Cold-Shock Domains—Abundance, Structure, Properties, and Nucleic-Acid Binding
Abstract
:Simple Summary
Abstract
1. Introduction
2. Definition, Abundance and Discovery of Cold-Shock Domains
2.1. Definition and Basic Properties of Cold-Shock Domains
2.2. Abundance of Cold-Shock Domains
2.3. Discovery of Cold-Shock Domains
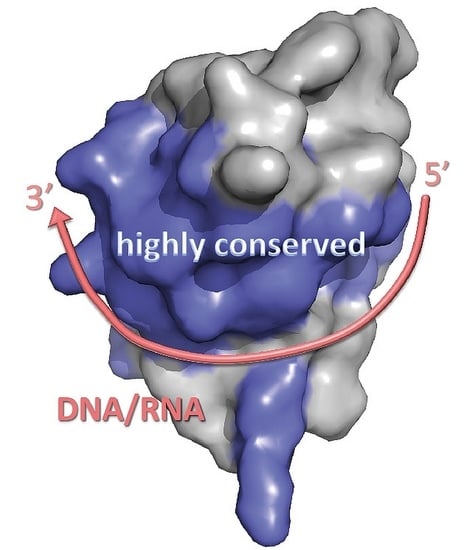
3. Structure of Cold-Shock Domains
CSD β-Barrel Stability and Formation of Domain-Swapped Dimers
4. Biophysical Properties of Cold-Shock Proteins
5. Cold-Shock Domain-Binding to Nucleic Acids
5.1. Bacterial Cold-Shock Proteins
5.1.1. DNA Binding
5.1.2. RNA Binding
5.2. Eukaryotic Cold-Shock Domains
5.2.1. DNA Binding
5.2.2. RNA Binding
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chothia, C. Proteins. One thousand families for the molecular biologist. Nature 1992, 357, 543–544. [Google Scholar] [CrossRef]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007, 8, 479–490. [Google Scholar] [CrossRef] [Green Version]
- Ray, D.; Kazan, H.; Cook, K.B.; Weirauch, M.T.; Najafabadi, H.S.; Li, X.; Gueroussov, S.; Albu, M.; Zheng, H.; Yang, A.; et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 2013, 499, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [Green Version]
- Baltz, A.G.; Munschauer, M.; Schwanhausser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef] [Green Version]
- Castello, A.; Fischer, B.; Frese, C.K.; Horos, R.; Alleaume, A.M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol. Cell 2016, 63, 696–710. [Google Scholar] [CrossRef] [Green Version]
- Budkina, K.S.; Zlobin, N.E.; Kononova, S.V.; Ovchinnikov, L.P.; Babakov, A.V. Cold Shock Domain Proteins: Structure and Interaction with Nucleic Acids. Biochemistry 2020, 85, S1–S19. [Google Scholar] [CrossRef]
- Zou, F.; Tu, R.; Duan, B.; Yang, Z.; Ping, Z.; Song, X.; Chen, S.; Price, A.; Li, H.; Scott, A.; et al. Drosophila YBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs. Proc. Natl. Acad. Sci. USA 2020, 117, 3603–3609. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, J.S.; Li, S.; Yang, Y.; Sun, P.; Zhu, Q.; Wang, J.; Jiang, B.; Yang, D.; Liu, M. Structural basis of DNA binding to human YB-1 cold shock domain regulated by phosphorylation. Nucleic Acids Res. 2020, 48, 9361–9371. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Han, X.; Yang, W.L.; Zhang, M.; Ma, H.L.; Sun, B.F.; Li, A.; Xia, J.; Chen, J.; et al. RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay. Mol. Cell 2019, 75, 1188–1202.e11. [Google Scholar] [CrossRef]
- Yang, X.J.; Zhu, H.; Mu, S.R.; Wei, W.J.; Yuan, X.; Wang, M.; Liu, Y.; Hui, J.; Huang, Y. Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA. J. Biol. Chem. 2019, 294, 10998–11010. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, A.; Sun, B.F.; Yang, Y.; Han, Y.N.; Yuan, X.; Chen, R.X.; Wei, W.S.; Liu, Y.; Gao, C.C.; et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019, 21, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Nam, Y.; Lee, A.K.; Yu, C.; Roth, K.; Chen, C.; Ransey, E.M.; Sliz, P. LIN28 Zinc Knuckle Domain Is Required and Sufficient to Induce let-7 Oligouridylation. Cell Rep. 2017, 18, 2664–2675. [Google Scholar] [CrossRef] [PubMed]
- Hennig, J.; Militti, C.; Popowicz, G.M.; Wang, I.; Sonntag, M.; Geerlof, A.; Gabel, F.; Gebauer, F.; Sattler, M. Structural basis for the assembly of the Sxl-Unr translation regulatory complex. Nature 2014, 515, 287–290. [Google Scholar] [CrossRef]
- Murzin, A.G. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993, 12, 861–867. [Google Scholar] [CrossRef]
- Theobald, D.L.; Mitton-Fry, R.M.; Wuttke, D.S. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 115–133. [Google Scholar] [CrossRef] [Green Version]
- Amir, M.; Kumar, V.; Dohare, R.; Rehman, M.T.; Hussain, A.; Alajmi, M.F.; El-Seedi, H.R.; Hassan, H.M.A.; Islam, A.; Ahmad, F.; et al. Investigating architecture and structure-function relationships in cold shock DNA-binding domain family using structural genomics-based approach. Int. J. Biol. Macromol. 2019, 133, 484–494. [Google Scholar] [CrossRef]
- Deryusheva, E.I.; Machulin, A.V.; Selivanova, O.M.; Galzitskaya, O.V. Taxonomic distribution, repeats, and functions of the S1 domain-containing proteins as members of the OB-fold family. Proteins 2017, 85, 602–613. [Google Scholar] [CrossRef]
- Lim, C.J.; Barbour, A.T.; Zaug, A.J.; Goodrich, K.J.; McKay, A.E.; Wuttke, D.S.; Cech, T.R. The structure of human CST reveals a decameric assembly bound to telomeric DNA. Science 2020, 368, 1081–1085. [Google Scholar] [CrossRef]
