Genetic Variants in pre-miR-146a, pre-miR-499, pre-miR-125a, pre-miR-605, and pri-miR-182 Are Associated with Breast Cancer Susceptibility in a South American Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Families
2.2. Control Population
2.3. Mutation Analysis
2.4. Statistical Analysis
3. Results
3.1. Association Study between rs2910164, rs4541843, rs3746444, rs12975333, and rs2043556 with Familial Breast Cancer and Early-Onset Non-Familial Breast Cancer in Non-Carriers of BRCA1/2 Mutations
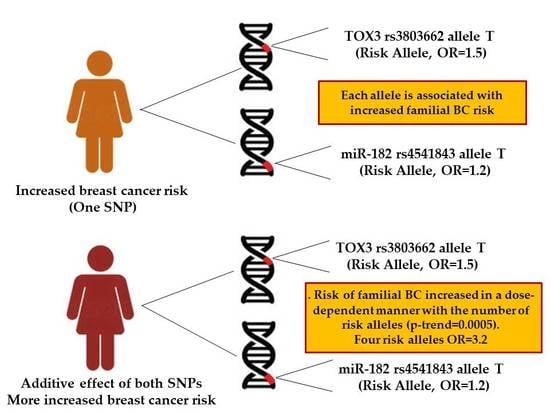
3.2. Combined Effect between TOX3 rs3803662-T and pri-miR-182 rs4541843-T Alleles with Breast Cancer Risk
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Approval and Informed Consent
References
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer statistics, 2009. CA Cancer J. Clin. 2009, 59, 225–249. [Google Scholar] [CrossRef] [PubMed]
- De Salud, M. Guía Clínica AUGE Cáncer de Mama; Manejo Integral del Cáncer y otros Tumores, Ed.; Ministerio de Salud—Gobierno de Chile: Santiago, Chile, 2015. [Google Scholar]
- Stratton, M.R.; Rahman, N. The emerging landscape of breast cancer susceptibility. Nat. Genet. 2008, 40, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Couch, F.J.; Hart, S.N.; Sharma, P.; Toland, A.E.; Wang, X.; Miron, P.; Olson, J.E.; Godwin, A.K.; Pankratz, V.S.; Olswold, C.; et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015, 33, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br. J. Cancer 2000, 83, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.R.; Bilgili, E.P.; Merner, N.D. A review of whole-exome sequencing efforts toward hereditary breast cancer susceptibility gene discovery. Hum. Mutat. 2016, 37, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Leyton, Y.; Gonzalez-Hormazabal, P.; Blanco, R.; Bravo, T.; Fernandez-Ramires, R.; Morales, S.; Landeros, N.; Reyes, J.M.; Peralta, O.; Tapia, J.C.; et al. Association of PALB2 sequence variants with the risk of familial and early-onset breast cancer in a South-American population. BMC Cancer 2015, 15, 30. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hormazabal, P.; Reyes, J.M.; Blanco, R.; Bravo, T.; Carrera, I.; Peralta, O.; Gomez, F.; Waugh, E.; Margarit, S.; Ibañez, G.; et al. The BARD1 Cys557Ser variant and risk of familial breast cancer in a South-American population. Mol. Biol. Rep. 2012, 39, 8091–8098. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hormazabal, P.; Bravo, T.; Blanco, R.; Valenzuela, C.Y.; Gomez, F.; Wauhg, E.; Peralta, O.; Ortuzar, W.; Reyes, J.M.; Jara, L. Association of common ATM variants with familial breast cancer in a South American population. BMC Cancer 2008, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hormazabal, P.; Castro, V.G.; Blanco, R.; Gomez, F.; Peralta, O.; Waugh, E.; Bravo, T.; Reyes, J.M.; Jara, L. Absence of CHEK2 1100delC mutation in familial breast cancer cases from a South American population. Breast Cancer Res. Treat. 2008, 110, 543–545. [Google Scholar] [CrossRef] [PubMed]
- Jara, L.; Gonzalez-Hormazabal, P.; Cerceno, K.; Di Capua, G.A.; Reyes, J.M.; Blanco, R.; Bravo, T.; Peralta, O.; Gomez, F.; Waugh, E.; et al. Genetic variants in FGFR2 and MAP3K1 are associated with the risk of familial and early-onset breast cancer in a South-American population. Breast Cancer Res. Treat. 2013, 137, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Elematore, I.; Gonzalez-Hormazabal, P.; Reyes, J.M.; Blanco, R.; Bravo, T.; Peralta, O.; Gomez, F.; Waugh, E.; Margarit, S.; Ibanez, G.; et al. Association of genetic variants at TOX3, 2q35 and 8q24 with the risk of familial and early-onset breast cancer in a South-American population. Mol. Biol. Rep. 2014, 41, 3715–3722. [Google Scholar] [CrossRef] [PubMed]
