A Cleared View on Retinal Organoids
Abstract
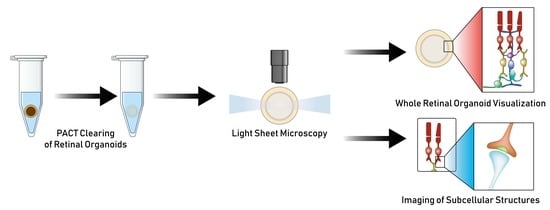
:1. Introduction
2. Materials and Methods
2.1. hiPSC Culture
2.2. hiPSCs-Derived Retinal Organoids Culture
2.3. Lentiviral Transfection of Retinal Organoids
2.4. Tissue Clearing
2.5. Immunocytochemistry
- Antibodies used in immunocytochemistry:
- Primary antibodies:
- - Arrestin 3 (1:100, sc54355, Santa Cruz Biotechnologies, Dallas, TX, USA)
- - CRALBP [B2] (1:250, ab15051, Abcam, Cambridge, UK)
- - CtBP2 (Ribeye) (1:200, 612044, BD Biosciences)
- - Glutamine Synthase (1:500, MAB302, Merck Millipore)
- - Opsin, blue (1:200, AB5407, Merck Millipore)
- - PKCα (1:500, sc208, Santa Cruz Biotechnologies)
- - Recoverin (1:1000, sc20353, Santa Cruz Biotechnologies)
- - Rhodopsin (1:200, sc57432, Santa Cruz Biotechnologies)
- - ROM1 (1:200, 21984-1-AP, Proteintech, Rosemont, IL, USA)
- - ZO-1 (1:100, 33-9100, Thermo Fisher Scientific, USA)
- Secondary antibodies:
- Donkey anti-Mouse Alexa Fluor® 488/568 (1:250, Thermo Fisher Scientific)
- Donkey anti-Rabbit IgG (H + L) Alexa Fluor® 488/568 (1:250, Thermo Fisher Scientific)
- Donkey anti-Goat IgG (H + L) Alexa Fluor® 647 (1:250, Thermo Fisher Scientific)
- Antibodies used in whole-mount immunocytochemistry:
- Primary antibodies:
- Arrestin 3 (1:50, sc54355, Santa Cruz Biotechnologies)
- CtBP2 (Ribeye) (1:100, 612044, BD Biosciences)
- GFP (1:500, A-6455, Thermo Fisher Scientific)
- PKCα (1:250, sc208, Santa Cruz Biotechnologies)
- Recoverin (1:500, sc20353, Santa Cruz Biotechnologies)
- Rhodopsin (1:100, sc57432, Santa Cruz Biotechnologies)
- β-III-Tubulin (1:200, 802001, BioLegend, San Diego, CA, USA)
- Secondary antibodies:
- Donkey anti-Mouse Alexa Fluor® 488/568/647 (1:1000, Thermo Fisher Scientific)
- Donkey anti-Rabbit IgG (H + L) Alexa Fluor® 488/568/647 (1:1000, Thermo Fisher Scientific)
- Donkey anti-Goat IgG (H + L) Alexa Fluor® 488/568/647 (1:1000, Thermo Fisher Scientific)
2.6. Fluorescence Microscopy
2.7. Electron Microscopy
3. Results
3.1. Passive Clarity Technique (PACT) Protocol Optimized for the Retinal Organoid
3.2. PACT Greatly Improves Retinal Organoid Immunocytochemistry and High-Resolution Imaging
3.3. Visualization of Photoreceptor Morphology
3.4. Visualization of the Photoreceptor Ribbon Synapse
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Huch, M.; Bonfanti, P.; Boj, S.F.; Sato, T.; Loomans, C.J.M.; van de Wetering, M.; Sojoodi, M.; Li, V.S.W.; Schuijers, J.; Gracanin, A.; et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013, 32, 2708–2721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taguchi, A.; Kaku, Y.; Ohmori, T.; Sharmin, S.; Ogawa, M.; Sasaki, H.; Nishinakamura, R. Redefining the In Vivo Origin of Metanephric Nephron Progenitors Enables Generation of Complex Kidney Structures from Pluripotent Stem Cells. Cell Stem Cell 2014, 14, 53–67. [Google Scholar] [CrossRef] [Green Version]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef]
- Zhong, X.; Gutierrez, C.; Xue, T.; Hampton, C.; Vergara, M.N.; Cao, L.-H.; Peters, A.; Park, T.S.; Zambidis, E.T.; Meyer, J.S.; et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014, 5, 4047. [Google Scholar] [CrossRef]
- Hallam, D.; Hilgen, G.; Dorgau, B.; Zhu, L.; Yu, M.; Bojic, S.; Hewitt, P.; Schmitt, M.; Uteng, M.; Kustermann, S.; et al. Human-Induced Pluripotent Stem Cells Generate Light Responsive Retinal Organoids with Variable and Nutrient-Dependent Efficiency. Stem Cells 2018, 36, 1535–1551. [Google Scholar] [CrossRef]
- Wahlin, K.J.; Maruotti, J.A.; Sripathi, S.R.; Ball, J.; Angueyra, J.M.; Kim, C.; Grebe, R.; Li, W.; Jones, B.W.; Zack, D.J. Photoreceptor Outer Segment-like Structures in Long-Term 3D Retinas from Human Pluripotent Stem Cells. Sci. Rep. 2017, 7, 766. [Google Scholar] [CrossRef] [Green Version]