- Landsman, D. RNP-1, an RNA-binding motif is conserved in the DNA-binding cold shock domain. Nucleic Acids Res. 1992, 20, 2861–2864. [Google Scholar] [CrossRef]
- Nagai, K.; Oubridge, C.; Jessen, T.H.; Li, J.; Evans, P.R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature 1990, 348, 515–520. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020. [Google Scholar] [CrossRef]
- Schindelin, H.; Marahiel, M.A.; Heinemann, U. Universal Nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein. Nature 1993, 364, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Sigrist, C.J.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreeva, A.; Kulesha, E.; Gough, J.; Murzin, A.G. The SCOP database in 2020: Expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2020, 48, D376–D382. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2020, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S.; Alsina, J.; Inouye, M. Cold-shock response and cold-shock proteins. Curr Opin Microbiol. 1999, 2, 175–180. [Google Scholar] [CrossRef]
- Schindler, T.; Graumann, P.L.; Perl, D.; Ma, S.; Schmid, F.X.; Marahiel, M.A. The family of cold shock proteins of Bacillus subtilis. Stability and dynamics in vitro and in vivo. J. Biol. Chem. 1999, 274, 3407–3413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amir, M.; Kumar, V.; Dohare, R.; Islam, A.; Ahmad, F.; Hassan, M.I. Sequence, structure and evolutionary analysis of cold shock domain proteins, a member of OB fold family. J. Evol. Biol. 2018, 31, 1903–1917. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2020, 49, D344–D354. [Google Scholar] [CrossRef]
- Jones, P.G.; VanBogelen, R.A.; Neidhardt, F.C. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 1987, 169, 2092–2095. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, J.; Pollitt, N.S.; Inouye, M. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 283–287. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Jones, P.; Inouye, M. Chloramphenicol induces the transcription of the major cold shock gene of Escherichia coli, cspA. J. Bacteriol. 1993, 175, 5824–5828. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Xie, A.; Jiang, W.; Etchegaray, J.P.; Jones, P.G.; Inouye, M. Family of the major cold-shock protein, CspA (CS7.4), of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Mol. Microbiol. 1994, 11, 833–839. [Google Scholar] [CrossRef]
- Jones, P.G.; Inouye, M. The cold-shock response—A hot topic. Mol. Microbiol. 1994, 11, 811–818. [Google Scholar] [CrossRef]
- Charles, J.; Masnoddin, M.; Nazaie, F.; Yusof, N.A. Structure and function of a novel cold regulated cold shock domain containing protein from an obligate psychrophilic yeast, Glaciozyma antarctica. Adv. Polar Sci. 2020, 31, 137–145. [Google Scholar]
- Catalan-Moreno, A.; Caballero, C.J.; Irurzun, N.; Cuesta, S.; Lopez-Sagaseta, J.; Toledo-Arana, A. One evolutionarily selected amino acid variation is sufficient to provide functional specificity in the cold shock protein paralogs of Staphylococcus aureus. Mol. Microbiol. 2020, 113, 826–840. [Google Scholar] [CrossRef] [Green Version]
- Caballero, C.J.; Menendez-Gil, P.; Catalan-Moreno, A.; Vergara-Irigaray, M.; Garcia, B.; Segura, V.; Irurzun, N.; Villanueva, M.; Ruiz de Los Mozos, I.; Solano, C.; et al. The regulon of the RNA chaperone CspA and its auto-regulation in Staphylococcus aureus. Nucleic Acids Res. 2018, 46, 1345–1361. [Google Scholar] [CrossRef] [Green Version]
- Kondo, K.; Inouye, M. TIP 1, a cold shock-inducible gene of Saccharomyces cerevisiae. J. Biol. Chem. 1991, 266, 17537–17544. [Google Scholar] [CrossRef]
- Kondo, K.; Kowalski, L.R.; Inouye, M. Cold shock induction of yeast NSR1 protein and its role in pre-rRNA processing. J. Biol. Chem. 1992, 267, 16259–16265. [Google Scholar] [CrossRef]
- Morf, J.; Rey, G.; Schneider, K.; Stratmann, M.; Fujita, J.; Naef, F.; Schibler, U. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science 2012, 338, 379–383. [Google Scholar] [CrossRef]
- Coburn, K.; Melville, Z.; Aligholizadeh, E.; Roth, B.M.; Varney, K.M.; Carrier, F.; Pozharski, E.; Weber, D.J. Crystal structure of the human heterogeneous ribonucleoprotein A18 RNA-recognition motif. Acta Crystallogr. F Struct. Biol. Commun. 2017, 73, 209–214. [Google Scholar] [CrossRef] [Green Version]
- Didier, D.K.; Schiffenbauer, J.; Woulfe, S.L.; Zacheis, M.; Schwartz, B.D. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc. Natl. Acad. Sci. USA 1988, 85, 7322–7326. [Google Scholar] [CrossRef] [Green Version]
- Wistow, G. Cold shock and DNA binding. Nature 1990, 344, 823–824. [Google Scholar] [CrossRef] [Green Version]
