.C.; Deveshwar, N. Metabolically-driven maturation of hiPSC-cell derived heart-on-a-chip. bioRxiv 2018, 485169. [Google Scholar] [CrossRef]
- Caluori, G.; Pribyl, J.; Pesl, M.; Jelinkova, S.; Rotrekl, V.; Skladal, P.; Raiteri, R. Non-invasive electromechanical cell-based biosensors for improved investigation of 3D cardiac models. Biosens. Bioelectron. 2019, 124–125, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Inácio, P.; Mestre, A.; de Medeiros, M.; Asgarifar, S.; Canudo, J.; Elamine, Y.; Santos, J.; Morgado, J.; Braganca, J.; Biscarini, F.; et al. Bioelectrical Signal Detection Using Conducting Polymer Electrodes and the Displacement Current Method. IEEE Sens. J. 2017, 17, 3961–3966. [Google Scholar] [CrossRef]
- Koutsouras, D.A.; Perrier, R.; Villarroel Marquez, A.; Pirog, A.; Pedraza, E.; Cloutet, E.; Renaud, S.; Raoux, M.; Malliaras, G.G.; Lang, J. Simultaneous monitoring of single cell and of micro-organ activity by PEDOT:PSS covered multi-electrode arrays. Mater. Sci. Eng. C 2017, 81, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.R.; Zhang, Y.S.; Kim, D.-J.; Manbohi, A.; Avci, H.; Silvestri, A.; Aleman, J.; Hu, N.; Kilic, T.; Keung, W.; et al. Aptamer-Based Microfluidic Electrochemical Biosensor for Monitoring Cell-Secreted Trace Cardiac Biomarkers. Anal. Chem. 2016, 88, 10019–10027. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.R.; Kilic, T.; Zhang, Y.S.; Avci, H.; Hu, N.; Kim, D.; Branco, C.; Aleman, J.; Massa, S.; Silvestri, A.; et al. Label-Free and Regenerative Electrochemical Microfluidic Biosensors for Continual Monitoring of Cell Secretomes. Adv. Sci. 2017, 4, 1600522. [Google Scholar] [CrossRef] [PubMed]
- Riahi, R.; Shaegh, S.A.M.; Ghaderi, M.; Zhang, Y.S.; Shin, S.R.; Aleman, J.; Massa, S.; Kim, D.; Dokmeci, M.R.; Khademhosseini, A. Automated microfluidic platform of bead-based electrochemical immunosensor integrated with bioreactor for continual monitoring of cell secreted biomarkers. Sci. Rep. 2016, 6, 24598. [Google Scholar] [CrossRef] [PubMed]
- van Der Helm, M.W.; Van Der Meer, A.D.; Eijkel, J.C.; van den Berg, A.; Segerink, L.I. Microfluidic organ-on-chip technology for blood-brain barrier research. Tissue Barriers 2016, 4, e1142493. [Google Scholar] [CrossRef]
- Yi, Y.; Park, J.; Lim, J.; Lee, C.J.; Lee, S.-H. Central nervous system and its disease models on a chip. Trends Biotechnol. 2015, 33, 762–776. [Google Scholar] [CrossRef]
- Ahadian, S.; Civitarese, R.; Bannerman, D.; Mohammadi, M.H.; Lu, R.; Wang, E.; Davenport-Huyer, L.; Lai, B.; Zhang, B.; Zhao, Y. Organ-on-a-chip platforms: A convergence of advanced materials, cells, and microscale technologies. Adv. Healthc. Mater. 2018, 7, 1700506. [Google Scholar] [CrossRef]
- Uzel, S.G.; Platt, R.J.; Subramanian, V.; Pearl, T.M.; Rowlands, C.J.; Chan, V.; Boyer, L.A.; So, P.T.; Kamm, R.D. Microfluidic device for the formation of optically excitable, 3D, compartmentalized motor units. Sci. Adv. 2016, 2, e1501429. [Google Scholar] [CrossRef]
- Sticker, D.; Rothbauer, M.; Ehgartner, J.; Steininger, C.; Liske, O.; Liska, R.; Neuhaus, W.; Mayr, T.; Haraldsson, T.; Kutter, J.P. Oxygen Management at the Microscale: A Functional Biochip Material with Long-Lasting and Tunable Oxygen Scavenging Properties for Cell Culture Applications. ACS Appl. Mater. Interfaces 2019, 11, 9730–9739. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Kankala, R.K.; Wang, S.-B.; Chen, A.-Z. Multi-organs-on-chips: Towards long-term biomedical investigations. Molecules 2019, 24, 675. [Google Scholar] [CrossRef] [PubMed]
