Roles for the IKK-Related Kinases TBK1 and IKKε in Cancer
Abstract
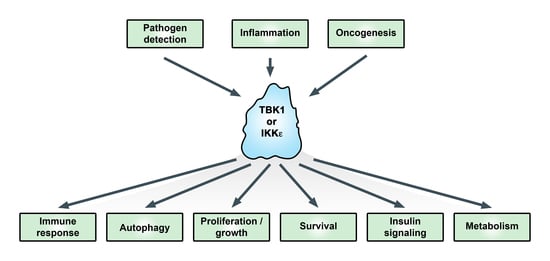
:1. Introduction
2. Cancers Controlled by TBK1 and/or IKKε
3. Control of IKKε and TBK1, and Downstream Signaling
4. Subcellular Localization and Target Specificity
5. TBK1 and Mitosis
6. Autophagy Regulation by TBK1
7. Promotion of KRAS-Induced Oncogenesis and Control of Akt
8. TBK1 and IKKε Control of mTORC1 and Metabolism
9. TBK1 and Antitumor Immunity
10. DNA Damage and Cancer: Is TBK1 Involved?
11. Therapeutic Potential and Future Considerations
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abe, T.; Barber, G.N. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 2014, 88, 5328–5341. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chiu, Y.-H.; Chen, Z.J. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell 2014, 54, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Imamura, Y.; Jenkins, R.W.; Cañadas, I.; Kitajima, S.; Aref, A.; Brannon, A.; Oki, E.; Castoreno, A.; Zhu, Z.; et al. Autophagy Inhibition Dysregulates TBK1 Signaling and Promotes Pancreatic Inflammation. Cancer Immunol. Res. 2016, 4, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Weidberg, H.; Elazar, Z. TBK1 mediates crosstalk between the innate immune response and autophagy. Sci. Signal. 2011, 4, pe39. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.R.; Hahn, W.C. Emerging roles for the non-canonical IKKs in cancer. Oncogene 2011, 30, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, J.L.; Baltimore, D. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 1999, 18, 6694–6704. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Kawai, T.; Takeda, K.; Matsumoto, M.; Inoue, J.; Tatsumi, Y.; Kanamaru, A.; Akira, S. IKK-i, a novel lipopolysaccharide-inducible kinase that is related to IkappaB kinases. Int. Immunol. 1999, 11, 1357–1362. [Google Scholar] [CrossRef] [PubMed]
- Peters, R.T.; Liao, S.M.; Maniatis, T. IKKepsilon is part of a novel PMA-inducible IkappaB kinase complex. Mol. Cell 2000, 5, 513–522. [Google Scholar] [CrossRef]
- Bonnard, M.; Mirtsos, C.; Suzuki, S.; Graham, K.; Huang, J.; Ng, M.; Itié, A.; Wakeham, A.; Shahinian, A.; Henzel, W.J.; et al. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 2000, 19, 4976–4985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenoever, B.R.; Ng, S.-L.; Chua, M.A.; McWhirter, S.M.; García-Sastre, A.; Maniatis, T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science 2007, 315, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.-H.; Bazuine, M.; Lumeng, C.N.; Geletka, L.M.; Mowers, J.; White, N.M.; Ma, J.-T.; Zhou, J.; Qi, N.; Westcott, D.; et al. The protein kinase IKKepsilon regulates energy balance in obese mice. Cell 2009, 138, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 2012, 26, 203–234. [Google Scholar] [CrossRef] [PubMed]
- Perkins, N.D. The diverse and complex roles of NF-κB subunits in cancer. Nat. Rev. Cancer 2012, 12, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Rinkenbaugh, A.L.; Baldwin, A.S. The NF-κB Pathway and Cancer Stem Cells. Cells 2016, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Wang, C.; Jiang, Q.; Lv, M.; Gao, P.; Yu, X.; Mu, P.; Zhang, R.; Bi, S.; Feng, J.-M.; et al. NEMO-IKKβ Are Essential for IRF3 and NF-κB Activation in the cGAS-STING Pathway. J. Immunol. 2017, 199, 3222–3233. [Google Scholar] [CrossRef] [PubMed]