- Evdokimova, V.; Ruzanov, P.; Imataka, H.; Raught, B.; Svitkin, Y.; Ovchinnikov, L.P.; Sonenberg, N. The major mRNA-associated protein YB-1 is a potent 5’ cap-dependent mRNA stabilizer. EMBO J. 2001, 20, 5491–5502. [Google Scholar] [CrossRef] [Green Version]
- Kleene, K.C. Y-box proteins combine versatile cold shock domains and arginine-rich motifs (ARMs) for pleiotropic functions in RNA biology. Biochem. J. 2018, 475, 2769–2784. [Google Scholar] [CrossRef]
- Mehta, S.; Algie, M.; Al-Jabry, T.; McKinney, C.; Kannan, S.; Verma, C.S.; Ma, W.; Zhang, J.; Bartolec, T.K.; Masamsetti, V.P.; et al. Critical Role for Cold Shock Protein YB-1 in Cytokinesis. Cancers 2020, 12, 2473. [Google Scholar] [CrossRef]
- Das, S.; Chattopadhyay, R.; Bhakat, K.K.; Boldogh, I.; Kohno, K.; Prasad, R.; Wilson, S.H.; Hazra, T.K. Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J. Biol. Chem. 2007, 282, 28474–28484. [Google Scholar] [CrossRef] [Green Version]
- Eliseeva, I.A.; Kim, E.R.; Guryanov, S.G.; Ovchinnikov, L.P.; Lyabin, D.N. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry 2011, 76, 1402–1433. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.A.; Hammond, S.M. Lin-28: An early embryonic sentinel that blocks Let-7 biogenesis. Int. J. Biochem. Cell Biol. 2010, 42, 1330–1333. [Google Scholar] [CrossRef] [PubMed]
- Jurchott, K.; Kuban, R.J.; Krech, T.; Bluthgen, N.; Stein, U.; Walther, W.; Friese, C.; Kielbasa, S.M.; Ungethum, U.; Lund, P.; et al. Identification of Y-box binding protein 1 as a core regulator of MEK/ERK pathway-dependent gene signatures in colorectal cancer cells. PLoS Genet. 2010, 6, e1001231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindquist, J.A.; Mertens, P.R. Cold shock proteins: From cellular mechanisms to pathophysiology and disease. Cell Commun. Signal. 2018, 16, 63. [Google Scholar] [CrossRef] [Green Version]
- Lindquist, J.A.; Brandt, S.; Bernhardt, A.; Zhu, C.; Mertens, P.R. The role of cold shock domain proteins in inflammatory diseases. J. Mol. Med. 2014, 92, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Sangermano, F.; Delicato, A.; Calabro, V. Y box binding protein 1 (YB-1) oncoprotein at the hub of DNA proliferation, damage and cancer progression. Biochimie 2020, 179, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Mordovkina, D.; Lyabin, D.N.; Smolin, E.A.; Sogorina, E.M.; Ovchinnikov, L.P.; Eliseeva, I. Y-Box Binding Proteins in mRNP Assembly, Translation, and Stability Control. Biomolecules 2020, 10, 591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frye, B.C.; Halfter, S.; Djudjaj, S.; Muehlenberg, P.; Weber, S.; Raffetseder, U.; En-Nia, A.; Knott, H.; Baron, J.M.; Dooley, S.; et al. Y-box protein-1 is actively secreted through a non-classical pathway and acts as an extracellular mitogen. EMBO Rep. 2009, 10, 783–789. [Google Scholar] [CrossRef]
- Shurtleff, M.J.; Yao, J.; Qin, Y.; Nottingham, R.M.; Temoche-Diaz, M.M.; Schekman, R.; Lambowitz, A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E8987–E8995. [Google Scholar] [CrossRef] [Green Version]
- Lyons, S.M.; Achorn, C.; Kedersha, N.L.; Anderson, P.J.; Ivanov, P. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016, 44, 6949–6960. [Google Scholar] [CrossRef]
- Ivanov, P.; O’Day, E.; Emara, M.M.; Wagner, G.; Lieberman, J.; Anderson, P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. USA 2014, 111, 18201–18206. [Google Scholar] [CrossRef] [Green Version]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef] [Green Version]
- Nie, M.; Balda, M.S.; Matter, K. Stress- and Rho-activated ZO-1-associated Nucleic acid binding protein binding to p21 mRNA mediates stabilization, translation, and cell survival. Proc. Natl. Acad. Sci. USA 2012, 109, 10897–10902. [Google Scholar] [CrossRef] [Green Version]
- Nie, M.; Aijaz, S.; Leefa Chong San, I.V.; Balda, M.S.; Matter, K. The Y-box factor ZONAB/DbpA associates with GEF-H1/Lfc and mediates Rho-stimulated transcription. EMBO Rep. 2009, 10, 1125–1131. [Google Scholar] [CrossRef]
- Balda, M.S.; Matter, K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 2000, 19, 2024–2033. [Google Scholar] [CrossRef]
- Ranjan, M.; Tafuri, S.R.; Wolffe, A.P. Masking mRNA from translation in somatic cells. Genes Dev. 1993, 7, 1725–1736. [Google Scholar] [CrossRef] [Green Version]
- Bouvet, P.; Wolffe, A.P. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell 1994, 77, 931–941. [Google Scholar] [CrossRef]
