Mir526b and Mir655 Promote Tumour Associated Angiogenesis and Lymphangiogenesis in Breast Cancer
Abstract
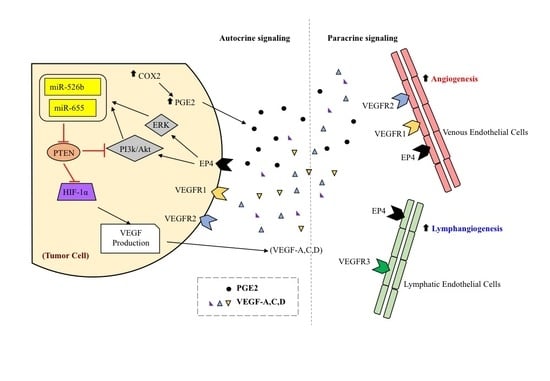
:1. Introduction
2. Results
2.1. Over-Expression of miR526b and miR655 in Poorly Metastatic (ER Positive) MCF7 Breast Cancer Cell Line Results in Upregulation of Angiogenesis and Lymphangiogenesis Markers
2.2. Human Umbilical Vein Endothelial Cells (HUVECs) Treated with MCF7-miR526b and MCF7-miR655 Conditioned Media Show Higher Expression of VEGF and EP4 Receptors
2.3. Cancer Cell Conditioned Media Induces Migration and Tube Formation of HUVEC Cells
2.4. Treatments with COX2 Inhibitor (COX-I) and EP4 Antagonist (EP4A) Significantly Inhibits miRNA Induced Functions
2.4.1. Inhibition of Cell Migration
2.4.2. Inhibition of Tube Formation
2.5. Expression of miR526b and miR655 in Human Breast Tumour Tissue Correlated with Angiogenesis and Lymphangiogenesis Markers
3. Discussion
4. Materials and Methods
4.1. Ethics Statements
4.2. Cell Culture
4.3. Collection of Conditioned Media
4.4. Drugs and Chemicals
4.5. Tube Formation Assay
4.6. HUVEC Migration Assay
4.7. Quantitative Real-Time PCR (qRT-PCR)
4.8. Western Blot
4.9. Human Breast Cancer Tissue Samples
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Global Health Risks: Mortality and Burden of Disease Attributable to Selected Major Risks; World Health Organization: Geneva, Switzerland, 2009; ISBN 9789241563871.
- Singh-Ranger, G.; Salhab, M.; Mokbel, K. The Role of Cyclooxygenase-2 in Breast Cancer: Review. Breast Cancer Res. Treat. 2008, 109, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Howe, L.R. Inflammation and Breast Cancer. Cyclooxygenase/Prostaglandin Signaling and Breast Cancer. Breast Cancer Res. BCR 2007, 9, e210. [Google Scholar] [CrossRef] [PubMed]
- Ristimaki, A.; Sivula, A.; Lundin, J.; Lundin, M.; Salminen, T.; Haglund, C.; Joensuu, H.; Isola, J. Prognostic Significance of Elevated Cyclooxygenase-2 Expression in Breast Cancer. Cancer Res. 2002, 62, e632. [Google Scholar]
- Kawahara, K.; Hohjoh, H.; Inazumi, T.; Tsuchiya, S.; Sugimoto, Y. Prostaglandin E2-Induced Inflammation: Relevance of Prostaglandin E Receptors. BBA Mol. Cell Biol. Lipids 2015, 1851, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Majumder, M.; Nandi, P.; Omar, A.; Ugwuagbo, K.C.; Lala, P.K. EP4 as a Therapeutic Target for Aggressive Human Breast Cancer. Int. J. Mol. Sci. 2018, 19, 1019. [Google Scholar] [CrossRef]
- Ugwuagbo, K.C.; Maiti, S.; Omar, A.; Hunter, S.; Nault, B.; Northam, C.; Majumder, M. Prostaglandin E2 Promotes Embryonic Vascular Development and Maturation in Zebrafish. Biol. Open 2019, 8, bio039768. [Google Scholar] [CrossRef]
- Majumder, M.; Xin, X.; Liu, L.; Tutunea-Fatan, E.; Rodriguez-Torres, M.; Vincent, K.; Postovit, L.; Hess, D.; Lala, P.K. COX-2 Induces Breast Cancer Stem Cells Via EP4/PI3K/AKT/NOTCH/WNT Axis. Stem Cells 2016, 34, 2290–2305. [Google Scholar] [CrossRef]
- Singh, R.; Mo, Y. Role of microRNAs in Breast Cancer. Cancer Biol. Ther. 2013, 14, 201–212. [Google Scholar] [CrossRef]
- Majumder, M.; Landman, E.; Liu, L.; Hess, D.; Lala, P.K. COX-2 Elevates Oncogenic miR-526b in Breast Cancer by EP4 Activation. Mol. Cancer Res. 2015, 13, 1022–1033. [Google Scholar] [CrossRef]
- Majumder, M.; Dunn, L.; Liu, L.; Hasan, A.; Vincent, K.; Brackstone, M.; Hess, D.; Lala, P.K. COX-2 Induces Oncogenic Micro RNA miR655 in Human Breast Cancer. Sci. Rep. 2018, 8, e3. [Google Scholar] [CrossRef]
