Non-Saccharomyces in Winemaking: Source of Mannoproteins, Nitrogen, Enzymes, and Antimicrobial Compounds
Abstract
:1. Introduction
2. Non-Saccharomyces Yeasts as a Source of Polysaccharides
2.1. Non-Saccharomyces Species as a Source of Polysaccharides
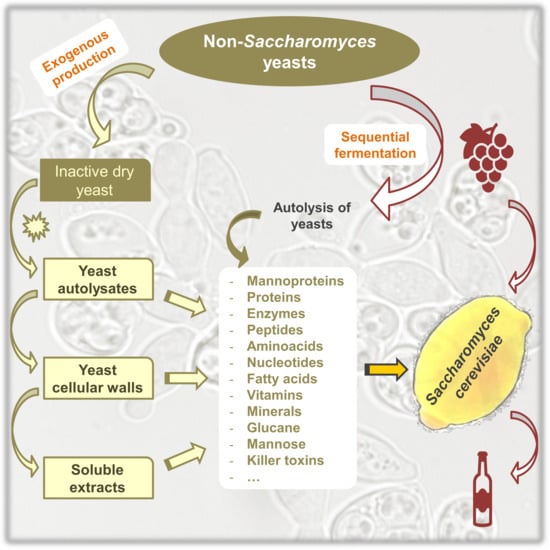
2.2. Accelerated Release of Polysaccharides
3. Non-Saccharomyces Yeasts as a Source of Nitrogen for Saccharomyces cerevisiae
3.1. Non-Saccharomyces Species as a Source of Nitrogen
3.2. Nitrogen Requirements for Sparkling Wines Production
4. Non-Saccharomyces Yeasts as a Source of Exogenous Enzymes
5. Non-Saccharomyces as Biocontrol Agents Against Contaminating Yeasts
5.1. Antimicrobial Peptides
5.2. Killer Toxins
5.3. Other Molecules as Biocontrol Agents
6. Future Perspectives
Funding
Conflicts of Interest
References
- Morata, A. Enological repercussions of non-Saccharomyces species in wine biotechnology. Fermentation 2019, 5, 72. [Google Scholar] [CrossRef] [Green Version]
- Vejarano, R. Saccharomycodes ludwigii, control and potential uses in winemaking processes. Fermentation 2018, 4, 71. [Google Scholar] [CrossRef] [Green Version]
- Ivit, N.N.; Kemp, B. The impact of non-Saccharomyces yeast on traditional method sparkling wine. Fermentation 2018, 4, 73. [Google Scholar] [CrossRef] [Green Version]
- Prior, K.J.; Bauer, F.F.; Divol, B. The utilisation of nitrogenous compounds by commercial non-Saccharomyces yeasts associated with wine. Food Microbiol. 2019, 79, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Del Barrio-Galán, R.; Ortega-Heras, M.; Sánchez-Iglesias, M.; Pérez-Magariño, S. Interactions of phenolic and volatile compounds with yeast lees, commercial yeast derivatives and non toasted chips in model solutions and young red wines. Eur. Food Res. Technol. 2012, 234, 231–244. [Google Scholar] [CrossRef]
- Suárez-Lepe, J.A.; Morata, A. Nuevo Método de Crianza Sobre Lías. Oficina Española de Patentes y Marcas ES2311372B2, 7 October 2009. [Google Scholar]
- Alexandre, H.; Guilloux-Benatier, M. Yeast autolysis in sparkling wine—A review. Aust. J. Grape Wine Res. 2006, 12, 119–127. [Google Scholar] [CrossRef]
- Vidal, S.; Francis, L.; Williams, P.; Kwiatkowski, M.; Gawel, R.; Cheynier, V.; Waters, E. The mouth-feel properties of polysaccharides and anthocyanins in a wine like medium. Food Chem. 2004, 85, 519–525. [Google Scholar] [CrossRef]
- Guadalupe, Z.; Palacios, A.; Ayestarán, B. Maceration enzymes and mannoproteins: A possible strategy to increase colloidal stability and color extraction in red wines. J. Agric. Food Chem. 2007, 55, 4854–4862. [Google Scholar] [CrossRef]
- Chalier, P.; Angot, B.; Delteil, D.; Doco, T.; Gunata, Z. Interactions between aroma compounds and whole mannoprotein isolated from Saccharomyces cerevisiae strains. Food Chem. 2007, 100, 22–30. [Google Scholar] [CrossRef]
- Gonzalez-Ramos, D.; Cebollero, E.; Gonzalez, R. A recombinant Saccharomyces cerevisiae strain overproducing mannoproteins stabilizes wine against protein haze. Appl. Environ. Microbiol. 2008, 74, 5533–5540. [Google Scholar] [CrossRef] [Green Version]
- Lubbers, S.; Leger, B.; Charpentier, C.; Feuillat, M. Effet colloide-protecteur d’extraits de parois de levures sur la stabilité tartrique d’une solution hydro-alcoolique modèle. OENO One 1993, 27, 13. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, A.; Ricardo-Da-Silva, J.M.; Lucas, C.; Laureano, O. Effect of commercial mannoproteins on wine colour and tannins stability. Food Chem. 2012, 131, 907–914. [Google Scholar] [CrossRef]
- Morata, A.; Loira, I.; Suárez-Lepe, J.A. Influence of yeasts in wine colour. In Grape and Wine Biotechnology; Morata, A., Loira, I., Eds.; IntechOpen: London, UK, 2016; pp. 285–305. [Google Scholar]
- Vejarano, R.; Gil-Calderón, A.; Díaz-Silva, V.; León-Vargas, J. Improvement of the bioactive profile in wines and its incidence on human health: Technological strategies. In Advances in Grape and Wine Biotechnology; Morata, A., Loira, I., Eds.; IntechOpen: London, UK, 2019. [Google Scholar]
- Loira, I.; Vejarano, R.; Morata, A.; Ricardo-Da-Silva, J.M.; Laureano, O.; González, M.C.; Suárez-Lepe, J.A. Effect of Saccharomyces strains on the quality of red wines aged on lees. Food Chem. 2013, 139, 1044–1051. [Google Scholar] [CrossRef]
- Rosi, I.; Gheri, A.; Domizio, P.; Fia, G. Production de macromolecules parietals de Saccharomyces cerevisiae au cours de la fermentation et leur influence sur la fermentation malolactique. Rev. des Œnologues 2000, 94, 18–20. [Google Scholar]
- Moruno, E.G.; Sanlorenzo, C.; Boccaccino, B.; Di Stefano, R. Treatment with yeast to reduce the concentration of ochratoxin A in red wine. Am. J. Enol. Vitic. 2005, 56, 73–76. [Google Scholar]