- Arnold, A.; Rahman, M.M.; Lee, M.C.; Muehlhaeusser, S.; Katic, I.; Gaidatzis, D.; Hess, D.; Scheckel, C.; Wright, J.E.; Stetak, A.; et al. Functional characterization of C. elegans Y-box-binding proteins reveals tissue-specific functions and a critical role in the formation of polysomes. Nucleic Acids Res. 2014, 42, 13353–13369. [Google Scholar] [CrossRef] [Green Version]
- Jacquemin-Sablon, H.; Triqueneaux, G.; Deschamps, S.; le Maire, M.; Doniger, J.; Dautry, F. Nucleic acid binding and intracellular localization of unr, a protein with five cold shock domains. Nucleic Acids Res. 1994, 22, 2643–2650. [Google Scholar] [CrossRef] [Green Version]
- Guo, A.X.; Cui, J.J.; Wang, L.Y.; Yin, J.Y. The role of CSDE1 in translational reprogramming and human diseases. Cell Commun. Signal. 2020, 18, 14. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Bartsch, D.; Xiao, C.; Guerrero, S.; Ahuja, G.; Schindler, C.; Moresco, J.J.; Yates, J.R., 3rd; Gebauer, F.; Bazzi, H.; et al. A post-transcriptional program coordinated by CSDE1 prevents intrinsic neural differentiation of human embryonic stem cells. Nat. Commun. 2017, 8, 1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollmann, N.M.; Jagtap, P.K.A.; Masiewicz, P.; Guitart, T.; Simon, B.; Provaznik, J.; Stein, F.; Haberkant, P.; Sweetapple, L.J.; Villacorta, L.; et al. Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras. Cell Rep. 2020, 32, 107930. [Google Scholar] [CrossRef]
- Doniger, J.; Landsman, D.; Gonda, M.A.; Wistow, G. The product of unr, the highly conserved gene upstream of N-ras, contains multiple repeats similar to the cold-shock domain (CSD), a putative DNA-binding motif. New Biol. 1992, 4, 389–395. [Google Scholar] [PubMed]
- Nastasi, T.; Scaturro, M.; Bellafiore, M.; Raimondi, L.; Beccari, S.; Cestelli, A.; di Liegro, I. PIPPin is a brain-specific protein that contains a cold-shock domain and binds specifically to H1 degrees and H3.3 mRNAs. J. Biol. Chem. 1999, 274, 24087–24093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Place, R.F.; Li, L.C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef] [Green Version]
- Pfeiffer, J.R.; McAvoy, B.L.; Fecteau, R.E.; Deleault, K.M.; Brooks, S.A. CARHSP1 is required for effective tumor necrosis factor alpha mRNA stabilization and localizes to processing bodies and exosomes. Mol. Cell Biol. 2011, 31, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y. A mirror of two faces: Lin28 as a master regulator of both miRNA and mRNA. Wiley Interdiscip. Rev. RNA 2012, 3, 483–494. [Google Scholar] [CrossRef]
- Viswanathan, S.R.; Daley, G.Q. Lin28: A microRNA regulator with a macro role. Cell 2010, 140, 445–449. [Google Scholar] [CrossRef] [Green Version]
- Peters, D.T.; Fung, H.K.; Levdikov, V.M.; Irmscher, T.; Warrander, F.C.; Greive, S.J.; Kovalevskiy, O.; Isaacs, H.V.; Coles, M.; Antson, A.A. Human Lin28 Forms a High-Affinity 1:1 Complex with the 106~363 Cluster miRNA miR-363. Biochemistry 2016, 55, 5021–5027. [Google Scholar] [CrossRef] [Green Version]
- Mayr, F.; Heinemann, U. Mechanisms of Lin28-mediated miRNA and mRNA regulation--a structural and functional perspective. Int. J. Mol. Sci. 2013, 14, 16532–16553. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Kim, Y.K.; Ha, M.; Yoon, M.J.; Cho, J.; Yeom, K.H.; Han, J.; Kim, V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.M.; Triboulet, R.; Thornton, J.E.; Gregory, R.I. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013, 497, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Balzeau, J.; Menezes, M.R.; Cao, S.; Hagan, J.P. The LIN28/let-7 Pathway in Cancer. Front. Genet. 2017, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powers, J.T.; Tsanov, K.M.; Pearson, D.S.; Roels, F.; Spina, C.S.; Ebright, R.; Seligson, M.; de Soysa, Y.; Cahan, P.; Theissen, J.; et al. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 2016, 535, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Mendell, J.T.; Olson, E.N. MicroRNAs in stress signaling and human disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef] [Green Version]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef]
- Zhu, H.; Shyh-Chang, N.; Segre, A.V.; Shinoda, G.; Shah, S.P.; Einhorn, W.S.; Takeuchi, A.; Engreitz, J.M.; Hagan, J.P.; Kharas, M.G.; et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef] [Green Version]
- Shyh-Chang, N.; Zhu, H.; Yvanka de Soysa, T.; Shinoda, G.; Seligson, M.T.; Tsanov, K.M.; Nguyen, L.; Asara, J.M.; Cantley, L.C.; Daley, G.Q. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 2013, 155, 778–792. [Google Scholar] [CrossRef] [Green Version]