- Tordjman, J.; Majumder, M.; Amirr, M.; Hasan, A.; Hess, D.; Lala, P.K. Tumor suppressor role of cytoplasmic polyadenylation element binding protein 2 (CPEB2) in human mammary epithelial cells. BMC Cancer 2019, 19, e561. [Google Scholar] [CrossRef] [PubMed]
- Urbich, C.; Kuehbacher, A.; Dimmeler, S. Role of microRNAs in Vascular Diseases, Inflammation, and Angiogenesis. Cardiovasc. Res. 2008, 79, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Lala, P.K.; Nandi, P.; Majumder, M. Roles of Prostaglandins in Tumor-Associated Lymphangiogenesis with Special Reference to Breast Cancer. Cancer Metastasis Rev. 2018, 37, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Otrock, Z.K.; Mahfouz, R.A.R.; Makarem, J.A.; Shamseddine, A.I. Understanding the Biology of Angiogenesis: Review of the most Important Molecular Mechanisms. Blood Cells Mol. Dis. 2007, 39, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Lou, W.; Liu, J.; Gao, Y.; Zhong, G.; Chen, D.; Shen, J.; Bao, C.; Xu, L.; Pan, J.; Cheng, J.; et al. MicroRNAs in Cancer Metastasis and Angiogenesis. Oncotarget 2017, 8, e115787. [Google Scholar] [CrossRef]
- Ran, S.; Volk, L.; Hall, K.; Flister, M.J. Lymphangiogenesis and Lymphatic Metastasis in Breast Cancer. Pathophysiology 2010, 17, 229–251. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M. Vascular Endothelial Growth Factor (VEGF) and its Receptor (VEGFR) Signaling in Angiogenesis: A Crucial Target for Anti- and Pro-Angiogenic Therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef]
- Park, S.; Sorenson, C.M.; Sheibani, N. PECAM-1 Isoforms, eNOS and Endoglin Axis in Regulation of Angiogenesis. Clin. Sci. 2015, 129, 217–234. [Google Scholar] [CrossRef]
- Rao, R.; Redha, R.; Macias-Perez, I.; Su, Y.; Hao, C.; Zent, R.; Breyer, M.D.; Pozzi, A. Prostaglandin E2-EP4 Receptor Promotes Endothelial Cell Migration Via ERK Activation and Angiogenesis in Vivo. J. Biol. Chem. 2007, 282, 16959–16968. [Google Scholar] [CrossRef]
- Tutunea-Fatan, E.; Majumder, M.; Xin, X.; Lala, P.K. The Role of CCL21/CCR7 Chemokine Axis in Breast Cancer-Induced Lymphangiogenesis. Mol. Cancer 2015, 14, 35. [Google Scholar] [CrossRef]
- Cascio, S.; D’Andrea, A.; Ferla, R.; Surmacz, E.; Gulotta, E.; Amodeo, V.; Bazan, V.; Gebbia, N.; Russo, A. miR-20b Modulates VEGF Expression by Targeting HIF-1 Alpha and STAT3 in MCF-7 Breast Cancer Cells. J. Cell. Physiol. 2010, 224, 242–249. [Google Scholar] [PubMed]
- Liu, X.; Guan, Y.; Wang, L.; Niu, Y. MicroRNA-10b Expression in Node-Negative Breast Cancer-Correlation with Metastasis and Angiogenesis. Oncol. Lett. 2017, 14, 5845–5852. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Chu, P.; Hou, M.; Hung, W. MiR-182 Promotes Proliferation and Invasion and Elevates the HIF-1α-VEGF-A Axis in Breast Cancer Cells by Targeting FBXW7. Am. J. Cancer Res. 2016, 6, 1785. [Google Scholar] [PubMed]
- Luengo-Gil, G.; Gonzalez-Billalabeitia, E.; Perez-Henarejos, S.A.; Manzano, E.N.; Chaves-Benito, A.; Garcia-Martinez, E.; Garcia-Garre, E.; Vicente, V.; Ayala La Peña, F. Angiogenic Role of miR-20a in Breast Cancer. PLoS ONE 2018, 13, e0194638. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cheng, Y.; Li, Y.; Jin, Z.; Pan, Y.; Liu, G.; Fu, S.; Zhang, Y.; Feng, K.; Feng, Y. microRNA-128 Plays a Critical Role in Human Non-Small Cell Lung Cancer Tumourigenesis, Angiogenesis and Lymphangiogenesis by Directly Targeting Vascular Endothelial Growth Factor-C. Eur. J. Cancer 2014, 50, 2336–2350. [Google Scholar] [CrossRef] [PubMed]