- Moreno-Arribas, V.; Pueyo, E.; Nieto, F.J.; Martín-Álvarez, P.J.; Polo, M.C. Influence of the polysaccharides and the nitrogen compounds on foaming properties of sparkling wines. Food Chem. 2000, 70, 309–317. [Google Scholar] [CrossRef]
- Gallardo-Chacón, J.J.; Vichi, S.; López-Tamames, E.; Buxaderas, S. Changes in the sorption of diverse volatiles by Saccharomyces cerevisiae lees during sparkling wine aging. J. Agric. Food Chem. 2010, 58, 12426–12430. [Google Scholar] [CrossRef]
- Palomero, F.; Morata, A.; Benito, S.; Calderón, F.; Suárez-Lepe, J.A. New genera of yeasts for over-lees aging of red wine. Food Chem. 2009, 112, 432–441. [Google Scholar] [CrossRef]
- Kopecká, M.; Fleet, G.H.; Phaff, H.J. Ultrastructure of the cell wall of Schizosaccharomyces pombe following treatment with various glucanases. J. Struct. Biol. 1995, 114, 140–152. [Google Scholar] [CrossRef]
- Hidalgo, J. Tratado de Enología, Tomo I, 2nd ed.; Mundiprensa: Madrid, Spain, 2011; ISBN 84-8476-119-3. [Google Scholar]
- Domizio, P.; Liu, Y.; Bisson, L.F.; Barile, D. Cell wall polysaccharides released during the alcoholic fermentation by Schizosaccharomyces pombe and S. japonicus: Quantification and characterization. Food Microbiol. 2017, 61, 136–149. [Google Scholar] [CrossRef] [Green Version]
- Giovani, G.; Rosi, I.; Bertuccioli, M. Quantification and characterization of cell wall polysaccharides released by non-Saccharomyces yeast strains during alcoholic fermentation. Int. J. Food Microbiol. 2012, 160, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Domizio, P.; Romani, C.; Lencioni, L.; Comitini, F.; Gobbi, M.; Mannazzu, I.; Ciani, M. Outlining a future for non-Saccharomyces yeasts: Selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int. J. Food Microbiol. 2011, 147, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Wightman, J.D.; Wrolstad, R.E. β-glucosidase activity in juice-processing enzymes based on anthocyanin analysis. J. Food Sci. 1996, 61, 544–548. [Google Scholar] [CrossRef]
- Morata, A.; Loira, I.; Escott, C.; del Fresno, J.M.; Bañuelos, M.A.; Suárez-Lepe, J.A. Applications of Metschnikowia pulcherrima in wine biotechnology. Fermentation 2019, 5, 63. [Google Scholar] [CrossRef] [Green Version]
- Morata, A.; Gómez-Cordovés, M.C.; Calderón, F.; Suárez, J.A. Effects of pH, temperature and SO2 on the formation of pyranoanthocyanins during red wine fermentation with two species of Saccharomyces. Int. J. Food Microbiol. 2006, 106, 123–129. [Google Scholar] [CrossRef]
- Ivit, N.N.; Loira, I.; Morata, A.; Benito, S.; Palomero, F.; Suárez-Lepe, J.A. Making natural sparkling wines with non-Saccharomyces yeasts. Eur. Food Res. Technol. 2018, 244, 925–935. [Google Scholar] [CrossRef]
- Granchi, L.; Romano, P.; Mangani, S.; Guerrini, S.; Vincenzini, M. Production of biogenic amines by wine microorganisms. Bull. OIV 2005, 78, 595–609. [Google Scholar]
- Marcobal, A.; Martín-Álvarez, P.J.; Polo, M.C.; Muñoz, R.; Moreno-Arribas, M.V. Formation of biogenic amines throughout the industrial manufacture of red wine. J. Food Prot. 2006, 69, 397–404. [Google Scholar] [CrossRef]
- Martin, V. Hanseniaspora Vineae: Caracterización y su Uso en la Vinificación; Universidad de la Republica: Montevideo, Uruguay, 2016. [Google Scholar]
- Domizio, P.; Liu, Y.; Bisson, L.F.; Barile, D. Use of non-Saccharomyces wine yeasts as novel sources of mannoproteins in wine. Food Microbiol. 2014, 43, 5–15. [Google Scholar] [CrossRef]
- Lencioni, L.; Romani, C.; Gobbi, M.; Comitini, F.; Ciani, M.; Domizio, P. Controlled mixed fermentation at winery scale using Zygotorulaspora florentina and Saccharomyces cerevisiae. Int. J. Food Microbiol. 2016, 234, 36–44. [Google Scholar] [CrossRef]
- Zha, Y.; Hossain, A.H.; Tobola, F.; Sedee, N.; Havekes, M.; Punt, P.J. Pichia anomala 29X: A resistant strain for lignocellulosic biomass hydrolysate fermentation. FEMS Yeast Res. 2013, 13, 609–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fugelsang, K.C.; Edwards, C.G. Wine Microbiology: Practical Applications and Procedures; Springer Science & Business Media: Berlin, Germany, 2007; ISBN 038733341X. [Google Scholar]
- Loira, I.; Morata, A.; Palomero, F.; González, C.; Suárez-Lepe, J.A. Schizosaccharomyces pombe: A promising biotechnology for modulating wine composition. Fermentation 2018, 4, 70. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, P.; Loira, I.; Morata, A.; Tesfaye, W.; González, M.C.; Suárez-Lepe, J.A. Use of non-Saccharomyces yeast strains coupled with ultrasound treatment as a novel technique to accelerate ageing on lees of red wines and its repercussion in sensorial parameters. LWT 2015, 64, 1262. [Google Scholar] [CrossRef]