- Maoz, B.M.; Herland, A.; Henry, O.Y.F.; Leineweber, W.D.; Yadid, M.; Doyle, J.; Mannix, R.; Kujala, V.J.; FitzGerald, E.A.; Parker, K.K.; et al. Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 2017, 17, 2294–2302. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.F.L.; Huyer, L.D.; Lu, R.X.Z.; Drecun, S.; Radisic, M.; Zhang, B. InVADE: Integrated Vasculature for Assessing Dynamic Events. Adv. Funct. Mater. 2017, 27, 1703524. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Shaegh, S.A.M.; Massa, S.; Riahi, R.; Chae, S.; Hu, N. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302. [Google Scholar] [CrossRef] [PubMed]
- Oleaga, C.; Riu, A.; Rothemund, S.; Lavado, A.; McAleer, C.W.; Long, C.J.; Persaud, K.; Narasimhan, N.S.; Tran, M.; Roles, J. Investigation of the effect of hepatic metabolism on off-target cardiotoxicity in a multi-organ human-on-a-chip system. Biomaterials 2018, 182, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Murphy, S.V.; Devarasetty, M.; Mead, I.; Kang, H.-W.; Seol, Y.-J.; Zhang, Y.S.; Shin, S.-R.; Zhao, L.; Aleman, J. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017, 7, 8837. [Google Scholar] [CrossRef] [PubMed]
- Oleaga, C.; Bernabini, C.; Smith, A.S.T.; Srinivasan, B.; Jackson, M.; McLamb, W.; Platt, V.; Bridges, R.; Cai, Y.; Santhanam, N.; et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci. Rep. 2016, 6, 20030. [Google Scholar] [CrossRef] [PubMed]
- Oleaga, C.; Lavado, A.; Riu, A.; Rothemund, S.; Carmona-Moran, C.A.; Persaud, K.; Yurko, A.; Lear, J.; Narasimhan, N.S.; Long, C.J.; et al. Long-Term Electrical and Mechanical Function Monitoring of a Human-on-a-Chip System. Adv. Funct. Mater. 2018, 29, 1805792. [Google Scholar] [CrossRef]
- Gaio, N.; Waafi, A.; Vlaming, M.L.H.; Boschman, E.; Dijkstra, P.; Nacken, P.; Braam, S.R.; Boucsein, C.; Sarro, P.M.; Dekker, R. A multiwell plate Organ-on-Chip (OOC) device for in-vitro cell culture stimulation and monitoring. In Proceedings of the 2018 IEEE Micro Electro Mechanical Systems (MEMS), Belfast, UK, 21–25 January 2018; pp. 314–317. [Google Scholar]













| Organ | Simulated Organ Function | Cell Type (+Primary, -Cancer, * Stem Cell) | Tissue Architecture | Sensing Principle | References |
|---|---|---|---|---|---|
| Skeletal muscle | Tissue morphogenesis and maturation and effects to cardiotoxins | C2C12 mouse murine myoblast (-) | 3D cell-laden hydrogel structures | Pillar deformation (fluorescence microscopy) and finite element method | [14] |
| Vascular network | Oxygen gradients in vascular networks | HUVEC (human umbilical vein endothelial cells) (+) and ASC (adipose-derived stem cells) (+) | 3D cell-laden hydrogel structures | Oxygen sensing by fluorescence measurements of oxygen sensitive platinum-based dye (PtTPTBPF) | [29] |
| Pancreatic islets | Glucose concentration-dependent micro-organ activity | Β-cells of pancreatic islets adult male C57BL/6 mice (+) | 3D islets | Electrical activity sensing of pancreatic islets by multielectrode array | [40] |
| Liver | Mitochondrial respiration | HepG2/C3A (-) | 3D cell-laden hydrogel structures | Oxygen sensing by phosphorescence of a ruthenium dye and glucose and lactate sensing by oxidation of platinum electrodes | [41] |
| Cancer (colon) microtissue | Glucose and lactate metabolism | Fluorescent human colon carcinoma cell line HCT116 eGFP (-) | 3D spheroid | Glucose and lactate sensing by using electrodes functionalized with oxidase enzymes and amperometry | [42] |