- Mehrgou, A.; Akouchekian, M. The importance of BRCA1 and BRCA2 genes mutations in breast cancer development. Med. J. Islam. Repub. Iran 2016, 30, 369. [Google Scholar] [PubMed]
- Rothe, F.; Ignatiadis, M.; Chaboteaux, C.; Haibe-Kains, B.; Kheddoumi, N.; Majjaj, S.; Badran, B.; Fayyad-Kazan, H.; Desmedt, C.; Harris, A.L.; et al. Global microRNA expression profiling identifies miR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS ONE 2011, 6, e20980. [Google Scholar] [CrossRef] [PubMed]
- Song, F.J.; Chen, K.X. Single-nucleotide polymorphisms among microRNA: Big effects on cancer. Chin. J. Cancer 2011, 30, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. microRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Erson, A.E.; Petty, E.M. microRNAs in development and disease. Clin. Genet. 2008, 74, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. microRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- O’Day, E.; Lal, A. microRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010, 12, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.H.; Wang, Q.B.; Zhang, B. Ethnicity modifies the association between functional microRNA polymorphisms and breast cancer risk: A HuGE meta-analysis. Tumour Biol. 2014, 35, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Catucci, I.; Yang, R.; Verderio, P.; Pizzamiglio, S.; Heesen, L.; Hemminki, K.; Sutter, C.; Wappenschmidt, B.; Dick, M.; Arnold, N.; et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum. Mutat. 2010, 31, E1052–E1057. [Google Scholar] [CrossRef] [PubMed]
- Pastrello, C.; Polesel, J.; Della Puppa, L.; Viel, A.; Maestro, R. Association between hsa-miR-146a genotype and tumor age-of-onset in BRCA1/BRCA2-negative familial breast and ovarian cancer patients. Carcinogenesis 2010, 31, 2124–2126. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Schlehe, B.; Hemminki, K.; Sutter, C.; Bugert, P.; Wappenschmidt, B.; Volkmann, J.; Varon, R.; Weber, B.H.; et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res. Treat. 2010, 121, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.I.; Cox, D.G.; Barjhoux, L.; Verny-Pierre, C.; Barnes, D.; Antoniou, A.C.; Stoppa-Lyonnet, D.; Sinilnikova, O.M.; Mazoyer, S. The rs2910164:G>C SNP in the MIR146A gene is not associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Hum. Mutat. 2011, 32, 1004–1007. [Google Scholar] [CrossRef] [PubMed]
- Catucci, I.; Verderio, P.; Pizzamiglio, S.; Bernard, L.; Dall’olio, V.; Sardella, D.; Ravagnani, F.; Galastri, L.; Barile, M.; Peissel, B.; et al. The SNP rs895819 in miR-27a is not associated with familial breast cancer risk in Italians. Breast Cancer Res. Treat. 2012, 133, 805–807. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.; Wang, L.; Zhang, J. Increased risk of breast cancer associated with CC genotype of Has-miR-146a Rs2910164 polymorphism in Europeans. PLoS ONE 2012, 7, e31615. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Liang, J.; Wang, Z.; Tian, T.; Zhou, X.; Chen, J.; Miao, R.; Wang, Y.; Wang, X.; Shen, H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 2009, 30, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jin, M.; Yu, Y.; Zhang, S.; Wu, Y.; Liu, H.; Chen, B.; Li, Q.; Ma, X.; Chen, K. Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur. J. Cancer Care 2012, 21, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Alshatwi, A.A.; Shafi, G.; Hasan, T.N.; Syed, N.A.; Al-Hazzani, A.A.; Alsaif, M.A.; Alsaif, A.A. Differential expression profile and genetic variants of microRNAs sequences in breast cancer patients. PLoS ONE 2012, 7, e30049. [Google Scholar] [CrossRef] [PubMed]
- Kontorovich, T.; Levy, A.; Korostishevsky, M.; Nir, U.; Friedman, E. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int. J. Cancer 2010, 127, 589–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabirizadeh, S.; Azadeh, M.; Mirhosseini, M.; Ghaedi, K.; Tanha, H.M. The SNP rs3746444 within miR-499a is associated with breast cancer risk in Iranian population. J. Cell. Immnother. 2016, 2, 95–97. [Google Scholar] [CrossRef]
- Linhares, J.J.; Azevedo, M., Jr.; Siufi, A.A.; de Carvalho, C.V.; Wolgien Mdel, C.; Noronha, E.C.; Bonetti, T.C.; da Silva, I.D. Evaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancer. BMC Med. Genet. 2012, 13, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales, S.; Gulppi, F.; Gonzalez-Hormazabal, P.; Fernandez-Ramires, R.; Bravo, T.; Reyes, J.M.; Gomez, F.; Waugh, E.; Jara, L. Association of single nucleotide polymorphisms in pre-miR-27a, pre-miR-196a2, pre-miR-423, miR-608 and pre-miR-618 with breast cancer susceptibility in a South American population. BMC Genet. 2016, 17, 109. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.Y.; Gao, Z.H.; Jiang, Z.H.; Li, X.X.; Jiang, B.F.; Xie, S.Y. The associations of single nucleotide polymorphisms in miR-146a, miR-196a and miR-499 with breast cancer susceptibility. PLoS ONE 2013, 8, e70656. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Duan, R.; Kooy, F.; Sherman, S.L.; Zhou, W.; Jin, P. Germline mutation of microRNA-125a is associated with breast cancer. J. Med. Genet. 2009, 46, 358–360. [Google Scholar] [CrossRef] [PubMed]
- Peterlongo, P.; Caleca, L.; Cattaneo, E.; Ravagnani, F.; Bianchi, T.; Galastri, L.; Bernard, L.; Ficarazzi, F.; Dall'olio, V.; Marme, F.; et al. The rs12975333 variant in the miR-125a and breast cancer risk in Germany, Italy, Australia and Spain. J. Med. Genet. 2011, 48, 703–704. [Google Scholar] [CrossRef] [PubMed]
- Quann, K.; Jing, Y.; Rigoutsos, I. Post-transcriptional regulation of BRCA1 through its coding sequence by the miR-15/107 group of miRNAs. Front. Genet. 2015, 6, 242. [Google Scholar] [CrossRef] [PubMed]
- Moskwa, P.; Buffa, F.M.; Pan, Y.; Panchakshari, R.; Gottipati, P.; Muschel, R.J.; Beech, J.; Kulshrestha, R.; Abdelmohsen, K.; Weinstock, D.M.; et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 2011, 41, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Georges, S.A.; Biery, M.C.; Kim, S.Y.; Schelter, J.M.; Guo, J.; Chang, A.N.; Jackson, A.L.; Carleton, M.O.; Linsley, P.S.; Cleary, M.A.; et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008, 68, 10105–10112. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Ambrosone, C.B.; DiCioccio, R.A.; Odunsi, K.; Lele, S.B.; Zhao, H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis 2008, 29, 1963–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, K.; Wu, Z.Z.; Yu, J.P.; Guo, W.; Wu, N.; Wei, L.J.; Zhang, H.; Zhao, J.; Liu, J.T. Meta-analysis of the association between three microRNA polymorphisms and breast cancer susceptibility. Oncotarget 2017, 8, 68809–68824. [Google Scholar] [CrossRef] [PubMed]