- Todd, B.E.N.; Fleet, G.H.; Henschke, P.A. Promotion of autolysis through the interaction of killer and sensitive yeasts: Potential application in sparkling wine production. Am. J. Enol. Vitic. 2000, 51, 65–72. [Google Scholar]
- Englezos, V.; Rantsiou, K.; Giacosa, S.; Río Segade, S.; Rolle, L.; Cocolin, L. Cell-to-cell contact mechanism modulates Starmerella bacillaris death in mixed culture fermentations with Saccharomyces cerevisiae. Int. J. Food Microbiol. 2019, 289, 106–114. [Google Scholar] [CrossRef]
- Palomero, F.; Morata, A.; Benito, S.; González, M.C.; Suárez-Lepe, J.A. Conventional and enzyme-assisted autolysis during ageing over lees in red wines: Influence on the release of polysaccharides from yeast cell walls and on wine monomeric anthocyanin content. Food Chem. 2007, 105, 838–846. [Google Scholar] [CrossRef]
- Del Fresno, J.M.; Loira, I.; Morata, A.; González, C.; Suárez-Lepe, J.A.; Cuerda, R. Application of ultrasound to improve lees ageing processes in red wines. Food Chem. 2018, 261, 157–163. [Google Scholar] [CrossRef]
- Del Fresno, J.M.; Morata, A.; Escott, C.; Loira, I.; Cuerda, R.; Suárez-Lepe, J.A. Sonication of yeast biomasses to improve the ageing on lees technique in red wines. Molecules 2019, 24, 635. [Google Scholar] [CrossRef] [Green Version]
- Jackson, R.S. Fermentation. In Wine Science: Principles and Applications; Jackson, R.S., Ed.; Academic Press/Elsevier: Hoboken, NJ, USA, 2014; ISBN 9780123814692. [Google Scholar]
- Bely, M.; Rinaldi, A.; Dubourdieu, D. Influence of assimilable nitrogen on volatile acidity production by Saccharomyces cerevisiae during high sugar fermentation. J. Biosci. Bioeng. 2003, 96, 507–512. [Google Scholar] [CrossRef]
- Gobert, A.; Tourdot-Maréchal, R.; Morge, C.; Sparrow, C.; Liu, Y.; Quintanilla-Casas, B.; Vichi, S.; Alexandre, H. Non-Saccharomyces yeasts nitrogen source preferences: Impact on sequential fermentation and wine volatile compounds profile. Front. Microbiol. 2017, 8, 2175. [Google Scholar] [CrossRef] [Green Version]
- Ribéreau-Gayon, P.; Glories, Y.; Maujean, A.; Dubourdieu, D. Nitrogen compounds. In Handbook of Enology: The Chemistry of Wine Stabilization and Treatments; John Wiley and Sons Ltd.: Hoboken, NJ, USA, 2006; ISBN 978-0-470-01037-2. [Google Scholar]
- Cooper, T.G. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: Connecting the dots. FEMS Microbiol. Rev. 2002, 26, 223–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Koker, S. Nitrogen Utilisation of Selected Non-Saccharomyces Yeasts and the Impact on Volatile Compound Production; Stellenbosch University: Stellenbosch, South Africa, 2015. [Google Scholar]
- Martin, V.; Giorello, F.; Fariña, L.; Minteguiaga, M.; Salzman, V.; Boido, E.; Aguilar, P.S.; Gaggero, C.; Dellacassa, E.; Mas, A.; et al. De novo synthesis of benzenoid compounds by the yeast Hanseniaspora vineae increases the flavor diversity of wines. J. Agric. Food Chem. 2016, 64, 4574–4583. [Google Scholar] [CrossRef] [Green Version]
- Martin, V.; Boido, E.; Giorello, F.; Mas, A.; Dellacassa, E.; Carrau, F. Effect of yeast assimilable nitrogen on the synthesis of phenolic aroma compounds by Hanseniaspora vineae strains. Yeast 2016, 33, 323–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burin, V.M.; Gomes, T.M.; Caliari, V.; Rosier, J.P.; Bordignon Luiz, M.T. Establishment of influence the nitrogen content in musts and volatile profile of white wines associated to chemometric tools. Microchem. J. 2015, 122, 20–28. [Google Scholar] [CrossRef]
- Mendes-Ferreira, A.; Barbosa, C.; Lage, P.; Mendes-Faia, A. The impact of nitrogen on yeast fermentation and wine quality. Cienc. Tec. Vitivinic. 2011, 26, 17–32. [Google Scholar]
- Sturgeon, J.Q.; Bohlscheid, J.C.; Edwards, C.G. The effect of nitrogen source on yeast metabolism and H2S formation. J. Wine Res. 2013, 24, 182–194. [Google Scholar] [CrossRef]
- Ribereau-Gayon, P.; Dubourdieu, D.; Doneche, B.; Lonvaud, A. Handbook of Enology: The Microbiology of Wine and Vinifications, 2nd ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2006; Volume 1, ISBN 978-0-470-01034-1. [Google Scholar]
- Chi, Z.; Yan, K.; Gao, L.; Li, J.; Wang, X.; Wang, L. Diversity of marine yeasts with high protein content and evaluation of their nutritive compositions. J. Mar. Biol. Assoc. United Kingdom 2008, 88, 1347–1352. [Google Scholar] [CrossRef]
- Manitchotpisit, P.; Skory, C.D.; Leathers, T.D.; Lotrakul, P.; Eveleigh, D.E.; Prasongsuk, S.; Punnapayak, H. α-Amylase activity during pullulan production and α-amylase gene analyses of Aureobasidium pullulans. J. Ind. Microbiol. Biotechnol. 2011, 38, 1211–1218. [Google Scholar] [CrossRef]