| Lung | Mechanical strain of alveolar barrier during breathing | Human type II alveolar epithelial-like A549 cells (-) | Barrier model | Barrier movement and membrane permeabilization sensing by real-time measurement of resistivity changes in three impedimetric coplanar electrodes. | [45] |
| Lung and gut | Barrier function formation (by stem cell differentiation) | Primary human airway epithelial cells (hAECs) (+) and human Caco2 intestinal epithelial cells (-) | Barrier model | Barrier integrity sensing by TEER measurements | [46] |
| Skin | Allergic und irritant contact dermatitis | Immortalized human keratinocytes (HaCaT) (-) and human leukemic monocyte lymphoma cell line (U937) (-) | Barrier model | Barrier integrity and tight junction formation sensing by TEER measurements | [47] |
| Gastrointestinal human-microbe interface | Transcriptional, metabolic and immunological response | Caco-2 (-), CCD-18Co (+) and CD4+T cells (+) | Barrier model | Optodes for oxygen sensing and TEER measurements for cell growth and differentiation | [48] |
| Kidney | Barrier function | Canine epithelial kidney cells (MDCKII) (-) and human telomerase-immortalized fibroblasts (-) | 3D barrier model | Barrier integrity sensing by transconductance measurements | [49] |
| Skeletal muscle | Myokine secretion | Murine C2C12 skeletal myoblasts (-) | 3D cell-laden hydrogel structures | Myokine concentration measurement by functionalized gold electrodes | [50] |
| Heart | Formation of 3D cardiac microphysiological system | Human induced pluripotent stem cell derived cardiomyocytes (hiPSC-CMs) (*) | 3D cell-laden PDMS structure | Cardiac cell contraction sensing by micropillar deformation | [51] |
| Heart | Cardiac beat rate | Human embryoid stem cell line CCTL14 (*) and human induced pluripotent stem cells (*) | 3D organoid | Cardiomyocyte beating force sensing by multielectrode array and atomic force microscopy measurements | [52] |
| Embryoid body (cardiac cells) | Autonomous beat rate of embryoid bodies | Mouse embryonic stem cells (mESC) differentiated cardiomyocytes (*) | 3D embryoid body | Cardiac beat rate sensing by voltage and displacement current measurement by large area electrodes | [53] |
| Pancreatic Islets | Electrical Activity of single cells and islets | Pancreatic islets of mice and human (+) | 3D islets | Action potential local field potential measurement by multielectrode array | [54] |
| Heart | Cardiac biomarker secretion | Human embryonic stem cell-derived cardiomyocytes (ESC-CMs) (*) | 3D cell-laden hydrogel structures | Creatine kinase (CK)-MB sensing by impedance measurements using an aptamer functionalized microelectrode | [55] |
| Liver | Hepatic biomarker secretion | Human primary hepatocytes (+) | 3D cell-laden hydrogel structures | Biomarker sensing by impedance measurements regeneratable gold electrodes | [56] |
| Liver | Hepatic biomarker secretion | HepG2 (-) and primary human hepatocytes (+) | 3D cell-laden hydrogel structures | Immobilization of recognition molecules by magnetic microbeads and subsequent electrochemical measurement | [57] |
| Skeletal muscle and lower motor neurons | Neuromuscular junction | Mouse embryonic stem cell (mESC) line HBG3 (Hb9-GFP) (*) and mouse myoblasts C2C12 (-) | 3D cell-laden hydrogel structures | Muscle contraction sensing by passive force transducers (pillar deformation) | [61] |
| Blood brain barrier | Disease model of ischemic stroke | Murine brain endothelial cells (cerebEND) (-) | Barrier model | Oxygen sensing by fluorescence measurements of palladium-based dyes (PdTPTBFP) | [62] |