- Sasahira, T.; Kurihara, M.; Bhawal, U.K.; Ueda, N.; Shimomoto, T.; Yamamoto, K.; Kirita, T.; Kuniyasu, H. Downregulation of miR-126 Induces Angiogenesis and Lymphangiogenesis by Activation of VEGF-A in Oral Cancer. Br. J. Cancer 2012, 107, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Majumder, M.; Girish, G.V.; Mohindra, V.; Maruyama, T.; Lala, P.K. Targeting COX-2 and EP4 to Control Tumor Growth, Angiogenesis, Lymphangiogenesis and Metastasis to the Lungs and Lymph Nodes in a Breast Cancer Model. Lab. Investig. 2012, 92, 1115–1128. [Google Scholar] [CrossRef]
- Majumder, M.; Xin, X.; Liu, L.; Girish, G.V.; Lala, P.K. Prostaglandin E2 Receptor EP4 as the Common Target on Cancer Cells and Macrophages to Abolish Angiogenesis, Lymphangiogenesis, Metastasis, and Stem-like Cell Functions. Cancer Sci. 2014, 105, 1142–1151. [Google Scholar] [CrossRef]
- Perrot-Applanat, M.; Di Benedetto, M. Autocrine Functions of VEGF in Breast Tumor Cells. Cell Adh. Migr. 2012, 6, 547–553. [Google Scholar] [CrossRef]
- Ning, Q.; Liu, C.; Hou, L.; Meng, M.; Zhang, X.; Luo, M.; Shao, S.; Zuo, X.; Zhao, X. Vascular Endothelial Growth Factor Receptor-1 Activation Promotes Migration and Invasion of Breast Cancer Cells through Epithelial-Mesenchymal Transition. PLoS ONE 2013, 8, e65217. [Google Scholar] [CrossRef]
- Weigand, M.; Hantel, P.; Kreienberg, R.; Waltenberger, J. Autocrine Vascular Endothelial Growth Factor Signalling in Breast Cancer. Evidence from Cell Lines and Primary Breast Cancer Cultures in Vitro. Angiogenesis 2005, 8, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Timoshenko, A.V.; Chakraborty, C.; Wagner, G.F.; Lala, P.K. COX-2-Mediated Stimulation of the Lymphangiogenic Factor VEGF-C in Human Breast Cancer. Br. J. Cancer 2006, 94, 1154–1163. [Google Scholar] [CrossRef] [PubMed]
- Nandi, P.; Girish, G.V.; Majumder, M.; Xin, X.; Tutunea-Fatan, E.; Lala, P.K. PGE2 Promotes Breast Cancer-Associated Lymphangiogenesis by Activation of EP4 Receptor on Lymphatic Endothelial Cells. BMC Cancer 2017, 17, e11. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; He, L.; Richards, E.J.; Challa, S.; Xu, C.X.; Permuth-Wey, J.; Lancaster, J.M.; Coppola, D.; Sellers, T.A.; Djeu, J.Y.; et al. Upregulation of miRNA-155 Promotes Tumour Angiogenesis by Targeting VHL and is Associated with Poor Prognosis and Triple-Negative Breast Cancer. Oncogene 2014, 33, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Liu, Y.; Fang, X.; Liu, Y.; Fang, L.; Lin, L.; Liu, X.; Wang, N. Tumor-Derived microRNA-494 Promotes Angiogenesis in Non-Small Cell Lung Cancer. Angiogenesis 2015, 18, 373–382. [Google Scholar] [CrossRef]
- He, X.; Ping, J.; Wen, D. MicroRNA-186 Regulates the Invasion and Metastasis of Bladder Cancer Via Vascular Endothelial Growth Factor, C. Exp. Ther. Med. 2017, 14, 3253–3258. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Meng, X.; Bao, Y.; Wang, S.; Li, T. Evidence for the Involvement of COX-2/VEGF and PTEN/Pl3K/AKT Pathway the Mechanism of Oroxin B Treated Liver Cancer. Pharmacogn. Mag. 2018, 14, 207–213. [Google Scholar]
- Forsythe, J.A.; Jiang, B.H.; Iyer, N.V.; Agani, F.; Leung, S.W.; Koos, R.D.; Semenza, G.L. Activation of Vascular Endothelial Growth Factor Gene Transcription by Hypoxia-Inducible Factor 1. Mol. Cell. Biol. 1996, 16, 4604–4613. [Google Scholar] [CrossRef]
- Hirota, K.; Semenza, G.L. Regulation of Angiogenesis by Hypoxia-Inducible Factor 1. Crit. Rev. Oncol. Hematol. 2006, 59, 15–26. [Google Scholar] [CrossRef]