- Leite, R.S.R.; Bocchini, D.A.; Martins, E.S.; Silva, D.; Gomes, E.; Da Silva, R. Production of cellulolytic and hemicellulolytic enzymes from Aureobasidium pullulans on solid state fermentation. Appl. Biochem. Biotechnol. 2007, 137–140, 281–288. [Google Scholar]
- Leathers, T.D.; Rich, J.O.; Anderson, A.M.; Manitchotpisit, P. Lipase production by diverse phylogenetic clades of Aureobasidium pullulans. Biotechnol. Lett. 2013, 35, 1701–1706. [Google Scholar] [CrossRef]
- Manitchotpisit, P.; Leathers, T.D.; Peterson, S.W.; Kurtzman, C.; Li, X.L.; Eveleigh, D.E.; Lotrakul, P.; Prasongsuk, S.; Dunlap, C.A.; Vermillion, K.E.; et al. Multilocus phylogenetic analyses, pullulan production and xylanase activity of tropical isolates of Aureobasidium pullulans. Mycol. Res. 2009, 113, 1107–1120. [Google Scholar] [CrossRef]
- Rich, J.O.; Leathers, T.D.; Anderson, A.M.; Bischoff, K.M.; Manitchotpisit, P. Laccases from Aureobasidium pullulans. Enzyme Microb. Technol. 2013, 53, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Wang, F.; Chi, Z.; Yue, L.; Liu, G.; Zhang, T. Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast. Appl. Microbiol. Biotechnol. 2009, 82, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Chi, Z.; Ma, C.; Madzak, C. Cloning, characterization, and expression of the gene encoding alkaline protease in the marine yeast Aureobasidium pullulans 10. Mar. Biotechnol. 2008, 10, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Bozoudi, D.; Tsaltas, D. The multiple and versatile roles of Aureobasidium pullulans in the vitivinicultural sector. Fermentation 2018, 4, 85. [Google Scholar] [CrossRef] [Green Version]
- Huyben, D.; Nyman, A.; Vidaković, A.; Passoth, V.; Moccia, R.; Kiessling, A.; Dicksved, J.; Lundh, T. Effects of dietary inclusion of the yeasts Saccharomyces cerevisiae and Wickerhamomyces anomalus on gut microbiota of rainbow trout. Aquaculture 2017, 473, 528–537. [Google Scholar] [CrossRef]
- Medina, K.; Boido, E.; Dellacassa, E.; Carrau, F. Growth of non-Saccharomyces yeasts affects nutrient availability for Saccharomyces cerevisiae during wine fermentation. Int. J. Food Microbiol. 2012, 157, 245–250. [Google Scholar] [CrossRef]
- Bataillon, M.; Rico, A.; Sablayrolles, J.M.; Salmon, J.M.; Barre, P. Early thiamin assimilation by yeasts under enological conditions: Impact on alcoholic fermentation kinetics. J. Ferment. Bioeng. 1996, 82, 145–150. [Google Scholar] [CrossRef]
- Wang, X.D.; Bohlscheid, J.C.; Edwards, C.G. Fermentative activity and production of volatile compounds by Saccharomyces grown in synthetic grape juice media deficient in assimilable nitrogen and/or pantothenic acid. J. Appl. Microbiol. 2003, 94, 349–359. [Google Scholar] [CrossRef]
- Romano, P.; Capece, A.; Jespersen, L. Taxonomic and ecological diversity of food and beverage yeasts. In Yeasts in Food and Beverages; Springer: Berlin/Heidelberg, Germany, 2006; pp. 13–53. [Google Scholar]
- Barbosa, C.; Lage, P.; Esteves, M.; Chambel, L.; Mendes-Faia, A.; Mendes-Ferreira, A. Molecular and phenotypic characterization of Metschnikowia pulcherrima strains from Douro Wine Region. Fermentation 2018, 4, 8. [Google Scholar] [CrossRef] [Green Version]
- Vincenzi, S.; Crapisi, A.; Curioni, A. Foamability of Prosecco wine: Cooperative effects of high molecular weight glycocompounds and wine PR-proteins. Food Hydrocoll. 2014, 34, 202–207. [Google Scholar] [CrossRef]
- Martínez-Rodríguez, A.J.; Carrascosa, A.V.; Martín-Álvarez, P.J.; Moreno-Arribas, V.; Polo, M.C. Influence of the yeast strain on the changes of the amino acids, peptides and proteins during sparkling wine production by the traditional method. J. Ind. Microbiol. Biotechnol. 2002, 29, 314–322. [Google Scholar] [CrossRef]
- Lemos Junior, W.J.F.; Nadai, C.; Rolle, L.; da Silva Gulao, E.; Miguez da Rocha Leão, M.H.; Giacomini, A.; Corich, V.; Vincenzi, S. Influence of the mannoproteins of different strains of Starmenella bacillaris used in single and sequential fermentations on foamability, tartaric and protein stabilities of wines. OENO One 2020, 54, 231–243. [Google Scholar] [CrossRef] [Green Version]
- Del Fresno, J.M.; Morata, A.; Loira, I.; Bañuelos, M.A.; Escott, C.; Benito, S.; González Chamorro, C.; Suárez-Lepe, J.A. Use of non-Saccharomyces in single-culture, mixed and sequential fermentation to improve red wine quality. Eur. Food Res. Technol. 2017, 243, 2175–2185. [Google Scholar] [CrossRef]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef]