| Heart | Barrier function and electrical activity of endothelialized myocardium | Human umbilical cord vascular endothelial cells (HUVECs) (+) and human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) (*) | Barrier model | Barrier integrity and electrical activity sensing by TEER-multielectrode array measurements | [64] |
| Blood vessel, heart, liver | Cancer metastasis | Human umbilical vein endothelial cells (HUVEC) and human hepatocellular carcinoma (HepG2) (-) and human cardiomyocytes differentiation of human pluripotent stem cell (hPSC) line BJ1D (*) | 3D cell-laden hydrogel structures | Cardiac beat frequency sensing by fluorescence microscopy and computational analysis of microcantilevers. | [65] |
| Heart and liver | Organ toxicity | Human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) (*), primary hepatocytes (+) and HepG2/C3A hepatocellular carcinoma cells (-) | 3D organoids | pH sensing by light absorption of phenol red, oxygen sensing by fluorescence measurements of quenching effects of oxygen sensitive ruthenium dye and immunosensing by functionable electrodes | [66] |
| Heart and liver | Cardiotoxicity (primarily from hepatic cytochrome P450 (CYP) metabolism) | Human induced pluripotent stem cell (iPSc) derived cardiomyocytes (*) and human primary hepatocytes (+) | 2D monolayers | Multielectrode array for electrical activity sensing and cantilever array for sensing of cardiac mechanical function | [67] |
| Heart, liver and lung | Organ toxicity | Hepatic stellate cells (HSCs) (+), primary human hepatocytes (+), Kupffer cells (+), induced pluripotent stem cell-derived cardiomyocytes (iPSC CMs) (*), human primary cardiac fibroblasts (+), lung microvasculature endothelial cells (+), airway stromal mesenchymal cells (+), bronchial epithelial cells (+) | 3D organoids | Cardiac beat rate measurement by real-time imaging and computational analysis, antibody-binding by impedance measurement and barrier function by TEER measurement | [68] |
| Heart, liver, skeletal muscle and neuronal network | Organ toxicity | Human hepatocellular carcinoma HepG2/C3A (-), human induced pluripotent stem cell (iPSc) differentiated cardiomyocytes (*), human skeletal myofibers (+), human motoneurons differentiated from human spinal cord stem cell line (hSCSC) (*) and human iPSc differentiated cortical-like neurons (*) | 2D monolayers | Cardiomyocyte contraction (force) sensing by cantilever deflection (laser beam reflection) (69, 70) and electrical activity of cardiomyocytes or motoneurons by a multielectrode array (70) | [69,70] |
| Heart | Immune cell chemotaxis, stretching characteristics | Human induced pluripotent stem cells (hiPSCs) (*) and human induced pluripotent stem cell (hiPSCs)-derived cardiomyocytes (*) | 2D monolayers | Electrical field potential sensing of cardiomyocytes under membrane stretch by multielectrode array and membrane stretching sensing by measurement of electrical resistance change in strain gauges | [71] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kratz, S.R.A.; Höll, G.; Schuller, P.; Ertl, P.; Rothbauer, M. Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems. Biosensors 2019, 9, 110. https://0-doi-org.brum.beds.ac.uk/10.3390/bios9030110
Kratz SRA, Höll G, Schuller P, Ertl P, Rothbauer M. Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems. Biosensors. 2019; 9(3):110. https://0-doi-org.brum.beds.ac.uk/10.3390/bios9030110
Chicago/Turabian StyleKratz, Sebastian Rudi Adam, Gregor Höll, Patrick Schuller, Peter Ertl, and Mario Rothbauer. 2019. "Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems" Biosensors 9, no. 3: 110. https://0-doi-org.brum.beds.ac.uk/10.3390/bios9030110