Ectopic Expression of AhGLK1b (GOLDEN2-like Transcription Factor) in Arabidopsis Confers Dual Resistance to Fungal and Bacterial Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Microarray Analysis
2.3. Full-Length cDNA Cloning
2.4. Sequence and Phylogenetic Analysis
2.5. Subcellular Localization
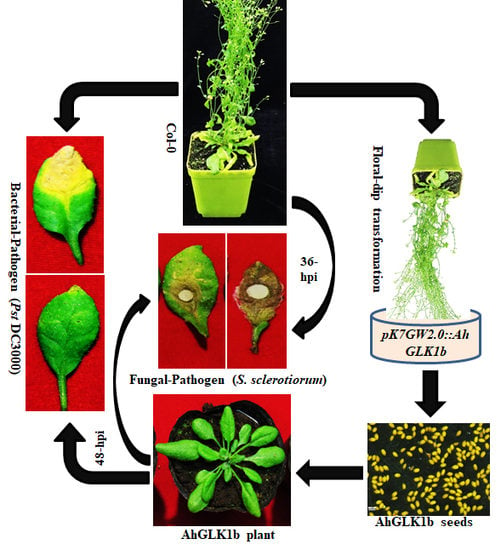
2.6. Development of Arabidopsis Transgenic Lines
2.7. PCR and Semi-Quantitative Real-Time PCR (sqRT-PCR) Genotyping for Transgene Confirmation
2.8. S. sclerotiorum Bioassays
2.9. Pst DC3000 Bioassays in Transgenic Arabidopsis
2.10. Quantitative Real-Time PCR (qRT-PCR)
2.11. Statistical Analysis
3. Results
3.1. Cloning and Sequence Analysis of AhGLK1b
3.2. Phylogenetic Analysis
3.3. Localization of AhGLK1b in the Nucleus
3.4. Expression Pattern of AhGLK1b in Different Peanut Tissues
3.5. AhGLK1b Response to Biotic and Abiotic Stresses
3.6. Overexpression of AhGLK1b in Arabidopsis Enhances Resistance to Fungal Pathogen S. sclerotiorum
3.7. AhGLK1b Confers Tolerance to Bacterial Pathogen Pst DC3000
3.8. Defense-Related Genes Response to Overexpression of AhGLK1b
4. Discussion
4.1. AhGLK1bAhGLK1b Conferes Resistance to Biotic and Abiotic Stresses
4.2. Defense-Related Gene Responded to AhGLK1b in Arabidopsis
4.3. AhGLK1b Enhace Resistance to Fungal Pathogen S. sclerotiorum
4.4. AhGLK1b Confer Resistance to Bacterial Pathogen Pst DC3000
4.5. AhGLK1b May be Involved in Embryo Development
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

Appendix B

Appendix C

Appendix D
| S. No. | Primer | F/R | Primer sequences (5′to 3′) |
|---|---|---|---|
| Cloning (ORF) | |||
| 1 | AhGLK-20269 | F | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGCTTGCGGTGTCACCTTTG |
| R | GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAATTAAGCACAGGAGTTG | ||
| 2 | AhGLK-20269-GFP | F | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGCTTGCGGTGTCACCTTTG |
| R | GGGGACCACTTTGTACAAGAAAGCTGGGTCATTAAGCACAGGAGTTGAG | ||
| Transgenes ID (P35s) | |||
| 3 | AhGLK-trans ID-35s | F | TGATGTGATATCTCCACTGACGTAAG |
| R | GATGGATTCACTACGATGGATCCATC | ||
| Semi-quantitative RT-PCR | |||
| 4 | qRT-AhGLK | F | TCCCTAGTCTTGAATGGTTG |
| R | TGATTGACATGATGAGGAGG | ||
| 5 | AtUBC21 (AT5G25760)1 | F | GGCATCAAGAGCGCGACTGT |
| R | GGCGAGGCGTGTATACATTT | ||
| 6 | At1g09500 Cinnamyl alcohol dehydrogenase | F | CAACCACATGCACACTAATTC |
| R | CCATTGGCTGAAGGAGTCTCG | ||
| 7 | At1g35310 PR10 | F | CAACATGCCTCCAAAGCCACTC |
| R | ACGCATACAATAACTCTCCCACAC | ||
| 8 | At1g73260 Trypsin protease inhibitor | F | CTATCAAGCCGCCTCACCTA |
| R | CTCACCGACCCGCCAGTA | ||
| 9 | At1g77960 Phox/Bem 1p domain protein | F | ATCGCCGTTGTTGAGTCCTTTATG |
| R | AGTATTTGTGCCCCTTTGTTCAGC | ||
| 10 | At1g80160 Lactoylglutathione lyase | F | GATGGCGCGTGGTTGTTT |
| R | GGGGAGGCTATCGCAGTTG | ||
| 11 | At3g28740 Cytochrome P450 | F | CGGCCGCGTAAACTAAACCTA |
| R | TCCCGGCAAGTATCATAACAAGTA | ||
| 12 | At4g37990 ELI3-2 | F | CGCCGCACCGCTCCTCTG |
| R | GCGGTACCTAACGTCGGCTTTCTC | ||
| 13 | At1g18870 Isochorismate synthase | F | AGTGCCCCAGGTCGAGTTTGATG |
| R | TCCCGCCTTTCTTTAGTTGATTGG | ||
| 14 | At4g21910 MATE efflux protein | F | GTTGGCACTACCGGCGATACTTA |
| R | ACACCCAACCGGAATACCAACAAC | ||
| 15 | At2g14610 PR-1 | F | CTCAAGATAGCCCACAAGA |
| R | AAGGCCACATATTTTACAT | ||
| Quantitative RT-PCR | |||
| 16 | qRT-AhGLK | F | TCCCTAGTCTTGAATGGTTG |
| R | TGATTGACATGATGAGGAGG | ||
| 17 | AtUBC21 (AT5G25760)2 | F | CTGAGCCGGACAGTCCTCTT |