- Munitic, I.; Giardino Torchia, M.L.; Meena, N.P.; Zhu, G.; Li, C.C.; Ashwell, J.D. Optineurin insufficiency impairs IRF3 but not NF-κB activation in immune cells. J. Immunol. 2013, 191, 6231–6240. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sun, H.; You, F.; Sun, W.; Zhou, X.; Chen, L.; Yang, J.; Wang, Y.; Tang, H.; Guan, Y.; et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 2011, 147, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Hsia, H.-C.; Hutti, J.E.; Baldwin, A.S. Cytosolic DNA Promotes Signal Transducer and Activator of Transcription 3 (STAT3) Phosphorylation by TANK-binding Kinase 1 (TBK1) to Restrain STAT3 Activity. J. Biol. Chem. 2017, 292, 5405–5417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helgason, E.; Phung, Q.T.; Dueber, E.C. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: The emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 2013, 587, 1230–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasan, M.; Yan, N. Therapeutic potential of targeting TBK1 in autoimmune diseases and interferonopathies. Pharmacol. Res. 2016, 111, 336–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, L.; Zhang, S.-Y.; Casanova, J.-L.; Sancho-Shimizu, V. Human TBK1: A Gatekeeper of Neuroinflammation. Trends Mol. Med. 2016, 22, 511–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oakes, J.A.; Davies, M.C.; Collins, M.O. TBK1: A new player in ALS linking autophagy and neuroinflammation. Mol. Brain 2017, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, S.E.; Hammaker, D.; Boyle, D.L.; Firestein, G.S. Regulation of c-Jun phosphorylation by the I kappa B kinase-epsilon complex in fibroblast-like synoviocytes. J. Immunol. 2005, 174, 6424–6430. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zou, Q.; Xie, X.; Liu, T.; Li, H.S.; Jie, Z.; Jin, J.; Hu, H.; Manyam, G.; Zhang, L.; et al. The kinase TBK1 functions in dendritic cells to regulate T cell homeostasis, autoimmunity, and antitumor immunity. J. Exp. Med. 2017, 214, 1493–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Zhou, X.; Chang, M.; Nakaya, M.; Chang, J.-H.; Xiao, Y.; Lindsey, J.W.; Dorta-Estremera, S.; Cao, W.; Zal, A.; et al. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat. Commun. 2015, 6, 6074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Feng, H.; Zhao, J.; Feldman, E.R.; Chen, S.-Y.; Yuan, W.; Huang, C.; Akbari, O.; Tibbetts, S.A.; Feng, P. IκB Kinase ε Is an NFATc1 Kinase that Inhibits T Cell Immune Response. Cell Rep. 2016, 16, 405–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lork, M.; Kreike, M.; Staal, J.; Beyaert, R. Importance of Validating Antibodies and Small Compound Inhibitors Using Genetic Knockout Studies-T Cell Receptor-Induced CYLD Phosphorylation by IKKε/TBK1 as a Case Study. Front. Cell Dev. Biol. 2018, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Lo, Y.-C.; Li, Q.; Napolitano, G.; Wu, X.; Jiang, X.; Dreano, M.; Karin, M.; Wu, H. Crystal structure of inhibitor of κB kinase β. Nature 2011, 472, 325–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, D.; Zhu, Z.; Zhou, A.Y.; Yun, C.-H.; Lee, K.-E.; Toms, A.V.; Li, Y.; Dunn, G.P.; Chan, E.; Thai, T.; et al. Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Rep. 2013, 3, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Larabi, A.; Devos, J.M.; Ng, S.-L.; Nanao, M.H.; Round, A.; Maniatis, T.; Panne, D. Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell Rep. 2013, 3, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Boehm, J.S.; Zhao, J.J.; Yao, J.; Kim, S.Y.; Firestein, R.; Dunn, I.F.; Sjostrom, S.K.; Garraway, L.A.; Weremowicz, S.; Richardson, A.L.; et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 2007, 129, 1065–1079. [Google Scholar] [CrossRef] [PubMed]