- Wilbert, M.L.; Huelga, S.C.; Kapeli, K.; Stark, T.J.; Liang, T.Y.; Chen, S.X.; Yan, B.Y.; Nathanson, J.L.; Hutt, K.R.; Lovci, M.T.; et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell 2012, 48, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Gerarden, K.P.; Fuchs, A.M.; Koch, J.M.; Mueller, M.M.; Graupner, D.R.; O’Rorke, J.T.; Frost, C.D.; Heinen, H.A.; Lackner, E.R.; Schoeller, S.J.; et al. Solution structure of the cold-shock-like protein from Rickettsia rickettsii. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 1284–1288. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Li, C.; Sako, Y.; Imai, A.; Nishiyama, T.; Thompson, K.; Kubo, M.; Hiwatashi, Y.; Kabeya, Y.; Karlson, D.; Wu, S.H.; et al. A Lin28 homologue reprograms differentiated cells to stem cells in the moss Physcomitrella patens. Nat. Commun. 2017, 8, 14242. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, H.; Herrler, M.; Willimsky, G.; Marahiel, M.A.; Heinemann, U. Overproduction, crystallization, and preliminary X-ray diffraction studies of the major cold shock protein from Bacillus subtilis, CspB. Proteins 1992, 14, 120–124. [Google Scholar] [CrossRef]
- Schnuchel, A.; Wiltscheck, R.; Czisch, M.; Herrler, M.; Willimsky, G.; Graumann, P.; Marahiel, M.A.; Holak, T.A. Structure in solution of the major cold-shock protein from Bacillus subtilis. Nature 1993, 364, 169–171. [Google Scholar] [CrossRef]
- Schindelin, H.; Jiang, W.; Inouye, M.; Heinemann, U. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1994, 91, 5119–5123. [Google Scholar] [CrossRef] [Green Version]
- Newkirk, K.; Feng, W.; Jiang, W.; Tejero, R.; Emerson, S.D.; Inouye, M.; Montelione, G.T. Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: Identification of a binding epitope for DNA. Proc. Natl. Acad. Sci. USA 1994, 91, 5114–5118. [Google Scholar] [CrossRef] [Green Version]
- Mueller, U.; Perl, D.; Schmid, F.X.; Heinemann, U. Thermal stability and atomic-resolution crystal structure of the Bacillus caldolyticus cold shock protein. J. Mol. Biol. 2000, 297, 975–988. [Google Scholar] [CrossRef]
- Kremer, W.; Schuler, B.; Harrieder, S.; Geyer, M.; Gronwald, W.; Welker, C.; Jaenicke, R.; Kalbitzer, H.R. Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima. Eur. J. Biochem. 2001, 268, 2527–2539. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Jeong, K.W.; Jin, B.; Ryu, K.S.; Kim, E.H.; Ahn, J.H.; Kim, Y. Structural and dynamic features of cold-shock proteins of Listeria monocytogenes, a psychrophilic bacterium. Biochemistry 2013, 52, 2492–2504. [Google Scholar] [CrossRef]
- Mayr, F.; Schutz, A.; Doge, N.; Heinemann, U. The Lin28 cold-shock domain remodels pre-let-7 microRNA. Nucleic Acids Res. 2012, 40, 7492–7506. [Google Scholar] [CrossRef]
- PyMOL, version 2.0; The PyMOL Molecular Graphics System; Schrödinger, LLC: New York, NY, USA, 2017.
- Laskowski, R.A.; Jablonska, J.; Pravda, L.; Varekova, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kloks, C.P.; Spronk, C.A.; Lasonder, E.; Hoffmann, A.; Vuister, G.W.; Grzesiek, S.; Hilbers, C.W. The solution structure and DNA-binding properties of the cold-shock domain of the human Y-box protein YB-1. J. Mol. Biol. 2002, 316, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Goroncy, A.K.; Koshiba, S.; Tochio, N.; Tomizawa, T.; Inoue, M.; Watanabe, S.; Harada, T.; Tanaka, A.; Ohara, O.; Kigawa, T.; et al. The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein. J. Struct. Funct. Genomics. 2010, 11, 181–188. [Google Scholar] [CrossRef]
- Hou, H.; Wang, F.; Zhang, W.; Wang, D.; Li, X.; Bartlam, M.; Yao, X.; Rao, Z. Structure-functional analyses of CRHSP-24 plasticity and dynamics in oxidative stress response. J. Biol. Chem. 2011, 286, 9623–9635. [Google Scholar] [CrossRef] [Green Version]
- Sawyer, A.L.; Landsberg, M.J.; Ross, I.L.; Kruse, O.; Mobli, M.; Hankamer, B. Solution structure of the RNA-binding cold-shock domain of the Chlamydomonas reinhardtii NAB1 protein and insights into RNA recognition. Biochem. J. 2015, 469, 97–106. [Google Scholar] [CrossRef]
- Morgan, H.P.; Wear, M.A.; McNae, I.; Gallagher, M.P.; Walkinshaw, M.D. Crystallization and X-ray structure of cold-shock protein E from Salmonella typhimurium. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 1240–1245. [Google Scholar] [CrossRef] [Green Version]
- Makhatadze, G.I.; Marahiel, M.A. Effect of pH and phosphate ions on self-association properties of the major cold-shock protein from Bacillus subtilis. Protein Sci. 1994, 3, 2144–2147. [Google Scholar] [CrossRef] [Green Version]