| R | TAGCGGCGAGGCGTGTATAC |
References
- Pandey, M.K.; Monyo, E.; Ozias-Akins, P.; Liang, X.; Guimarães, P.; Nigam, S.N.; Upadhyaya, H.D.; Janila, P.; Zhang, X.; Guo, B.; et al. Advances in Arachis genomics for peanut improvement. Biotechnol. Adv. 2012, 30, 639–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oerke, E.-C. Crop losses to pests. J. Agric. Sci. 2005, 144, 31–43. [Google Scholar] [CrossRef]
- Gimenez-Ibanez, S.; Solano, R. Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front. Plant Sci. 2013, 4, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazan, K.; Lyons, R. Intervention of Phytohormone Pathways by Pathogen Effectors. Plant Cell 2014, 26, 2285–2309. [Google Scholar] [CrossRef] [Green Version]
- Boland, G.; Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Bolton, M.D.; Thomma, B.P.H.J.; Nelson, B.D. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 2006, 7, 1–16. [Google Scholar] [CrossRef]
- Katagiri, F.; Thilmony, R.; He, S.Y. The Arabidopsis Thaliana-Pseudomonas Syringae Interaction. Arab. Book 2002, 1, e0039. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Huang, L.; Dai, Y.; Liu, S.; Hong, Y.; Tian, L.; Huang, L.; Cao, Z.; Li, D.; Song, F. Arabidopsis AtERF15 positively regulates immunity against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; -Z., C.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Li, G.; Chen, D.; Tang, X.; Liu, Y.S. Heterologous expression of kiwifruit (Actinidia chinensis) GOLDEN2-LIKE homolog elevates chloroplast level and nutritional quality in tomato (Solanum lycopersicum). Planta 2018, 247, 1351–1362. [Google Scholar] [CrossRef]
- Hall, L.N.; Rossini, L.; Cribb, L.; Langdale, J.A. GOLDEN 2: A novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 1998, 10, 925–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossini, L.; Cribb, L.; Martin, D.J.; Langdale, J.A. The maize golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 2001, 13, 1231–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosoda, K.; Imamura, A.; Katoh, E.; Hatta, T.; Tachiki, M.; Yamada, H.; Mizuno, T.; Yamazaki, T. Molecular Structure of the GARP Family of Plant Myb-Related DNA Binding Motifs of the Arabidopsis Response Regulators. Plant Cell 2002, 14, 2015–2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitter, D.W.; Martin, D.J.; Copley, M.J.; Scotland, R.W.; Langdale, J.A. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002, 31, 713–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasumura, Y.; Moylan, E.C.; Langdale, J.A. A Conserved Transcription Factor Mediates Nuclear Control of Organelle Biogenesis in Anciently Diverged Land Plants. Plant Cell 2005, 17, 1894–1907. [Google Scholar] [CrossRef] [Green Version]
- Bravo-Garcia, A.; Yasumura, Y.; Langdale, J.A. Specialization of the Golden2-like regulatory pathway during land plant evolution. New Phytol. 2009, 183, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Muramatsu, M.; Hakata, M.; Ueno, O.; Nagamura, Y.; Hirochika, H.; Takano, M.; Ichikawa, H. Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant Cell Physiol. 2009, 50, 1933–1949. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Fouracre, J.P.; Kelly, S.; Karki, S.; Gowik, U.; Aubry, S.; Shaw, M.K.; Westhoff, P.; Slamet-Loedin, I.H.; Quick, W.P.; et al. Evolution of GOLDEN2-LIKE gene function in C3 and C4 plants. Planta 2012, 237, 481–495. [Google Scholar] [CrossRef] [Green Version]