- Barbie, T.U.; Alexe, G.; Aref, A.R.; Li, S.; Zhu, Z.; Zhang, X.; Imamura, Y.; Thai, T.C.; Huang, Y.; Bowden, M.; et al. Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. J. Clin. Invest. 2014, 124, 5411–5423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, H.; Zhang, H.; Cai, J.; Wu, J.; Yuan, J.; Li, J.; Huang, Z.; Li, M. IKBKE is over-expressed in glioma and contributes to resistance of glioma cells to apoptosis via activating NF-κB. J. Pathol. 2011, 223, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Guo, J.; Henderson-Jackson, E.; Kim, D.; Malafa, M.; Coppola, D. IκB Kinase ε expression in pancreatic ductal adenocarcinoma. Am. J. Clin.l Pathol. 2011, 136, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-P.; Shu, S.-K.; He, L.; Lee, Y.-C.; Kruk, P.A.; Grenman, S.; Nicosia, S.V.; Mor, G.; Schell, M.J.; Coppola, D.; et al. Deregulation of IKBKE is associated with tumor progression, poor prognosis, and cisplatin resistance in ovarian cancer. Am. J. Pathol. 2009, 175, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.; Kim, M.; Hernandez, L.; Grajales, V.; Noonan, A.; Anver, M.; Davidson, B.; Annunziata, C.M. IKK-ε coordinates invasion and metastasis of ovarian cancer. Cancer Res. 2012, 72, 5494–5504. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Qu, Y.; Tan, B.; Jia, Y.; Wang, N.; Hu, P.; Wang, J. Prognostic significance of preoperative IKBKE expression in esophageal squamous cell carcinoma. Onco Targets Ther. 2018, 11, 1305–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajurkar, M.; Dang, K.; Fernandez-Barrena, M.G.; Liu, X.; Fernandez-Zapico, M.E.; Lewis, B.C.; Mao, J. IKBKE Is Required during KRAS-Induced Pancreatic Tumorigenesis. Cancer Res. 2017, 77, 320–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Péant, B.; Gilbert, S.; Le Page, C.; Poisson, A.; L’Ecuyer, E.; Boudhraa, Z.; Bienz, M.N.; Delvoye, N.; Saad, F.; Mes-Masson, A.-M. IκB-Kinase-epsilon (IKKε) over-expression promotes the growth of prostate cancer through the C/EBP-β dependent activation of IL-6 gene expression. Oncotarget 2017, 8, 14487–14501. [Google Scholar] [CrossRef] [PubMed]
- Challa, S.; Guo, J.-P.; Ding, X.; Xu, C.-X.; Li, Y.; Kim, D.; Smith, M.A.; Cress, D.W.; Coppola, D.; Haura, E.B.; et al. IKBKE Is a Substrate of EGFR and a Therapeutic Target in Non-Small Cell Lung Cancer with Activating Mutations of EGFR. Cancer Res. 2016, 76, 4418–4429. [Google Scholar] [CrossRef] [PubMed]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eskiocak, B.; McMillan, E.A.; Mendiratta, S.; Kollipara, R.K.; Zhang, H.; Humphries, C.G.; Wang, C.; Garcia-Rodriguez, J.; Ding, M.; Zaman, A.; et al. Biomarker Accessible and Chemically Addressable Mechanistic Subtypes of BRAF Melanoma. Cancer Discov. 2017, 7, 832–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, J.M.; Ou, Y.-H.; McMillan, E.A.; Vaden, R.M.; Zaman, A.; Bodemann, B.O.; Makkar, G.; Posner, B.A.; White, M.A. TBK1 Provides Context-Selective Support of the Activated AKT/mTOR Pathway in Lung Cancer. Cancer Res. 2017, 77, 5077–5094. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, L.; Cai, S.-H.; Cheng, H. Identification of TBK1 and IKKε, the non-canonical IκB kinases, as crucial pro-survival factors in HTLV-1-transformed T lymphocytes. Leuk. Res. 2016, 46, 37–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, C.; Cao, Y.; Yang, X.; Zheng, Z.; Guan, K.; Wang, Q.; Tai, Y.; Zhang, Y.; Ma, S.; Cao, Y.; et al. Elevated expression of TANK-binding kinase 1 enhances tamoxifen resistance in breast cancer. Proc. Natl. Acad. Sci. USA 2014, 111, E601–610. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Helgason, E.; Phung, Q.T.; Quan, C.L.; Iyer, R.S.; Lee, M.W.; Bowman, K.K.; Starovasnik, M.A.; Dueber, E.C. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc. Natl. Acad. Sci. USA 2012, 109, 9378–9383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, K.; Plater, L.; Peggie, M.; Cohen, P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: A distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 2009, 284, 14136–14146. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.Y.; Shen, R.R.; Kim, E.; Lock, Y.J.; Xu, M.; Chen, Z.J.; Hahn, W.C. IKKε-mediated tumorigenesis requires K63-linked polyubiquitination by a cIAP1/cIAP2/TRAF2 E3 ubiquitin ligase complex. Cell Rep. 2013, 3, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Saul, V.V.; Niedenthal, R.; Pich, A.; Weber, F.; Schmitz, M.L. SUMO modification of TBK1 at the adaptor-binding C-terminal coiled-coil domain contributes to its antiviral activity. Biochim. Biophys. Acta 2015, 1853, 136–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, R.R.; Zhou, A.Y.; Kim, E.; Lim, E.; Habelhah, H.; Hahn, W.C. IκB kinase ε phosphorylates TRAF2 to promote mammary epithelial cell transformation. Mol. Cell Biol. 2012, 32, 4756–4768. [Google Scholar] [CrossRef] [PubMed]
- Hutti, J.E.; Shen, R.R.; Abbott, D.W.; Zhou, A.Y.; Sprott, K.M.; Asara, J.M.; Hahn, W.C.; Cantley, L.C. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol. Cell 2009, 34, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-P.; Tian, W.; Shu, S.; Xin, Y.; Shou, C.; Cheng, J.Q. IKBKE phosphorylation and inhibition of FOXO3a: A mechanism of IKBKE oncogenic function. PLoS ONE 2013, 8, e63636. [Google Scholar] [CrossRef] [PubMed]
- Delhase, M.; Kim, S.-Y.; Lee, H.; Naiki-Ito, A.; Chen, Y.; Ahn, E.-R.; Murata, K.; Kim, S.-J.; Lautsch, N.; Kobayashi, K.S.; et al. TANK-binding kinase 1 (TBK1) controls cell survival through PAI-2/serpinB2 and transglutaminase 2. Proc. Natl. Acad. Sci. USA 2012, 109, E177–E186. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Xiao, Y.; Chang, J.-H.; Yu, J.; Hu, H.; Starr, R.; Brittain, G.C.; Chang, M.; Cheng, X.; Sun, S.-C. The kinase TBK1 controls IgA class switching by negatively regulating noncanonical NF-κB signaling. Nat. Immunol. 2012, 13, 1101–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adli, M.; Baldwin, A.S. IKK-i/IKKepsilon controls constitutive, cancer cell-associated NF-kappaB activity via regulation of Ser-536 p65/RelA phosphorylation. J. Biol. Chem. 2006, 281, 26976–26984. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.; Olière, S.; Sharma, S.; Sun, Q.; Lin, R.; Hiscott, J.; Grandvaux, N. Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK epsilon. J. Immunol. 2006, 177, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, I.; Geng, H.; Sebald, A.; Hodel, M.; Bucher, C.; Kracht, M.; Schmitz, M.L. Inducible phosphorylation of NF-kappa B p65 at serine 468 by T cell costimulation is mediated by IKK epsilon. J. Biol. Chem. 2006, 281, 6175–6183. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, A.; Bürckstümmer, T.; Dixit, E.; Scheicher, R.; Górna, M.W.; Karayel, E.; Sugar, C.; Stukalov, A.; Berg, T.; Kralovics, R.; et al. Functional dissection of the TBK1 molecular network. PLoS ONE 2011, 6, e23971. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.