- González-Royo, E.; Pascual, O.; Kontoudakis, N.; Esteruelas, M.; Esteve-Zarzoso, B.; Mas, A.; Canals, J.M.; Zamora, F. Oenological consequences of sequential inoculation with non-Saccharomyces yeasts (Torulaspora delbrueckii or Metschnikowia pulcherrima) and Saccharomyces cerevisiae in base wine for sparkling wine production. Eur. Food Res. Technol. 2015, 240, 999–1012. [Google Scholar] [CrossRef]
- Medina-Trujillo, L.; González-Royo, E.; Sieczkowski, N.; Heras, J.; Canals, J.M.; Zamora, F. Effect of sequential inoculation (Torulaspora delbrueckii/Saccharomyces cerevisiae) in the first fermentation on the foaming properties of sparkling wine. Eur. Food Res. Technol. 2017, 243, 681–688. [Google Scholar] [CrossRef]
- Perpetuini, G.; Di Gianvito, P.; Arfelli, G.; Schirone, M.; Corsetti, A.; Tofalo, R.; Suzzi, G. Biodiversity of autolytic ability in flocculent Saccharomyces cerevisiae strains suitable for traditional sparkling wine fermentation. Yeast 2016, 33, 303–312. [Google Scholar] [CrossRef] [Green Version]
- Loira, I.; Morata, A.; Escott, C.; Del Fresno, J.M.; Tesfaye, W.; Palomero, F.; Suárez-Lepe, J.A. Applications of nanotechnology in the winemaking process. Eur. Food Res. Technol. 2020, 246, 1533–1541. [Google Scholar] [CrossRef]
- Anbu, P.; Gopinath, S.C.B.; Chaulagain, B.P.; Lakshmipriya, T. Microbial enzymes and their applications in industries and medicine 2016. Biomed Res. Int. 2017, 2017, 2195808. [Google Scholar] [CrossRef]
- García, M.; Esteve-Zarzoso, B.; Cabellos, J.M.; Arroyo, T. Advances in the study of Candida stellata. Fermentation 2018, 4, 74. [Google Scholar] [CrossRef] [Green Version]
- Escribano, R.; González-Arenzana, L.; Garijo, P.; Berlanas, C.; López-Alfaro, I.; López, R.; Gutiérrez, A.R.; Santamaría, P. Screening of enzymatic activities within different enological non-Saccharomyces yeasts. J. Food Sci. Technol. 2017, 54, 1555–1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zott, K.; Thibon, C.; Bely, M.; Lonvaud-Funel, A.; Dubourdieu, D.; Masneuf-Pomarede, I. The grape must non-Saccharomyces microbial community: Impact on volatile thiol release. Int. J. Food Microbiol. 2011, 151, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Bueso, G.; Esteve-Zarzoso, B.; Cabellos, J.M.; Gil-Díaz, M.; Arroyo, T. Biotechnological potential of non-Saccharomyces yeasts isolated during spontaneous fermentations of Malvar (Vitis vinifera cv. L.). Eur. Food Res. Technol. 2013, 236, 193–207. [Google Scholar] [CrossRef]
- Swangkeaw, J.; Vichitphan, S.; Butzke, C.E.; Vichitphan, K. Characterization of β-glucosidases from Hanseniaspora sp. and Pichia anomala with potentially aroma-enhancing capabilities in juice and wine. World J. Microbiol. Biotechnol. 2011, 27, 423–430. [Google Scholar] [CrossRef]
- Fernández, M.; Úbeda, J.F.; Briones, A.I. Typing of non-Saccharomyces yeasts with enzymatic activities of interest in wine-making. Int. J. Food Microbiol. 2000, 59, 29–36. [Google Scholar] [CrossRef]
- Hock, R.; Benda, I.; Schreier, P. Formation of terpenes by yeasts during alcoholic fermentation. Z. Lebensm. Unters. Forsch. 1984, 179, 450–452. [Google Scholar] [CrossRef]
- Andorrà, I.; Berradre, M.; Rozès, N.; Mas, A.; Guillamón, J.M.; Esteve-Zarzoso, B. Effect of pure and mixed cultures of the main wine yeast species on grape must fermentations. Eur. Food Res. Technol. 2010, 231, 215–224. [Google Scholar] [CrossRef] [Green Version]
- Hu, K.; Jin, G.J.; Xu, Y.H.; Tao, Y.S. Wine aroma response to different participation of selected Hanseniaspora uvarum in mixed fermentation with Saccharomyces cerevisiae. Food Res. Int. 2018, 108, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Bovo, B.; Carlot, M.; Lombardi, A.; Lomolino, G.; Lante, A.; Giacomini, A.; Corich, V. Exploring the use of Saccharomyces cerevisiae commercial strain and Saccharomycodes ludwigii natural isolate for grape marc fermentation to improve sensory properties of spirits. Food Microbiol. 2014, 41, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Merín, M.G.; Morata de Ambrosini, V.I. Highly cold-active pectinases under wine-like conditions from non-Saccharomyces yeasts for enzymatic production during winemaking. Lett. Appl. Microbiol. 2015, 60, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Onetto, C.A.; Borneman, A.R.; Schmidt, S.A. Investigating the effects of Aureobasidium pullulans on grape juice composition and fermentation. Food Microbiol. 2020, 90, 103451. [Google Scholar] [CrossRef]