- Mayr, B.; Kaplan, T.; Lechner, S.; Scherer, S. Identification and purification of a family of dimeric major cold shock protein homologs from the psychrotrophic Bacillus cereus WSBC 10201. J. Bacteriol. 1996, 178, 2916–2925. [Google Scholar] [CrossRef] [Green Version]
- Johnston, D.; Tavano, C.; Wickner, S.; Trun, N. Specificity of DNA binding and dimerization by CspE from Escherichia coli. J. Biol. Chem. 2006, 281, 40208–40215. [Google Scholar] [CrossRef] [Green Version]
- Yamanaka, K.; Zheng, W.; Crooke, E.; Wang, Y.H.; Inouye, M. CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol. Microbiol. 2001, 39, 1572–1584. [Google Scholar] [CrossRef] [PubMed]
- Max, K.E.; Zeeb, M.; Bienert, R.; Balbach, J.; Heinemann, U. Common mode of DNA binding to cold shock domains. Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus. FEBS J. 2007, 274, 1265–1279. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, A.I.; Vallejos, G.; Komives, E.A.; Castro-Fernandez, V.; Leonardo, D.A.; Garratt, R.C.; Ramirez-Sarmiento, C.A.; Babul, J. Unusual dimerization of a BcCsp mutant leads to reduced conformational dynamics. FEBS J. 2017, 284, 1882–1896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.; Nettleship, J.E.; Sainsbury, S.; Saunders, N.J.; Owens, R.J. Structure of the cold-shock domain protein from Neisseria meningitidis reveals a strand-exchanged dimer. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 247–251. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.; Kather, I.; Schmid, F.X. Origins of the high stability of an in vitro-selected cold-shock protein. J. Mol. Biol. 2002, 318, 1341–1349. [Google Scholar] [CrossRef]
- Bycroft, M.; Hubbard, T.J.; Proctor, M.; Freund, S.M.; Murzin, A.G. The solution structure of the S1 RNA binding domain: A member of an ancient Nucleic acid-binding fold. Cell 1997, 88, 235–242. [Google Scholar] [CrossRef] [Green Version]
- de Bono, S.; Riechmann, L.; Girard, E.; Williams, R.L.; Winter, G. A segment of cold shock protein directs the folding of a combinatorial protein. Proc. Natl. Acad. Sci. USA 2005, 102, 1396–1401. [Google Scholar] [CrossRef] [Green Version]
- Schindler, T.; Herrler, M.; Marahiel, M.A.; Schmid, F.X. Extremely rapid protein folding in the absence of intermediates. Nat. Struct. Biol. 1995, 2, 663–673. [Google Scholar] [CrossRef]
- Perl, D.; Welker, C.; Schindler, T.; Schroder, K.; Marahiel, M.A.; Jaenicke, R.; Schmid, F.X. Conservation of rapid two-state folding in mesophilic, thermophilic and hyperthermophilic cold shock proteins. Nat. Struct. Biol. 1998, 5, 229–235. [Google Scholar] [CrossRef]
- Perl, D.; Mueller, U.; Heinemann, U.; Schmid, F.X. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat. Struct. Biol. 2000, 7, 380–383. [Google Scholar] [CrossRef]
- Delbruck, H.; Mueller, U.; Perl, D.; Schmid, F.X.; Heinemann, U. Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability. J. Mol. Biol. 2001, 313, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Su, J.G.; Han, X.M.; Zhao, S.X.; Hou, Y.X.; Li, X.Y.; Qi, L.S.; Wang, J.H. Impacts of the charged residues mutation S48E/N62H on the thermostability and unfolding behavior of cold shock protein: Insights from molecular dynamics simulation with Go model. J. Mol. Model. 2016, 22, 91. [Google Scholar] [CrossRef]
- Tych, K.M.; Batchelor, M.; Hoffmann, T.; Wilson, M.C.; Paci, E.; Brockwell, D.J.; Dougan, L. Tuning protein mechanics through an ionic cluster graft from an extremophilic protein. Soft Matter 2016, 12, 2688–2699. [Google Scholar] [CrossRef] [PubMed]
- Perl, D.; Schmid, F.X. Electrostatic stabilization of a thermophilic cold shock protein. J. Mol. Biol. 2001, 313, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Max, K.E.; Wunderlich, M.; Roske, Y.; Schmid, F.X.; Heinemann, U. Optimized variants of the cold shock protein from in vitro selection: Structural basis of their high thermostability. J. Mol. Biol. 2007, 369, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, M.; Martin, A.; Schmid, F.X. Stabilization of the cold shock protein CspB from Bacillus subtilis by evolutionary optimization of Coulombic interactions. J. Mol. Biol. 2005, 347, 1063–1076. [Google Scholar] [CrossRef]
- Makhatadze, G.I.; Loladze, V.V.; Gribenko, A.V.; Lopez, M.M. Mechanism of thermostabilization in a designed cold shock protein with optimized surface electrostatic interactions. J. Mol. Biol. 2004, 336, 929–942. [Google Scholar] [CrossRef]
- Gribenko, A.V.; Makhatadze, G.I. Role of the charge-charge interactions in defining stability and halophilicity of the CspB proteins. J. Mol. Biol. 2007, 366, 842–856. [Google Scholar] [CrossRef]