- Akkiz, H.; Bayram, S.; Bekar, A.; Akgollu, E.; Uskudar, O.; Sandikci, M. No association of pre-microRNA-146a rs2910164 polymorphism and risk of hepatocellular carcinoma development in Turkish population: A case-control study. Gene 2011, 486, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.P.; Zhang, T.; Peng, B.; Yu, L.; Jiang, D.K. Association between microRNA polymorphisms and cancer risk based on the findings of 66 case-control studies. PLoS ONE 2013, 8, e79584. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Dou, T.H.; Geng, L.; Zhou, F.G.; Gu, X.; Wang, H.; Gao, C.F. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum. Immunol. 2010, 71, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, J.; Jia, Z.; Cui, Q.; Zhang, C.; Wang, W.; Chen, P.; Ma, K.; Zhou, C. MiR-499 regulates cell proliferation and apoptosis during late-stage cardiac differentiation via Sox6 and cyclin D1. PLoS ONE 2013, 8, e74504. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Shu, Y.; Chen, J.; Hu, Z.; Xu, L.; Jin, G.; Liang, J.; Liu, P.; Zhou, X.; Miao, R.; et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1183–1187. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, K.; Srivastava, A.; Mittal, B. Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J. Hum. Genet. 2010, 55, 495–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Li, G.; Wei, S.; Niu, J.; El-Naggar, A.K.; Sturgis, E.M.; Wei, Q. Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer 2010, 116, 4753–4760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Lv, R.; Song, X.; Li, D.; Hu, X.; Ying, B.; Wei, Y.; Wang, L. Association between two genetic variants in miRNA and primary liver cancer risk in the Chinese population. DNA Cell Biol. 2012, 31, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Min, K.T.; Kim, J.W.; Jeon, Y.J.; Jang, M.J.; Chong, S.Y.; Oh, D.; Kim, N.K. Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. Mol. Carcinog. 2012, 51 (Suppl. 1), E65–E73. [Google Scholar] [CrossRef] [PubMed]
- Schulman, B.R.; Esquela-Kerscher, A.; Slack, F.J. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev. Dyn. 2005, 234, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Belasco, J.G. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 2005, 25, 9198–9208. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.K.; Goga, A.; Bhaumik, D.; Berger, C.E.; Sullivan, C.S.; Benz, C.C. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 2007, 282, 1479–1486. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. microRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Pak, C.; Jin, P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet. 2007, 16, 1124–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Yu, C.Y.; Wang, J.L.; Guan, J.; Chen, H.Y.; Fang, J.Y. microRNA sequence polymorphisms and the risk of different types of cancer. Sci. Rep. 2014, 4, 3648. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hormazabal, P.; Gutierrez-Enriquez, S.; Gaete, D.; Reyes, J.M.; Peralta, O.; Waugh, E.; Gomez, F.; Margarit, S.; Bravo, T.; Blanco, R.; et al. Spectrum of BRCA1/2 point mutations and genomic rearrangements in high-risk breast/ovarian cancer Chilean families. Breast Cancer Res. Treat. 