- Schlander, M.; Distler, U.; Tenzer, S.; Thines, E.; Claus, H. Purification and properties of yeast proteases secreted by Wickerhamomyces anomalus 227 and Metschnikovia pulcherrima 446 during growth in a white grape juice. Fermentation 2017, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Theron, L.W.; Bely, M.; Divol, B. Monitoring the impact of an aspartic protease (MpAPr1) on grape proteins and wine properties. Appl. Microbiol. Biotechnol. 2018, 102, 5173–5183. [Google Scholar] [CrossRef] [PubMed]
- Reid, V.J.; Theron, L.W.; Toit, M.D.; Divol, B. Identification and partial characterization of extracellular aspartic protease genes from Metschnikowia pulcherrima IWBT Y1123 and Candida apicola IWBT Y1384. Appl. Environ. Microbiol. 2012, 78, 6838–6849. [Google Scholar] [CrossRef] [Green Version]
- Strauss, M.L.A.; Jolly, N.P.; Lambrechts, M.G.; Van Rensburg, P. Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyces wine yeasts. J. Appl. Microbiol. 2001, 91, 182–190. [Google Scholar] [CrossRef]
- Schwentke, J.; Sabel, A.; Petri, A.; König, H.; Claus, H. The yeast Wickerhamomyces anomalus AS1 secretes a multifunctional exo-β-1,3-glucanase with implications for winemaking. Yeast 2014, 31, 349–359. [Google Scholar] [CrossRef] [Green Version]
- Claus, H.; Mojsov, K. Enzymes for wine fermentation: Current and perspective applications. Fermentation 2018, 4, 52. [Google Scholar] [CrossRef] [Green Version]
- Izgü, F.; Altinbay, D.; Acun, T. Killer toxin of Pichia anomala NCYC 432; purification, characterization and its exo-β-1,3-glucanase activity. Enzyme Microb. Technol. 2006, 39, 669–676. [Google Scholar] [CrossRef]
- Morata, A.; Vejarano, R.; Ridolfi, G.; Benito, S.; Palomero, F.; Uthurry, C.; Tesfaye, W.; González, C.; Suárez-Lepe, J.A. Reduction of 4-ethylphenol production in red wines using HCDC+ yeasts and cinnamyl esterases. Enzyme Microb. Technol. 2013, 52, 99–104. [Google Scholar] [CrossRef]
- Medina, K.; Boido, E.; Dellacassa, E.; Carrau, F. Effects of non-Saccharomyces yeasts on color, anthocyanin, and anthocyanin-derived pigments of Tannat grapes during fermentation. Am. J. Enol. Vitic. 2018, 69, 148–156. [Google Scholar] [CrossRef]
- Benito, S.; Morata, A.; Palomero, F.; González, M.C.; Suárez-Lepe, J.A. Formation of vinylphenolic pyranoanthocyanins by Saccharomyces cerevisiae and Pichia guillermondii in red wines produced following different fermentation strategies. Food Chem. 2011, 124, 15–23. [Google Scholar] [CrossRef]
- Lubbers, M.W.; Rodriguez, S.B.; Honey, N.K.; Thornton, R.J. Purification and characterization of urease from Schizosaccharomyces pombe. Can. J. Microbiol. 1996, 42, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Uthurry, C.A.; Suárez Lepe, J.A.; Lombardero, J.; García Del Hierro, J.R. Ethyl carbamate production by selected yeasts and lactic acid bacteria in red wine. Food Chem. 2006, 94, 262–270. [Google Scholar] [CrossRef]
- De Felice, D.V.; Solfrizzo, M.; De Curtis, F.; Lima, G.; Visconti, A.; Castoria, R. Strains of Aureobasidium pullulans can lower ochratoxin a contamination in wine grapes. Phytopathology 2008, 98, 1261–1270. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Apaliya, M.T.; Mahunu, G.K.; Chen, L.; Li, W. Control of ochratoxin A-producing fungi in grape berry by microbial antagonists: A review. Trends Food Sci. Technol. 2016, 51, 88–97. [Google Scholar] [CrossRef]
- Belda, I.; Conchillo, L.B.; Ruiz, J.; Navascués, E.; Marquina, D.; Santos, A. Selection and use of pectinolytic yeasts for improving clarification and phenolic extraction in winemaking. Int. J. Food Microbiol. 2016, 223, 1–8. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Navascués, E.; Marquina, D.; Santos, A. Improvement of aromatic thiol release through the selection of yeasts with increased β-lyase activity. Int. J. Food Microbiol. 2016, 225, 1–8. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Alastruey-Izquierdo, A.; Navascués, E.; Marquina, D.; Santos, A. Unraveling the enzymatic basis of wine “Flavorome”: A phylo-functional study of wine related yeast species. Front. Microbiol. 2016, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.M.H.; Boss, P.K.; Walker, M.E.; Sumby, K.M.; Grbin, P.R.; Jiranek, V. Evaluation of indigenous non-Saccharomyces yeasts isolated from a South Australian vineyard for their potential as wine starter cultures. Int. J. Food Microbiol. 2020, 312, 108373. [Google Scholar] [CrossRef]
- Manzanares, P.; Rojas, V.; Genovés, S.; Vallés, S. A preliminary search for anthocyanin-β-D-glucosidase activity in non-Saccharomyces wine yeasts. Int. J. Food Sci. Technol. 2000, 35, 95–103. [Google Scholar] [CrossRef]
- Zhang, L.J.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef] [PubMed]