- Schonfelder, J.; Perez-Jimenez, R.; Munoz, V. A simple two-state protein unfolds mechanically via multiple heterogeneous pathways at single-molecule resolution. Nat. Commun. 2016, 7, 11777. [Google Scholar] [CrossRef]
- de Sancho, D.; Best, R.B. Reconciling Intermediates in Mechanical Unfolding Experiments with Two-State Protein Folding in Bulk. J. Phys. Chem. Lett. 2016, 7, 3798–3803. [Google Scholar] [CrossRef] [Green Version]
- Morgan, H.P.; Estibeiro, P.; Wear, M.A.; Max, K.E.; Heinemann, U.; Cubeddu, L.; Gallagher, M.P.; Sadler, P.J.; Walkinshaw, M.D. Sequence specificity of single-stranded DNA-binding proteins: A novel DNA microarray approach. Nucleic Acids Res. 2007, 35, e75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, M.M.; Yutani, K.; Makhatadze, G.I. Interactions of the cold shock protein CspB from Bacillus subtilis with single-stranded DNA. Importance of the T base content and position within the template. J. Biol. Chem. 2001, 276, 15511–15518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bienert, R.; Zeeb, M.; Dostal, L.; Feske, A.; Magg, C.; Max, K.; Welfle, H.; Balbach, J.; Heinemann, U. Single-stranded DNA bound to bacterial cold-shock proteins: Preliminary crystallographic and Raman analysis. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 755–757. [Google Scholar] [CrossRef] [PubMed]
- Max, K.E.; Zeeb, M.; Bienert, R.; Balbach, J.; Heinemann, U. T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB. J. Mol. Biol. 2006, 360, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Zeeb, M.; Max, K.E.; Weininger, U.; Low, C.; Sticht, H.; Balbach, J. Recognition of T-rich single-stranded DNA by the cold shock protein Bs-CspB in solution. Nucleic Acids Res. 2006, 34, 4561–4571. [Google Scholar] [CrossRef] [Green Version]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar] [CrossRef] [Green Version]
- von Konig, K.; Kachel, N.; Kalbitzer, H.R.; Kremer, W. RNA and DNA Binding Epitopes of the Cold Shock Protein TmCsp from the Hyperthermophile Thermotoga maritima. Protein J. 2020, 39, 487–500. [Google Scholar] [CrossRef]
- Sachs, R.; Max, K.E.; Heinemann, U.; Balbach, J. RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution. RNA 2012, 18, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Nam, Y.; Chen, C.; Gregory, R.I.; Chou, J.J.; Sliz, P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011, 147, 1080–1091. [Google Scholar] [CrossRef] [Green Version]
- Mehta, S.; McKinney, C.; Algie, M.; Verma, C.S.; Kannan, S.; Harfoot, R.; Bartolec, T.K.; Bhatia, P.; Fisher, A.J.; Gould, M.L.; et al. Dephosphorylation of YB-1 is Required for Nuclear Localisation During G2 Phase of the Cell Cycle. Cancers 2020, 12, 315. [Google Scholar] [CrossRef] [Green Version]
- Evdokimova, V.; Ruzanov, P.; Anglesio, M.S.; Sorokin, A.V.; Ovchinnikov, L.P.; Buckley, J.; Triche, T.J.; Sonenberg, N.; Sorensen, P.H. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol. Cell Biol. 2006, 26, 277–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Tao, T.; Liu, F.; Ni, R.; Lu, C.; Shen, A. Hyper-O-GlcNAcylation of YB-1 affects Ser102 phosphorylation and promotes cell proliferation in hepatocellular carcinoma. Exp. Cell Res. 2016, 349, 230–238. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, G.H.; Itoh-Lindstrom, Y.; Ting, J.P. The transcriptional regulatory protein, YB-1, promotes single-stranded regions in the DRA promoter. J. Biol. Chem. 1995, 270, 3527–3533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukada, T.; Tonks, N.K. Identification of YB-1 as a regulator of PTP1B expression: Implications for regulation of insulin and cytokine signaling. EMBO J. 2003, 22, 479–493. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Yao, B.; Shin, J.; Lin, L.; Kim, N.; Song, Q.; Liu, S.; Su, Y.; Guo, J.U.; Huang, L.; et al. Lin28A Binds Active Promoters and Recruits Tet1 to Regulate Gene Expression. Mol. Cell 2016, 61, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Tan, F.E.; Yeo, G.W. Blurred Boundaries: The RNA Binding Protein Lin28A Is Also an Epigenetic Regulator. Mol. Cell 2016, 61, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.J.; Mu, S.R.; Heiner, M.; Fu, X.; Cao, L.J.; Gong, X.F.; Bindereif, A.; Hui, J. YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res. 2012, 40, 8622–8636. [Google Scholar] [CrossRef] [Green Version]
- Jayavelu, A.K.; Schnoder, T.M.; Perner, F.; Herzog, C.; Meiler, A.; Krishnamoorthy, G.; Huber, N.; Mohr, J.; Edelmann-Stephan, B.; Austin, R.; et al. Splicing factor YBX1 mediates persistence of JAK2-mutated neoplasms. Nature 2020, 588, 157–163. [Google Scholar] [CrossRef]