- Capowski, E.E.; Samimi, K.; Mayerl, S.J.; Phillips, M.J.; Pinilla, I.; Howden, S.E.; Saha, J.; Jansen, A.D.; Edwards, K.L.; Jager, L.D.; et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 2019, 146, dev171686. [Google Scholar] [CrossRef]
- Reichman, S.; Slembrouck, A.; Gagliardi, G.; Chaffiol, A.; Terray, A.; Nanteau, C.; Potey, A.; Belle, M.; Rabesandratana, O.; Duebel, J.; et al. Generation of Storable Retinal Organoids and Retinal Pigmented Epithelium from Adherent Human iPS Cells in Xeno-Free and Feeder-Free Conditions. Stem Cells 2017, 35, 1176–1188. [Google Scholar] [CrossRef]
- Richardson, D.S.; Lichtman, J.W. Clarifying Tissue Clearing. Cell 2015, 162, 246–257. [Google Scholar] [CrossRef] [Green Version]
- Chung, K.; Deisseroth, K. CLARITY for mapping the nervous system. Nat. Methods 2013, 10, 508–513. [Google Scholar] [CrossRef]
- Gradinaru, V.; Treweek, J.; Overton, K.; Deisseroth, K. Hydrogel-Tissue Chemistry: Principles and Applications. Annu. Rev. Biophys. 2018, 47, 355–376. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Treweek, J.B.; Kulkarni, R.P.; Deverman, B.E.; Chen, C.-K.; Lubeck, E.; Shah, S.; Cai, L.; Gradinaru, V. Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing. Cell 2014, 158, 945–958. [Google Scholar] [CrossRef]
- Neckel, P.H.; Mattheus, U.; Hirt, B.; Just, L.; Mack, A.F. Large-scale tissue clearing (PACT): Technical evaluation and new perspectives in immunofluorescence, histology and ultrastructure. Sci. Rep. 2016, 6, 34331. [Google Scholar] [CrossRef]
- Raab, S.; Klingenstein, M.; Liebau, S.; Linta, L. A Comparative View on Human Somatic Cell Sources for iPSC Generation. Stem Cells Int. 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Frank, S.; Zhang, M.; Schöler, H.R.; Greber, B. Small Molecule-Assisted, Line-Independent Maintenance of Human Pluripotent Stem Cells in Defined Conditions. PLoS ONE 2012, 7, e41958. [Google Scholar] [CrossRef]
- Kolb, H. Organization of the Outer Plexiform Layer of the Primate Retina: Electron Microscopy of Golgi-Impregnated Cells. Philos. Trans. R. Soc. B Biol. Sci. 1970, 258, 261–283. [Google Scholar] [CrossRef]
- Achberger, K.; Probst, C.; Haderspeck, J.; Bolz, S.; Rogal, J.; Chuchuy, J.; Nikolova, M.; Cora, V.; Antkowiak, L.; Haq, W.; et al. Human Retina-on-a-Chip: Merging Organoid and Organ-on-a-Chip Technology to Generate Complex Multi-Layer Tissue Models. bioRxiv 2019, 552158. [Google Scholar]
- Stefaniuk, M.; Gualda, E.J.; Pawlowska, M.; Legutko, D.; Matryba, P.; Koza, P.; Konopka, W.; Owczarek, D.; Wawrzyniak, M.; Loza-Alvarez, P.; et al. Light-sheet microscopy imaging of a whole cleared rat brain with Thy1-GFP transgene. Sci. Rep. 2016, 6, 28209. [Google Scholar] [CrossRef] [Green Version]
- Achberger, K.; Haderspeck, J.C.; Kleger, A.; Liebau, S. Stem cell-based retina models. Adv. Drug Deliv. Rev. 2018, in press. [Google Scholar] [CrossRef]
- Livet, J.; Weissman, T.A.; Kang, H.; Draft, R.W.; Lu, J.; Bennis, R.A.; Sanes, J.R.; Lichtman, J.W. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 2007, 450, 56–62. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cora, V.; Haderspeck, J.; Antkowiak, L.; Mattheus, U.; Neckel, P.H.; Mack, A.F.; Bolz, S.; Ueffing, M.; Pashkovskaia, N.; Achberger, K.; et al. A Cleared View on Retinal Organoids. Cells 2019, 8, 391. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8050391
Cora V, Haderspeck J, Antkowiak L, Mattheus U, Neckel PH, Mack AF, Bolz S, Ueffing M, Pashkovskaia N, Achberger K, et al. A Cleared View on Retinal Organoids. Cells. 2019; 8(5):391. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8050391
Chicago/Turabian StyleCora, Virginia, Jasmin Haderspeck, Lena Antkowiak, Ulrich Mattheus, Peter H. Neckel, Andreas F. Mack, Sylvia Bolz, Marius Ueffing, Natalia Pashkovskaia, Kevin Achberger, and et al. 2019. "A Cleared View on Retinal Organoids" Cells 8, no. 5: 391. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8050391