- Khan, F.; Esnakula, A.; Ricks-Santi, L.J.; Zafar, R.; Kanaan, Y.; Naab, T. Loss of PTEN in high grade adnaced triple negative breast ductal cancers in African American women. Pathol. Res. Pract. 2018, 5, 673–678. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR and the 2 (-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.Y.; Shao, Z.M.; Li, D.Q. Tumor Microenvironment: Driving Forces and Potential Therapeutic Targets for Breast Cancer Metastasis. Chin. J. Cancer 2017, 36, e36. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, Q. Cancer Stem Cells, Lymphangiogenesis, and Lymphatic Metastasis. Cancer Lett. 2014, 357, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Weis, S.M.; Cheresh, D.A. Tumor Angiogenesis: Molecular Pathways and Therapeutic Targets. Nat. Med. 2011, 17, 1359–1370. [Google Scholar] [CrossRef] [PubMed]








| Subjects | Control n = 20 (%) | Cancer n = 105 (%) | |
|---|---|---|---|
| Sex | Male | 0 | 3 (2.8) |
| Female | 20 (100) | 102 (97.2) | |
| Age Distribution (years) | Range | 52–87 | 27–92 |
| Age (years) | Mean ± SD | 66 ± 11 | 64 ± 12 |
| Smoking Habit | Smokers | 1 (5) | 3 (2.8) |
| Pack Year (PY) | 40 | 56 ± 11 | |
| Alcohol Consumption | Social/Occasional Drinker | 5 (25) | 29 (27.62) |
| Regular Drinker | 0 | 3 (2.8) | |
| Estrogen Receptor (ER) Status | Positive | 80 (76) | |
| Negative | 19(18) | ||
| Progesteron Receptor Status (PR) Status | Positive | 66 (62.9) | |
| Negative | 33 (31) | ||
| Human epidermal growth factor receptor 2 (HER2) Status | Positive | 21 (20) | |
| Negative | 68 (64.8) | ||
| ER, PR, HER2 (Triple) Negative | Negative | 10 (9.5) | |
| Tumour Grade and High miRNA Expression in Cancer Samples | ||||
| Tumour Grade | n (%) | miR-526b High n (%) | miR-655 High n (%) | |
| I (low-well differentiated) | 7 (6.7) | 0 | 2 (28.5) | |
| II (intermediate-moderately differentiated) | 26 (24.76) | 2 (7.69) | 7 (26.9) | |
| III (high-poorly differentiated) | 63 (60) | 5 (7.94) | 13 (20.6) | |
| X (Unknown) | 9 (8.57) | 0 | 2 (28.6) | |
| Tumour Receptor Status and High miRNA Expression in Cancer Samples | ||||
| Receptor Status of Cancer Samples | n (%) | miR-526b High n (%) | miR-655 High n (%) | |
| ER Status | Positive | 40 (38.1) | 6 (15) | 10 (25) |
| Negative | 20 (19) | 0 | 4 (20) | |
| PR Status | Positive | 33 (31.4) | 3 (9.1) | 8 (24.2) |
| Negative | 33 (31.4) | 2 (6.1) | 7 (21.2) | |
| HER2 Status | Positive | 22 (21) | 2 (9.1) | 8 (36.4) |
| Negative | 68 (64.8) | 4 (5.9) | 13 (19.1) | |
| ER,PR, HER2 (Triple) Negative | Negative | 10 (9.5) | 0 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hunter, S.; Nault, B.; Ugwuagbo, K.C.; Maiti, S.; Majumder, M. Mir526b and Mir655 Promote Tumour Associated Angiogenesis and Lymphangiogenesis in Breast Cancer. Cancers 2019, 11, 938. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11070938
Hunter S, Nault B, Ugwuagbo KC, Maiti S, Majumder M. Mir526b and Mir655 Promote Tumour Associated Angiogenesis and Lymphangiogenesis in Breast Cancer. Cancers. 2019; 11(7):938. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11070938
Chicago/Turabian StyleHunter, Stephanie, Braydon Nault, Kingsley Chukwunonso Ugwuagbo, Sujit Maiti, and Mousumi Majumder. 2019. "Mir526b and Mir655 Promote Tumour Associated Angiogenesis and Lymphangiogenesis in Breast Cancer" Cancers 11, no. 7: 938. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11070938