- Branco, P.; Viana, T.; Albergaria, H.; Arneborg, N. Antimicrobial peptides (AMPs) produced by Saccharomyces cerevisiae induce alterations in the intracellular pH, membrane permeability and culturability of Hanseniaspora guilliermondii cells. Int. J. Food Microbiol. 2015, 205, 112–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peña, R.; Ganga, M.A. Novel antimicrobial peptides produced by Candida intermedia LAMAP1790 active against the wine-spoilage yeast Brettanomyces bruxellensis. Antonie van Leeuwenhoek 2019, 112, 297–304. [Google Scholar]
- Albergaria, H.; Francisco, D.; Gori, K.; Arneborg, N.; Gírio, F. Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 2010, 86, 965–972. [Google Scholar] [CrossRef]
- Peña, R.; Chávez, R.; Rodríguez, A.; Ganga, M.A. A control alternative for the hidden enemy in the wine cellar. Fermentation 2019, 5, 25. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, M.J.; Breinig, F. The viral killer system in yeast: From molecular biology to application. FEMS Microbiol. Rev. 2002, 26, 257–276. [Google Scholar] [CrossRef]
- Mehlomakulu, N.N.; Setati, M.E.; Divol, B. Characterization of novel killer toxins secreted by wine-related non-Saccharomyces yeasts and their action on Brettanomyces spp. Int. J. Food Microbiol. 2014, 188, 83–91. [Google Scholar] [CrossRef]
- Villalba, M.L.; Susana Sáez, J.; del Monaco, S.; Lopes, C.A.; Sangorrín, M.P. TdKT, a new killer toxin produced by Torulaspora delbrueckii effective against wine spoilage yeasts. Int. J. Food Microbiol. 2016, 217, 94–100. [Google Scholar] [CrossRef]
- Comitini, F.; Ingeniis De, J.; Pepe, L.; Mannazzu, I.; Ciani, M. Pichia anomala and Kluyveromyces wickerhamii killer toxins as new tools against Dekkera/Brettanomyces spoilage yeasts. FEMS Microbiol. Lett. 2004, 238, 235–240. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.; San Mauro, M.; Bravo, E.; Marquina, D. PMKT2, a new killer toxin from Pichia membranifaciens, and its promising biotechnological properties for control of the spoilage yeast Brettanomyces bruxellensis. Microbiology 2009, 155, 624–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oro, L.; Ciani, M.; Bizzaro, D.; Comitini, F. Evaluation of damage induced by Kwkt and Pikt zymocins against Brettanomyces/Dekkera spoilage yeast, as compared to sulphur dioxide. J. Appl. Microbiol. 2016, 121, 207–214. [Google Scholar] [CrossRef] [PubMed]
- De Ingeniis, J.; Raffaelli, N.; Ciani, M.; Mannazzu, I. Pichia anomala DBVPG 3003 secretes a ubiquitin-like protein that has antimicrobial activity. Appl. Environ. Microbiol. 2009, 75, 1129–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández de Ullivarri, M.; Mendoza, L.M.; Raya, R.R. Characterization of the killer toxin KTCf20 from Wickerhamomyces anomalus, a potential biocontrol agent against wine spoilage yeasts. Biol. Control 2018, 121, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Lopes, C.A.; Sáez, J.S.; Sangorrín, M.P. Differential response of Pichia guilliermondii spoilage isolates to biological and physicochemical factors prevailing in Patagonian wine fermentations. Can. J. Microbiol. 2009, 55, 801–809. [Google Scholar] [CrossRef]
- Csutak, O.; Vassu, T.; Corbu, V.; Cîrpici, I.; Ionescu, R. Killer activity of Pichia anomala CMGB 88. Biointerface Res. Appl. Chem. 2017, 7, 2085–2089. [Google Scholar]
- Villalba, M.L.; Mazzucco, M.B.; Lopes, C.A.; Ganga, M.A.; Sangorrín, M.P. Purification and characterization of Saccharomyces eubayanus killer toxin: Biocontrol effectiveness against wine spoilage yeasts. Int. J. Food Microbiol. 2020, 331, 108714. [Google Scholar] [CrossRef]
- Türkel, S.; Ener, B. Isolation and characterization of new Metschnikowia pulcherrima strains as producers of the antimicrobial pigment pulcherrimin. Z. Naturforsch. C 2009, 64, 405–410. [Google Scholar]
- Oro, L.; Ciani, M.; Comitini, F. Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J. Appl. Microbiol. 2014, 116, 1209–1217. [Google Scholar] [CrossRef]
- Charpentier, C.; Escot, S.; Gonzalez, E.; Dulau, L.; Feuillat, M. The influence of yeast glycosylated proteins on tannins aggregation in model solution. J. Int. Sci. Vigne Vin 2004, 38, 209–218. [Google Scholar] [CrossRef]
- Buerth, C.; Heilmann, C.J.; Klis, F.M.; de Koster, C.G.; Ernst, J.F.; Tielker, D. Growth-dependent secretome of Candida utilis. Microbiology 2011, 157, 2493–2503. [Google Scholar] [CrossRef] [PubMed]

| Yeast | Protein and Monosaccharide Content in Mannoproteins [25] | Nitrogen Requirement | |||
|---|---|---|---|---|---|
| Protein (%) a | Mannose (%) b | Glucose (%) b | Galactose (%) b | ||
| Saccharomyces cerevisiae | 24 | 88 | 12 | - | |
| Metschnikowia pulcherrima | 25 | 86 | 14 | - | Slow ammonium uptake [4]. Weak or no growth in nitrate agar, and unable to develop in YPD agar at 37 °C [28]. |