-H.; Torres, M.; Ram, R.; Formstecher, E.; Roland, C.; Cheng, T.; Brekken, R.; Wurz, R.; Tasker, A.; Polverino, T.; et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol. Cell 2011, 41, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, K.; Goto, H.; Izawa, I.; Kiyono, T.; Watanabe, N.; Elowe, S.; Nigg, E.A.; Inagaki, M. PI 3-kinase-dependent phosphorylation of Plk1-Ser99 promotes association with 14-3-3γ and is required for metaphase-anaphase transition. Nat. Commun. 2013, 4, 1882. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-Y.; Welsh, E.A.; Oguz, U.; Fang, B.; Bai, Y.; Kinose, F.; Bronk, C.; Remsing Rix, L.L.; Beg, A.A.; Rix, U.; et al. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proc. Natl. Acad. Sci. USA 2013, 110, 12414–12419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillai, S.; Nguyen, J.; Johnson, J.; Haura, E.; Coppola, D.; Chellappan, S. Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat. Commun. 2015, 6, 10072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karsli-Uzunbas, G.; Guo, J.Y.; Price, S.; Teng, X.; Laddha, S.V.; Khor, S.; Kalaany, N.Y.; Jacks, T.; Chan, C.S.; Rabinowitz, J.D.; et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014, 4, 914–927. [Google Scholar] [CrossRef] [PubMed]
- Amaravadi, R.; Kimmelman, A.C.; White, E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016, 30, 1913–1930. [Google Scholar] [CrossRef] [PubMed]
- Rybstein, M.D.; Bravo San Pedro, J.M.; Kroemer, G.; Galluzzi, L. The autophagic network and cancer. Nat. Cell Biol. 2018, 20, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011, 333, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Richter, B.; Sliter, D.A.; Herhaus, L.; Stolz, A.; Wang, C.; Beli, P.; Zaffagnini, G.; Wild, P.; Martens, S.; Wagner, S.A.; et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. USA 2016, 113, 4039–4044. [Google Scholar] [CrossRef] [PubMed]
- Pilli, M.; Arko-Mensah, J.; Ponpuak, M.; Roberts, E.; Master, S.; Mandell, M.A.; Dupont, N.; Ornatowski, W.; Jiang, S.; Bradfute, S.B.; et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012, 37, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, G.; Shimogori, T.; Hattori, N.; Nukina, N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Human Mol. Genet. 2015, 24, 4429–4442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniguchi, K.; Yamachika, S.; He, F.; Karin, M. p62/SQSTM1-Dr. Jekyll and Mr. Hyde that prevents oxidative stress but promotes liver cancer. FEBS Lett. 2016, 590, 2375–2397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabakaran, T.; Bodda, C.; Krapp, C.; Zhang, B.C.; Christensen, M.H.; Sun, C.; Reinert, L.; Cai, Y.; Jensen, S.B.; Skouboe, M.K.; et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018, 37, e97858. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, X.; Contino, G.; Liesa, M.; Sahin, E.; Ying, H.; Bause, A.; Li, Y.; Stommel, J.M.; Dell’antonio, G.; et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011, 25, 717–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, A.; Herter-Sprie, G.; Zhang, H.; Lin, E.Y.; Biancur, D.; Wang, X.; Deng, J.; Hai, J.; Yang, S.; Wong, K.-K.; et al. Autophagy Sustains Pancreatic Cancer Growth through Both Cell-Autonomous and Nonautonomous Mechanisms. Cancer Discov. 2018, 8, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Cruz, V.H.; Brekken, R.A. Assessment of TANK-binding kinase 1 as a therapeutic target in cancer. J. Cell Commun. Signal. 2018, 12, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016, 129, 1287–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chien, Y.; Kim, S.; Bumeister, R.; Loo, Y.