- Giorgini, F.; Davies, H.G.; Braun, R.E. MSY2 and MSY4 bind a conserved sequence in the 3’ untranslated region of protamine 1 mRNA in vitro and in vivo. Mol. Cell Biol. 2001, 21, 7010–7019. [Google Scholar] [CrossRef] [Green Version]
- Lyabin, D.N.; Eliseeva, I.A.; Smolin, E.A.; Doronin, A.N.; Budkina, K.S.; Kulakovskiy, I.V.; Ovchinnikov, L.P. YB-3 substitutes YB-1 in global mRNA binding. RNA Biol. 2020, 17, 487–499. [Google Scholar] [CrossRef] [Green Version]
- Kretov, D.A.; Clement, M.J.; Lambert, G.; Durand, D.; Lyabin, D.N.; Bollot, G.; Bauvais, C.; Samsonova, A.; Budkina, K.; Maroun, R.C.; et al. YB-1, an abundant core mRNA-binding protein, has the capacity to form an RNA nucleoprotein filament: A structural analysis. Nucleic Acids Res. 2019, 47, 3127–3141. [Google Scholar] [CrossRef] [PubMed]
- Graf, R.; Munschauer, M.; Mastrobuoni, G.; Mayr, F.; Heinemann, U.; Kempa, S.; Rajewsky, N.; Landthaler, M. Identification of LIN28B-bound mRNAs reveals features of target recognition and regulation. RNA Biol. 2013, 10, 1146–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafner, M.; Max, K.E.; Bandaru, P.; Morozov, P.; Gerstberger, S.; Brown, M.; Molina, H.; Tuschl, T. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA 2013, 19, 613–626. [Google Scholar] [CrossRef] [Green Version]
- Desjardins, A.; Bouvette, J.; Legault, P. Stepwise assembly of multiple Lin28 proteins on the terminal loop of let-7 miRNA precursors. Nucleic Acids Res. 2014, 42, 4615–4628. [Google Scholar] [CrossRef]
- Sharma, C.; Mohanty, D. Molecular Dynamics Simulations for Deciphering the Structural Basis of Recognition of Pre-let-7 miRNAs by LIN28. Biochemistry 2017, 56, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Ustianenko, D.; Chiu, H.S.; Treiber, T.; Weyn-Vanhentenryck, S.M.; Treiber, N.; Meister, G.; Sumazin, P.; Zhang, C. LIN28 Selectively Modulates a Subclass of Let-7 MicroRNAs. Mol. Cell 2018, 71, 271–283.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desjardins, A.; Yang, A.; Bouvette, J.; Omichinski, J.G.; Legault, P. Importance of the NCp7-like domain in the recognition of pre-let-7g by the pluripotency factor Lin28. Nucleic Acids Res. 2012, 40, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Rowe, R.G.; Jaimes, A.; Yu, C.; Nam, Y.; Pearson, D.S.; Zhang, J.; Xie, X.; Marion, W.; Heffron, G.J.; et al. Small-Molecule Inhibitors Disrupt let-7 Oligouridylation and Release the Selective Blockade of let-7 Processing by LIN28. Cell Rep. 2018, 23, 3091–3101. [Google Scholar] [CrossRef]
- Evdokimova, V.; Ovchinnikov, L.P.; Sorensen, P.H. Y-box binding protein 1: Providing a new angle on translational regulation. Cell Cycle 2006, 5, 1143–1147. [Google Scholar] [CrossRef]






| CSP/CSD | Organism | Sequence 1 | Method | PDB ID 2 | Reference |
|---|---|---|---|---|---|
| BsCspB | B. subtilis | TTT TTT | X-ray | 2es2 | [135] |
| BcCspB | B. caldolyticus | TTTTTT | X-ray | 2hax | [113] |
| LIN28B CSD | X. tropicalis | TTTTTT TTTTTTT | X-ray X-ray | 4a75 4a76 | [100] |
| YBX1 CSDex 3 | human | AACACCT | NMR | 6lmr | [9] |
| CSP/CSD | Organism | Sequence 1 | Method | PDB ID 2 | Reference |
|---|---|---|---|---|---|
| BsCspB | B. subtilis | UUU UUU GUC UUU A | X-ray | 3pf5 3pf4 | [139] |
| LIN28A CSD+ZKD | M. musculus | GGG CAG AGA UUU UGC CCG GAG 3 GGG GUA GUG AUU UUA CCC UGG AG 4 GGG GUC UAU GAU ACC ACC CCG GAG 5 | X-ray | 3trz 3ts0 3ts2 | [140] |
| LIN28B CSD | X. tropicalis | UUUUUU | X-ray | 4alp | [100] |
| CSDE1 CSD1 6 | D. melanogaster | UUU UUU UGA GCA CGU GAA | X-ray | 4qqb | [14] |
| LIN28A CSD+ZKD | human | GGG GUA GUG AUU UUA CCC UGG AGA U | X-ray | 5udz | [13] |
| YBX1 CSD | human | UCA UCU UCU UCU UCA ACU UCA UGU | X-ray | 5ytv 5yts 5ytx 5ytt | [11] |
| YBX1 CSD | human | UCA Um 5 CU | X-ray | 6a6l | [12] |
| YBX1 CSD | D. rerio | UCA Um 5 CU | X-ray | 6a6j | [10] |
| YPS CSD | D. melanogaster | ACC AGC CU ACC AGm 5 C CU | X-ray | 6kug 6ktc | [8] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heinemann, U.; Roske, Y. Cold-Shock Domains—Abundance, Structure, Properties, and Nucleic-Acid Binding. Cancers 2021, 13, 190. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13020190
Heinemann U, Roske Y. Cold-Shock Domains—Abundance, Structure, Properties, and Nucleic-Acid Binding. Cancers. 2021; 13(2):190. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13020190
Chicago/Turabian StyleHeinemann, Udo, and Yvette Roske. 2021. "Cold-Shock Domains—Abundance, Structure, Properties, and Nucleic-Acid Binding" Cancers 13, no. 2: 190. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13020190