| Wickerhamomyces anomalusc | 9 | 74 | 26 | - | Capable to uptake nitrate [36]. |
| Saccharomycodes ludwigiid | 12 | 93 | 7 | - | Unable to uptake nitrate [37]. Capable to uptake cadaverine and ethylamine [37]. |
| Schizosaccharomyces pombee | 11 | 55 | 22 | 23 | |
| Starmerella bombicolaf | 14 | 73 | 27 | - | |
| Pichia fermentans | 15 | 87 | 13 | - | |
| Hanseniaspora uvarumg | 23 | 81 | 19 | - | |
| Hanseniaspora valbyensis | 20 | 75 | 25 | - | |
| Lachancea thermotolerans | 16 | 82 | 18 | - | |
| Torulaspora delbrueckii | 18 | 85 | 15 | - | |
| Zygosaccharomyces bailii | 29 | 79 | 21 | - | |
| Brettanomyces bruxellensis | 16 | 88 | 12 | - | |
| Enzyme | Yeast | Application |
|---|---|---|
| β-glucosidase | Lachancea thermotolerans | Release of terpenes and thiols from their precursors: improvement of the aromatic profile [83,84,85]. |
| Torulaspora delbrueckii | Release of thiols from their cysteinylated precursors: improvement of the aromatic profile [84]. | |
| Wickerhamomyces anomalus |
| |
| Metschnikowia pulcherrima | ||
| Candida stellata | Release of terpenes (β-myrcene, limonene, linalool, α-terpineol, and farnesol) from their glycosylated precursors: improvement of the aromatic profile [88]. | |
| Hanseniaspora uvarum |
| |
| Saccharomycodes ludwigii | Activity up to 46% higher than S. cerevisiae at 30 °C [91]. | |
| Aureobasidium pullulans | Release of terpenes from their glycosylated precursors: improvement of the aromatic profile [92,93]. | |
| Protease | Wickerhamomyces anomalus | Aspartic protease WaAPr1 excreted by W. anomalus 227 [94]. |
| Metschnikowia pulcherrima | ||
| Candida stellata | ||
| Hanseniaspora uvarum | Degradation of proteins: improvement of clarification and stabilization [89,97]. | |
| Lachancea thermotolerans, Torulaspora delbrueckii, Zygosaccharomyces bailii, Pichia kluyveri | ||
| Glucanase | Wickerhamomyces anomalus | |
| Schizosaccharomyces pombe | High mannoproteins-releasing during AOL [21,38,99]. | |
| Saccharomycodes ludwigii | High mannoproteins-releasing during AOL [21,25]. | |
| Lachancea thermotolerans, Metschnikowia pulcherrima, Debaryomyces hansenii | Release of mannoproteins [25,83]. | |
| Hydroxycinnamate decarboxylase (HCDC) | Metschnikowia pulcherrima, Hanseniaspora guilliermondii, Hanseniaspora opuntiae, Hanseniaspora vineae, Hanseniaspora clermontiae, Pichia guillermondii | Involved in the synthesis of vinylphenolic pyranoanthocyanins: improvement of color stability in red wines [83,101,102,103]. |
| Urease | Schizosaccharomyces pombe | |
| Carboxypeptidase | Aureobasidium pullulans | |
| Pectinase | Wickerhamomyces anomalus | Degradation of pectins: improvement of clarification and turbidity reduction [85]. |
| Metschnikowia pulcherrima |
| |
| Candida stellata | Degradation of pectins: improvement of clarification and turbidity reduction [85,97]. | |
| Hanseniaspora uvarum | Degradation of pectins: improvement of clarification and turbidity reduction [89,97]. | |
| Aureobasidium pullulans |
| |
| Cellulase | Lachancea thermotolerans, Metschnikowia pulcherrima, Candida stellata, Hanseniaspora uvarum, Aureobasidium pullulans, Debaryomyces hansenii |
|
| Xylanase | Lachancea thermotolerans, Candida stellata, Hanseniaspora uvarum, Aureobasidium pullulans | Degradation of hemicellulose: improvement of wine aroma by increasing of monoterpenyl diglycoside precursors in the grape-must [83,92,97]. |
| β-lyase | Torulaspora delbrueckii, Kluyveromyces marxianus, Meyerozyma guilliermondii (formerly Pichia guilliermondii) | Release of thiols from their cysteinylated precursors: improvement of the aromatic profile [109,110]. |
| Lipase | Lachancea thermotolerans | Increase on free fatty acids concentration [83]. |
| Aureobasidium pullulans | Improvement of wine aroma: synthesis of ethyl esters and ethyl acetates from lipid cleavage [111]. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vejarano, R. Non-Saccharomyces in Winemaking: Source of Mannoproteins, Nitrogen, Enzymes, and Antimicrobial Compounds. Fermentation 2020, 6, 76. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation6030076
Vejarano R. Non-Saccharomyces in Winemaking: Source of Mannoproteins, Nitrogen, Enzymes, and Antimicrobial Compounds. Fermentation. 2020; 6(3):76. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation6030076
Chicago/Turabian StyleVejarano, Ricardo. 2020. "Non-Saccharomyces in Winemaking: Source of Mannoproteins, Nitrogen, Enzymes, and Antimicrobial Compounds" Fermentation 6, no. 3: 76. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation6030076