-M.; Kwon, S.W.; Johnson, C.L.; Balakireva, M.G.; Romeo, Y.; Kopelovich, L.; Gale, M.; et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 2006, 127, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Aref, A.R.; Cohoon, T.J.; Barbie, T.U.; Imamura, Y.; Yang, S.; Moody, S.E.; Shen, R.R.; Schinzel, A.C.; Thai, T.C.; et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov. 2014, 4, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Muvaffak, A.; Pan, Q.; Yan, H.; Fernandez, R.; Lim, J.; Dolinski, B.; Nguyen, T.T.; Strack, P.; Wu, S.; Chung, R.; et al. Evaluating TBK1 as a therapeutic target in cancers with activated IRF3. Mol. Cancer Res. 2014, 12, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhang, D.; Zhao, B.; Lu, M.-K.; You, M.; Condorelli, G.; Wang, C.-Y.; Guan, K.-L. IkappaB kinase epsilon and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proc. Natl. Acad. Sci. USA 2011, 108, 6474–6479. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Liu, J.C.; Chung, P.E.D.; Uehling, D.; Aman, A.; Joseph, B.; Ketela, T.; Jiang, Z.; Schachter, N.F.; Rottapel, R.; et al. shRNA kinome screen identifies TBK1 as a therapeutic target for HER2+ breast cancer. Cancer Res. 2014, 74, 2119–2130. [Google Scholar] [CrossRef] [PubMed]
- Cañadas, I.; Thummalapalli, R.; Kim, J.W.; Kitajima, S.; Jenkins, R.W.; Christensen, C.L.; Campisi, M.; Kuang, Y.; Zhang, Y.; Gjini, E.; et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med. 2018, 24, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodur, C.; Kazyken, D.; Huang, K.; Ekim Ustunel, B.; Siroky, K.A.; Tooley, A.S.; Gonzalez, I.E.; Foley, D.H.; Acosta Jaquez, H.A.; Barnes, T.M.; et al. The IKK-related kinase TBK1 activates mTORC1 directly in response to growth factors and innate immune agonists. EMBO J. 2018, 37, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Gonugunta, V.K.; Dobbs, N.; Ali, A.; Palchik, G.; Calvaruso, M.A.; DeBerardinis, R.J.; Yan, N. Chronic innate immune activation of TBK1 suppresses mTORC1 activity and dysregulates cellular metabolism. Proc. Natl. Acad. Sci. USA 2017, 114, 746–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.K.; Jung, Y.; Wang, J.; Joseph, J.; Mishra, A.; Hill, E.E.; Krebsbach, P.H.; Pienta, K.J.; Shiozawa, Y.; Taichman, R.S. TBK1 regulates prostate cancer dormancy through mTOR inhibition. Neoplasia 2013, 15, 1064–1074. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Guan, K.-L. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Invest. 2015, 125, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Gulen, M.F.; Bulek, K.; Xiao, H.; Yu, M.; Gao, J.; Sun, L.; Beurel, E.; Kaidanovich-Beilin, O.; Fox, P.L.; DiCorleto, P.E.; et al. Inactivation of the enzyme GSK3α by the kinase IKKi promotes AKT-mTOR signaling pathway that mediates interleukin-1-induced Th17 cell maintenance. Immunity 2012, 37, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Bulek, K.; Liu, C.; Swaidani, S.; Wang, L.; Page, R.C.; Gulen, M.F.; Herjan, T.; Abbadi, A.; Qian, W.; Sun, D.; et al. The inducible kinase IKKi is required for IL-17-dependent signaling associated with neutrophilia and pulmonary inflammation. Nat. Immunol. 2011, 12, 844–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, C.V. Links between metabolism and cancer. Genes Dev. 2012, 26, 877–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhm, M.; Bazuine, M.; Zhao, P.; Chiang, S.