2011, 126, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Moazzeni, H.; Najafi, A.; Khani, M. Identification of direct target genes of miR-7, miR-9, miR-96, and miR-182 in the human breast cancer cell lines MCF-7 and MDA-MB-231. Mol. Cell. Probes 2017, 34, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Ruan, K.; Fang, X.; Ouyang, G. MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Lett. 2009, 285, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic variation in microRNA networks: The implications for cancer research. Nat. Rev. Cancer 2010, 10, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Afsharzadeh, S.M.; Mohaddes Ardebili, S.M.; Seyedi, S.M.; Karimian Fathi, N.; Mojarrad, M. Association between rs11614913, rs3746444, rs2910164 and occurrence of breast cancer in Iranian population. Meta Gene 2017, 11, 20–25. [Google Scholar] [CrossRef]
- Bansal, C.; Sharma, K.L.; Misra, S.; Srivastava, A.N.; Mittal, B.; Singh, U.S. Common genetic variants in pre-microRNAs and risk of breast cancer in the North Indian population. Ecancermedicalscience 2014, 8, 473. [Google Scholar] [PubMed]
- He, B.; Pan, Y.; Xu, Y.; Deng, Q.; Sun, H.; Gao, T.; Wang, S. Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumour Biol. 2015, 36, 4575–4582. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, K.; Srivastava, A. Comprehensive review of genetic association studies and meta-analyses on miRNA polymorphisms and cancer risk. PLoS ONE 2012, 7, e50966. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.B.; Bai, P.; Pan, X.M.; Jia, J.; Li, L.J.; Liang, W.B.; Tang, M.; Zhang, L.S.; Wei, Y.G.; Zhang, L. The association between two polymorphisms in pre-miRNAs and breast cancer risk: A meta-analysis. Breast Cancer Res. Treat. 2011, 125, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Y.; Yan, W.; Wang, W.; Zhao, X.; Ma, X.; Gao, X.; Zhang, S. Association between three functional microRNA polymorphisms (miR-499 rs3746444, miR-196a rs11614913 and miR-146a rs2910164) and breast cancer risk: A meta-analysis. Oncotarget 2017, 8, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Coke, R. Ethnic origin and evolution of the Chilean population. Rev. Med. Chile 1976, 104, 365–368. [Google Scholar] [PubMed]
- Valenzuela, C.Y.; Acuna, M.P.; Harb, Z. Sociogenetic gradient in the Chilean population. Rev. Med. Chile 1987, 115, 295–299. [Google Scholar] [PubMed]
- Valenzuela, C.Y.; Harb, Z. Socioeconomic assortative mating in Santiago, Chile: A demonstration using stochastic matrices of mother-child relationships applied to ABO blood groups. Soc. Biol. 1977, 24, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Chen, C.; Wu, D. The association between common genetic variant of microRNA-499 and cancer susceptibility: A meta-analysis. Mol. Biol. Rep. 2013, 40, 3389–3394. [Google Scholar] [CrossRef] [PubMed]
- Omrani, M.; Hashemi, M.; Eskandari-Nasab, E.; Hasani, S.S.; Mashhadi, M.A.; Arbabi, F.; Taheri, M. hsa-miR-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomark. Med. 2014, 8, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.M.; Kang, H.F.; Zhang, W.G.; Li, H.B.; Zhang, S.Q.; Ma, X.B.; Lin, S.; Wang, M.; Feng, Y.J.; Liu, K.; et al. The associations of single nucleotide polymorphisms in miR196a2, miR-499, and miR-608 with breast cancer susceptibility: A STROBE-compliant observational study. Medicine 2016, 95, e2826. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhang, J.; Zhou, F. miR-499 rs3746444 polymorphism is associated with cancer development among Asians and related to breast cancer susceptibility. Mol. Biol. Rep. 2012, 39, 10433–10438. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, M.; Pulgar, I.; Gallo, C.; Bortolini, M.C.; Canizales-Quinteros, S.; Bedoya, G.; Gonzalez-Jose, R.; Ruiz-Linares, A.; Rothhammer, F. Gene geography of Chile: Regional distribution of American, European and African genetic contributions. Rev. Med. Chile 2014, 142, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Muinos-Gimeno, M.; Montfort, M.; Bayes, M.; Estivill, X.; Espinosa-Parrilla, Y. Design and evaluation of a panel of single-nucleotide polymorphisms in microRNA genomic regions for association studies in human disease. Eur. J. Hum. Genet. 2010, 18, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Wang, L.; Zhu, L.; Du, J.; Zhu, X.; Niu, Y.; Wang, R.; Hu, Z.; Chen, N.; Shen, H.; et al. Association of microRNA polymorphisms with the risk of head and neck squamous cell carcinoma in a Chinese population: A case-control study. Chin. J. Cancer 2016, 35, 77. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Zhang, L.; Wang, J.; Huang, Z.; Li, X.; Wu, M.; Li, S.; Tang, H.; Xie, X. High expression of microRNA-183/182/96 cluster as a prognostic biomarker for breast cancer. Sci. Rep. 2016, 6, 24502. [Google Scholar] [CrossRef] [PubMed]
- Dambal, S.; Shah, M.; Mihelich, B.; Nonn, L. The microRNA-183 cluster: The family that plays together stays together. Nucleic Acids Res. 2015, 43, 7173–7188. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Sheng, C.; Huang, L.; Zhang, H.; Cheng, Z.; Zhu, Q. miR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014, 16, 473. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, K.; Steptoe, A.L.; Martin, H.C.; Wani, S.; Nones, K.; Waddell, N.; Mariasegaram, M.; Simpson, P.T.; Lakhani, S.R.; Gabrielli, B.; et al. microRNA-182–5p targets a network of genes involved in DNA repair. RNA 2013, 19, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Guttilla, I.K.; White, B.A. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 2009, 284, 23204–23216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kong, X.; Li, J.; Luo, Q.; Li, X.; Shen, L.; Chen, L.; Fang, L. miR-96 promotes tumor proliferation and invasion by targeting RECK in breast cancer. Oncol. Rep. 2014, 31, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.H.; Qiu, Z.; Ghosh, A. TOX3 regulates calcium-dependent transcription in neurons. Proc. Natl. Acad. Sci. USA 2009, 106, 2909–2914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maia, A.T.; Spiteri, I.; Lee, A.J.; O'Reilly, M.; Jones, L.; Caldas, C.; Ponder, B.A. Extent of differential allelic expression of candidate breast cancer genes is similar in blood and breast. Breast Cancer Res. 2009, 11, R88. [Google Scholar] [CrossRef] [PubMed]
- Seksenyan, A.; Kadavallore, A.; Walts, A.E.; de la Torre, B.; Berel, D.; Strom, S.P.; Aliahmad, P.; Funari, V.A.; Kaye, J. TOX3 is expressed in mammary ER+ epithelial cells and regulates ER target genes in luminal breast cancer. BMC Cancer 2015, 15, 22. [Google Scholar] [CrossRef] [PubMed]