- Brand, A.; Borovsky, Y.; Hill, T.; Rahman, K.A.A.; Bellalou, A.; Van Deynze, A.; Paran, I. CaGLK2 regulates natural variation of chlorophyll content and fruit color in pepper fruit. Theor. Appl. Genet. 2014, 127, 2139–2148. [Google Scholar] [CrossRef]
- Nguyen, C.V.; Vrebalov, J.T.; Gapper, N.E.; Zheng, Y.; Zhong, S.; Fei, Z.; Giovannoni, J.J. Tomato GOLDEN2-LIKE Transcription Factors Reveal Molecular Gradients That Function during Fruit Development and Ripening. Plant Cell 2014, 26, 585–601. [Google Scholar] [CrossRef] [Green Version]
- Waters, M.; Moylan, E.C.; Langdale, J.A. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 2008, 56, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.; Wang, P.; Korkaric, M.; Capper, R.G.; Saunders, N.; Langdale, J.A. GLK Transcription Factors Coordinate Expression of the Photosynthetic Apparatus in Arabidopsis. Plant Cell 2009, 21, 1109–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tokumaru, M.; Adachi, F.; Toda, M.; Ito-Inaba, Y.; Yazu, F.; Hirosawa, Y.; Sakakibara, Y.; Suiko, M.; Kakizaki, T.; Inaba, T. Ubiquitin-Proteasome Dependent Regulation of the GOLDEN2-LIKE 1 Transcription Factor in Response to Plastid Signals. Plant Physiol. 2016, 173, 524–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, A.L.T.; Nguyen, C.V.; Hill, T.; Cheng, K.L.; Figueroa-Balderas, R.; Aktas, H.; Ashrafi, H.; Pons, C.; Fernandez-Munoz, R.; Vicente, A.R.; et al. Uniform ripening Encodes a Golden 2-like Transcription Factor Regulating Tomato Fruit Chloroplast Development. Science 2012, 336, 1711–1715. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Bradley, G.; Pyke, K.; Ball, G.; Lu, C.; Fray, R.; Marshall, A.; Jayasuta, S.; Baxter, C.; Van Wijk, R.; et al. Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 2013, 161, 1476–1485. [Google Scholar] [CrossRef] [Green Version]
- Savitch, L.V.; Allard, G.; Seki, M.; Robert, L.S.; Tinker, N.A.; Hüner, N.P.; Shinozaki, K.; Singh, J. The Effect of Overexpression of Two Brassica CBF/DREB1-like Transcription Factors on Photosynthetic Capacity and Freezing Tolerance in Brassica napus. Plant Cell Physiol. 2005, 46, 1525–1539. [Google Scholar] [CrossRef]
- Murmu, J.; Wilton, M.; Allard, G.; Pandeya, R.; Desveaux, D.; Singh, J.; Subramaniam, R. Arabidopsis GOLDEN2-LIKE (GLK) transcription factors activate jasmonic acid (JA)-dependent disease susceptibility to the biotrophic pathogen Hyaloperonospora arabidopsidis, as well as JA-independent plant immunity against the necrotrophic pathogen Botrytis cinerea. Mol. Plant Pathol. 2013, 15, 174–184. [Google Scholar]
- Townsend, P.D.; Dixon, C.H.; Slootweg, E.J.; Sukarta, O.C.A.; Yang, A.W.H.; Hughes, T.R.; Sharples, G.; Palsson, L.-O.; Takken, F.L.W.; Goverse, A.; et al. The intracellular immune receptor Rx1 regulates the DNA-binding activity of a Golden2-like transcription factor. J. Boil. Chem. 2017, 293, 3218–3233. [Google Scholar] [CrossRef] [Green Version]
- Han, X.-Y.; Li, P.-X.; Zou, L.-J.; Tan, W.-R.; Zheng, T.; Zhang, D.-W.; Lin, H. GOLDEN2-LIKE transcription factors coordinate the tolerance to Cucumber mosaic virus in Arabidopsis. Biochem. Biophys. Res. Commun. 2016, 477, 626–632. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, C.; Chen, H.; Yuan, M.; Nipper, R.; Prakash, C.S.; Zhuang, R.-R.; He, G. QTL mapping for bacterial wilt resistance in peanut (Arachis hypogaea L.). Mol. Breed. 2016, 36, 13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Lin, J.; Zhuang, W. Study on molecular basis of resistance to Ralstonia solanacearum in peanut. Master’s Thesis, Fujian Agriculture and forestry university, Fujian, China, 2010; pp. 22–28. [Google Scholar]