-H.; Xiong, T.; Karunanithi, S.; Chang, L.; Saltiel, A.R. Phosphorylation of the exocyst protein Exo84 by TBK1 promotes insulin-stimulated GLUT4 trafficking. Sci. Signal. 2017, 10, eaah5085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, M.C.; Giani, J.F.; Mayer, M.A.; Toblli, J.E.; Turyn, D.; Dominici, F.P. TANK-binding kinase 1 mediates phosphorylation of insulin receptor at serine residue 994: A potential link between inflammation and insulin resistance. J. Endocrinol. 2009, 201, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wong, K.I.; Sun, X.; Reilly, S.M.; Uhm, M.; Liao, Z.; Skorobogatko, Y.; Saltiel, A.R. TBK1 at the Crossroads of Inflammation and Energy Homeostasis in Adipose Tissue. Cell 2018, 172, 731–743.e12. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hu, D.; Chen, H.; Shi, G.; Fetahu, I.S.; Wu, F.; Rabidou, K.; Fang, R.; Tan, L.; Xu, S.; et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature 2018, 559, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Zubair, H.; Azim, S.; Srivastava, S.K.; Ahmad, A.; Bhardwaj, A.; Khan, M.A.; Patel, G.K.; Arora, S.; Carter, J.E.; Singh, S.; et al. Glucose Metabolism Reprogrammed by Overexpression of IKKε Promotes Pancreatic Tumor Growth. Cancer Res. 2016, 76, 7254–7264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welcker, M.; Orian, A.; Jin, J.; Grim, J.E.; Grim, J.A.; Harper, J.W.; Eisenman, R.N.; Clurman, B.E. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA 2004, 101, 9085–9090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.-H.; Xie, X.; Sun, S.-C. TBK1 as a regulator of autoimmunity and antitumor immunity. Nat. Publ. Group 2018, 5, 212. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, R.W.; Aref, A.R.; Lizotte, P.H.; Ivanova, E.; Stinson, S.; Zhou, C.W.; Bowden, M.; Deng, J.; Liu, H.; Miao, D.; et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018, 8, 196–215. [Google Scholar] [CrossRef] [PubMed]
- Manguso, R.T.; Pope, H.W.; Zimmer, M.D.; Brown, F.D.; Yates, K.B.; Miller, B.C.; Collins, N.B.; Bi, K.; LaFleur, M.W.; Juneja, V.R.; et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 2017, 547, 413–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, T.; Konno, H.; Barber, G.N. Recurrent Loss of STING Signaling in Melanoma Correlates with Susceptibility to Viral Oncolysis. Cancer Res. 2016, 76, 6747–6759. [Google Scholar] [CrossRef] [PubMed]
- Baird, J.R.; Friedman, D.; Cottam, B.; Dubensky, T.W.; Kanne, D.B.; Bambina, S.; Bahjat, K.; Crittenden, M.R.; Gough, M.J. Radiotherapy Combined with Novel STING-Targeting Oligonucleotides Results in Regression of Established Tumors. Cancer Res. 2016, 76, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Corrales, L.; McWhirter, S.M.; Dubensky, T.W.; Gajewski, T.F. The host STING pathway at the interface of cancer and immunity. J. Clin. Invest. 2016, 126, 2404–2411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harding, S.M.; Benci, J.L.; Irianto, J.; Discher, D.E.; Minn, A.J.; Greenberg, R.A. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017, 548, 466–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhanwani, R.; Takahashi, M.; Sharma, S. Cytosolic sensing of immuno-stimulatory DNA, the enemy within. Curr. Opin. Immunol. 2018, 50, 82–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.-A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef] [PubMed]
- McIver, E.G.; Bryans, J.; Birchall, K.; Chugh, J.; Drake, T.; Lewis, S.J.; Osborne, J.; Smiljanic-Hurley, E.; Tsang, W.; Kamal, A.; et al. Synthesis and structure-activity relationships of a novel series of pyrimidines as potent inhibitors of TBK1/IKKε kinases. Bioorg. Med. Chem. Lett. 2012, 22, 7169–7173. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, J.; Jeong, J.-H.; Park, S.-J.; Wei, R.; Peng, J.; Luo, Z.; Chen, Y.T.; Feng, Y.; Luo, J.-L. Selective TBK1/IKKi dual inhibitors with anticancer potency. Int. J. Cancer 2014, 134, 1972–1980. [Google Scholar] [CrossRef] [PubMed]
- Richters, A.; Basu, D.; Engel, J.; Ercanoglu, M.S.; Balke-Want, H.; Tesch, R.; Thomas, R.K.; Rauh, D. Identification and further development of potent TBK1 inhibitors. ACS Chem. Biol. 2015, 10, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Louis, C.; Ngo, D.; D’Silva, D.B.; Hansen, J.; Phillipson, L.; Jousset, H.; Novello, P.; Segal, D.; Lawlor, K.E.; Burns, C.J.; et al. Therapeutic effects of a TBK1 kinase inhibitor in germinal center-driven, autoantibody-mediated inflammatory arthritis. Arthritis Rheumatol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Oral, E.A.; Reilly, S.M.; Gomez, A.V.; Meral, R.; Butz, L.; Ajluni, N.; Chenevert, T.L.; Korytnaya, E.; Neidert, A.H.; Hench, R.; et al. Inhibition of IKKε and TBK1 Improves Glucose Control in a Subset of Patients with Type 2 Diabetes. Cell Metab. 2017, 26, 157–170.e7. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.-Y.; Chiu, C.-F.; Kapuriya, N.P.; Shieh, T.-M.; Tsai, Y.-C.; Wu, C.-Y.; Sargeant, A.M.; Weng, J.-R. BX795, a TBK1 inhibitor, exhibits antitumor activity in human oral squamous cell carcinoma through apoptosis induction and mitotic phase arrest. Eur. J. Pharmacol. 2015, 769, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Crew, A.P.; Raina, K.; Dong, H.; Qian, Y.; Wang, J.; Vigil, D.; Serebrenik, Y.V.; Hamman, B.D.; Morgan, A.; Ferraro, C.; et al. Identification and Characterization of von Hippel-Lindau-Recruiting Proteolysis Targeting Chimeras (PROTACs) of TANK-Binding Kinase 1. J. Med. Chem. 2018, 61, 583–598. [Google Scholar] [CrossRef] [PubMed]



| Biological Process | Kinase | Substrate | Protein Function |
|---|---|---|---|
| Immune response/inflammation | TBK1 | Act1 | E3 ubiquitin ligase |
| cRel | transcription factor | ||
| IRF3 | transcription factor | ||
| IRF7 | transcription factor | ||
| NFATc1 | transcription factor | ||
| RelA/p65 | transcription factor | ||
| PELI1 | E3 ubiquitin ligase | ||
| STAT3 | transcription factor | ||
| STAT6 | transcription factor | ||
| STING | receptor/adaptor | ||
| TANK | adaptor | ||
| TRAF2 | E3 ubiquitin ligase | ||
| XIAP | E3 ubiquitin ligase | ||
| IKKε | c-Jun | transcription factor | |
| Autophagy | TBK1 | OPTN | autophagy receptor |
| p62 | autophagy receptor | ||
| Proliferation/growth | TBK1 | Akt | kinase |
| CEP170 | centrosome associated protein | ||
| CYLD | deubiquitinase | ||
| mTOR | kinase | ||
| NuMA | centrosome associated protein | ||
| PLK | centrosome associated protein | ||
| Sec5 | exocyst component | ||
| Insulin signaling | TBK1 | IR | receptor kinase |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durand, J.K.; Zhang, Q.; Baldwin, A.S. Roles for the IKK-Related Kinases TBK1 and IKKε in Cancer. Cells 2018, 7, 139. https://0-doi-org.brum.beds.ac.uk/10.3390/cells7090139
Durand JK, Zhang Q, Baldwin AS. Roles for the IKK-Related Kinases TBK1 and IKKε in Cancer. Cells. 2018; 7(9):139. https://0-doi-org.brum.beds.ac.uk/10.3390/cells7090139
Chicago/Turabian StyleDurand, Joel K., Qing Zhang, and Albert S. Baldwin. 2018. "Roles for the IKK-Related Kinases TBK1 and IKKε in Cancer" Cells 7, no. 9: 139. https://0-doi-org.brum.beds.ac.uk/10.3390/cells7090139