- Chen, H.; Zhang, C.; Cai, T.c.; Deng, Y.; Zhou, S.; Zheng, Y.; Ma, S.; Tang, R.; Varshney, R.K.; Zhuang, W. Identification of low Ca2+ stress-induced embryo apoptosis response genes in Arachis hypogaea by SSH-associated library lift (SSHaLL). Plant Biotechnol. J. 2016, 14, 682–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, K.R.R.; Kirti, P.B. Differential gene expression in Arachis diogoi upon interaction with peanut late leaf spot pathogen, Phaeoisariopsis personata and characterization of a pathogen induced cyclophilin. Plant Mol. Boil. 2011, 75, 497–513. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.A.; Ali, N.; Gandeka, M.; Liu, Q.; Zhuang, R.; Chen, H. Extraction of purified plants gDNA free of Secondary metabolites. Int. J. Biosci. 2017, 10, 223–231. [Google Scholar]
- Pieterse, C.; Van Wees, S.; Hoffland, E.; Van Pelt, J.A.; Van Loon, L.C. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 1996, 8, 1225–1237. [Google Scholar]
- Fan, J.; Crooks, C.; Lamb, C. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 2007, 53, 393–399. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C T method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef]
- Misra, R.C.; Sandeep; Kamthan, M.; Kumar, S.; Ghosh, S. A thaumatin-like protein of Ocimum basilicum confers tolerance to fungal pathogen and abiotic stress in transgenic Arabidopsis. Sci. Rep. 2016, 6, 25340. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.; Scheible, W.-R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis1. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Savitch, L.V.; Subramaniam, R.; Allard, G.C.; Singh, J. The GLK1 ‘regulon’ encodes disease defense related proteins and confers resistance to Fusarium graminearum in Arabidopsis. Biochem. Biophys. Res. Commun. 2007, 359, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Lebeau, A.; Daunay, M.-C.; Frary, A.; Palloix, A.; Wang, J.-F.; Dintinger, J.; Chiroleu, F.; Wicker, E.; Prior, P. Bacterial Wilt Resistance in Tomato, Pepper, and Eggplant: Genetic Resources Respond to Diverse Strains in theRalstonia solanacearumSpecies Complex. Phytopathol. 2011, 101, 154–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, V.U.; Gopal, J.; Singh, B.P. Improvement for Bacterial Wilt Resistance in Potato By Conventional and Biotechnological Approaches. Agric. Res. 2012, 1, 299–316. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Li, L.; Li, M.; Su, L.; Lian, S.; Zhang, B.; Li, X.; Ge, K.; Li, L. AhGLK1 affects chlorophyll biosynthesis and photosynthesis in peanut leaves during recovery from drought. Sci. Rep. 2018, 8, 2250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagatoshi, Y.; Mitsuda, N.; Hayashi, M.; Inoue, S.-I.; Okuma, E.; Kubo, A.; Murata, Y.; Seo, M.; Saji, H.; Kinoshita, T.; et al. GOLDEN 2-LIKE transcription factors for chloroplast development affect ozone tolerance through the regulation of stomatal movement. Proc. Natl. Acad. Sci. USA 2016, 113, 4218–4223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabu, G.; Prasad, D.T. Functional characterization of sugarcane MYB transcription factor gene promoter (PScMYBAS1) in response to abiotic stresses and hormones. Plant Cell Rep. 2011, 31, 661–669. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, C.-M. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010, 186, 471–483. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate Starvation Responses and Gibberellic Acid Biosynthesis Are Regulated by the MYB62 Transcription Factor in Arabidopsis. Mol. Plant 2008, 2, 43–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mengiste, T.; Chen, X.; Salmeron, J.; Dietrich, R. The BOTRYTIS SUSCEPTIBLE1 Gene Encodes an R2R3MYB Transcription Factor Protein That Is Required for Biotic and Abiotic Stress Responses in Arabidopsis. Plant Cell 2003, 15, 2551–2565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, M.H.; Paulsen, I.T.; Skurray, R.A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 1999, 31, 394–395. [Google Scholar] [CrossRef]
- Nawrath, C.; Heck, S.; Parinthawong, N.; Métraux, J.-P. EDS5, an Essential Component of Salicylic Acid–Dependent Signaling for Disease Resistance in Arabidopsis, Is a Member of the MATE Transporter Family. Plant Cell 2002, 14, 275–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Reduced virulence of Gibberella zeae caused by disruption of a trichthecine toxin biosynthetic gene. Mol. Plant-Microbe Interact. 1995, 8, 593–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, L.; Gleddie, S.C. A modified Rpl3 gene from rice confers tolerance of the Fusarium graminearum mycotoxin deoxynivalenol to transgenic tobacco. Physiol. Mol. Plant Pathol. 2001, 58, 173–181. [Google Scholar] [CrossRef]
- Moiseyev, G.; Beintema, J.; Fedoreyeva, L.; Yakovlev, G. High sequence similarity between a ribonuclease from ginseng calluses and fungus-elicited proteins from parsley indicates that intracellular pathogenesis-related proteins are ribonucleases. Planta 1994, 193, 470–472. [Google Scholar] [CrossRef]
- Despres, C.; Subramaniam, R.; Matton, D.P.; Brisson, N. The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1. Plant Cell 1995, 7, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Marković-Housley, Z.; Degano, M.; Lamba, D.; Von Roepenack-Lahaye, E.; Clemens, S.; Susani, M.; Ferreira, F.; Scheiner, O.; Breiteneder, H. Crystal Structure of a Hypoallergenic Isoform of the Major Birch Pollen Allergen Bet v 1 and its Likely Biological Function as a Plant Steroid Carrier. J. Mol. Boil. 2003, 325, 123–133. [Google Scholar] [CrossRef]
- Park, C.-J.; Kim, K.-J.; Shin, R.; Park, J.M.; Shin, Y.-C.; Paek, K.-H. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 2004, 37, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Simon-Plas, F.; Elmayan, T.; Blein, J.-P. The plasma membrane oxidase NtrbohD is responsible for AOS production in elicited tobacco cells. Plant J. 2002, 31, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Narusaka, M.; Park, P.; Kubo, Y.; Hirayama, T.; Seki, M.; Shiraishi, T.; Ishida, J.; Nakashima, M.; Enju, A.; et al. RCH1, a Locus inArabidopsisThat Confers Resistance to the Hemibiotrophic Fungal PathogenColletotrichum higginsianum. Mol. Plant-Microbe Interactions 2004, 17, 749–762. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.A.; Jones, A.; Dangl, J.L. Pathogen-induced, NADPH oxidase–derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005, 37, 1130–1134. [Google Scholar] [CrossRef]
- Mitchell, H.J.; Hall, J.L.; Barber, M.S. Elicitor-Induced Cinnamyl Alcohol Dehydrogenase Activity in Lignifying Wheat (Triticum aestivum L.) Leaves. Plant Physiol. 1994, 104, 551–556. [Google Scholar] [CrossRef] [Green Version]
- Somssich, I.; Wernert, P.; Kiedrowski, S.; Hahlbrock, K. Arabidopsis thaliana defense-related protein ELI3 is an aromatic alcohol:NADP+ oxidoreductase. Proc. Natl. Acad. Sci. USA 1996, 93, 14199–14203. [Google Scholar] [CrossRef] [Green Version]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, K.J.; Nasmith, C.G.; Allard, G.; Singh, J.; Subramaniam, R.; Desveaux, D. Found in Translation: High-Throughput Chemical Screening in Arabidopsis thaliana Identifies Small Molecules That Reduce Fusarium Head Blight Disease in Wheat. Mol. Plant-Microbe Interactions 2011, 24, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Meyers, B.C.; Kozik, A.; West, M.A.L.; Morgante, M.; Clair, D.A.S.; Bent, A.; Michelmore, R. Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Boil. 2007, 7, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guyon, K.; Balagué, C.; Roby, D.; Raffaele, S. Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genom. 2014, 15, 336. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Seo, J.-K.; Gao, S.; Cui, X.; Jin, H. Silencing of AtRAP, a target gene of a bacteria-induced small RNA, triggers antibacterial defense responses through activation of LSU2 and down-regulation of GLK1. New Phytol. 2017, 215, 1144–1155. [Google Scholar] [CrossRef] [Green Version]
- Bennett, A.B.; Powell, A.L.; Hill, T.A.; Cheng, K.L.; Figueroa-Balderas, R.E. GLK genes for improved fruit quality. U.S. Patent No. 9,549,509, 24 January 2017. [Google Scholar]
- Liebsch, D.; Keech, O. Dark-induced leaf senescence: New insights into a complex light-dependent regulatory pathway. New Phytol. 2016, 212, 563–570. [Google Scholar] [CrossRef]
- Kobayashi, K.; Fujii, S.; Sasaki, D.; Baba, S.; Ohta, H.; Masuda, T.; Wada, H. Transcriptional regulation of thylakoid galactolipid biosynthesis coordinated with chlorophyll biosynthesis during the development of chloroplasts in Arabidopsis. Front. Plant Sci. 2014, 5, 272. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Pan, Y.; Qu, C.; Su, C.; Li, J.; Zhang, X. Identification and cloning of GOLDEN2-LIKE1 (GLK1), a transcription factor associated with chloroplast development in Brassica napus L. Genet. Mol. Res. 2017, 16, 16. [Google Scholar] [CrossRef]
- Chen, M.; Liu, X.; Jiang, S.; Wen, B.; Yang, C.; Xiao, W.; Fu, X.; Li, N.; Chen, X.; Gao, D.; et al. Transcriptomic and Functional Analyses Reveal That PpGLK1 Regulates Chloroplast Development in Peach (Prunus persica). Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, N.; Chen, H.; Zhang, C.; Khan, S.A.; Gandeka, M.; Xie, D.; Zhuang, W. Ectopic Expression of AhGLK1b (GOLDEN2-like Transcription Factor) in Arabidopsis Confers Dual Resistance to Fungal and Bacterial Pathogens. Genes 2020, 11, 343. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11030343
Ali N, Chen H, Zhang C, Khan SA, Gandeka M, Xie D, Zhuang W. Ectopic Expression of AhGLK1b (GOLDEN2-like Transcription Factor) in Arabidopsis Confers Dual Resistance to Fungal and Bacterial Pathogens. Genes. 2020; 11(3):343. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11030343
Chicago/Turabian StyleAli, Niaz, Hua Chen, Chong Zhang, Shahid Ali Khan, Mamadou Gandeka, Dongyang Xie, and Weijian Zhuang. 2020. "Ectopic Expression of AhGLK1b (GOLDEN2-like Transcription Factor) in Arabidopsis Confers Dual Resistance to Fungal and Bacterial Pathogens" Genes 11, no. 3: 343. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11030343