- Easton, D.F.; Pooley, K.A.; Dunning, A.M.; Pharoah, P.D.; Thompson, D.; Ballinger, D.G.; Struewing, J.P.; Morrison, J.; Field, H.; Luben, R.; et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007, 447, 1087–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Narvaez, E.A.; Rosenberg, L.; Cozier, Y.C.; Cupples, L.A.; Adams-Campbell, L.L.; Palmer, J.R. Polymorphisms in the TOX3/LOC643714 locus and risk of breast cancer in African-American women. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1320–1327. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; Baumgartner, K.B.; Giuliano, A.R.; Byers, T.; Herrick, J.S.; Wolff, R.K. Replication of five GWAS-identified loci and breast cancer risk among Hispanic and non-Hispanic white women living in the Southwestern United States. Breast Cancer Res. Treat. 2011, 129, 531–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stacey, S.N.; Manolescu, A.; Sulem, P.; Rafnar, T.; Gudmundsson, J.; Gudjonsson, S.A.; Masson, G.; Jakobsdottir, M.; Thorlacius, S.; Helgason, A.; et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007, 39, 865–869. [Google Scholar] [CrossRef] [PubMed]
- Udler, M.S.; Ahmed, S.; Healey, C.S.; Meyer, K.; Struewing, J.; Maranian, M.; Kwon, E.M.; Zhang, J.; Tyrer, J.; Karlins, E.; et al. Fine scale mapping of the breast cancer 16q12 locus. Hum. Mol. Genet. 2010, 19, 2507–2515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riaz, M.; Berns, E.M.; Sieuwerts, A.M.; Ruigrok-Ritstier, K.; de Weerd, V.; Groenewoud, A.; Uitterlinden, A.G.; Look, M.P.; Klijn, J.G.; Sleijfer, S.; et al. Correlation of breast cancer susceptibility loci with patient characteristics, metastasis-free survival, and mRNA expression of the nearest genes. Breast Cancer Res. Treat. 2012, 133, 843–851. [Google Scholar] [CrossRef] [PubMed]
| Inclusion Criteria | Families n |
|---|---|
| Three or more family members with breast and/or ovarian cancer | 121 (27.5%) |
| Two family members with breast and/or ovarian cancer | 148 (33.6%) |
| Single affected individual with breast cancer ≤35 years of age | 87 (19.8%) |
| Single affected individual with breast cancer between 36 and 50 years of age | 84 (19.1%) |
| Total | 440 (100%) |
| All BC Cases (n = 440) | Families with ≥2 BC and/or OC Cases (n = 269) | Families with a Single Case, Diagnosis at ≤50 Years of Age (n = 171) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype or Allele | Controls (n = 1048) (%) | BC Cases (%) | p-Value a | OR [95% CI] | BC Cases (%) | p-Value a | OR [95% CI] | BC Cases (%) | p-Value a | OR [95% CI] |
| rs3746444 (Pre-miR-499) | ||||||||||
| A/A | 772 (73.7) | 319 (72.5) | - | 1.0 (Ref) | 198 (73.6) | - | 1.0 (Ref) | 121 (70.8) | - | 1.0 (Ref) |
| A/G | 254 (24.2) | 111 (25.2) | 0.6 | 1.0 [0.8–1.3] | 64 (23.8) | 0.9 | 0.9 [0.7–1.3] | 47 (27.5) | 0.3 | 1.1 [0.8–1.7] |
| G/G | 22 (2.1) | 10 (2.3) | 0.8 | 1.1 [0.5–2.3] | 7 (2.6) | 0.6 | 1.2 [0.5–2.9] | 3 (1.7) | 1.0 | 0.8 [0.2–2.9] |
| A/G + G/G | 276 (26.3) | 121 (27.5) | 0.6 | 1.0 [0.8–1.3] | 71 (26.4) | 1.0 | 1.0 [0.7–1.3] | 50 (29.2) | 0.4 | 1.1 [0.8–1.6] |
| Allele A | 1798 (85.8) | 749 (85.1) | - | 1.0 (Ref) | 460 (85.5) | - | 1.0 (Ref) | 289 (84.5) | - | 1.0 (Ref) |
| Allele G | 298 (14.2) | 131 (14.9) | 0.6 | 1.0 [0.8–1.3] | 78 (14.5) | 0.9 | 1.0 [0.7–1.3] | 53 (15.5) | 0.5 | 1.1 [0.8–1.5] |
| rs2910164 (Pre-miR-146a) | ||||||||||
| G/G | 561 (53.5) | 236 (53.6) | - | 1.0 (Ref) | 149 (55.4) | - | 1.0 (Ref) | 87 (50.9) | - | 1.0 (Ref) |
| G/C | 410 (39.1) | 165 (37.5) | 0.7 | 0.9 [0.7–1.1] | 101 (37.5) | 0.6 | 0.9 [0.6–1.2] | 64 (37.4) | 1.0 | 1.0 [0.7–1.4] |
| C/C | 77 (7.4) | 39 (8.9) | 0.3 | 1.2 [0.7–1.8] | 19 (7.1) | 0.8 | 0.9 [0.5–1.5] | 20 (11.7) | 0.06 | 1.6 [0.9–2.8] |
| G/C + C/C | 487 (46.5) | 204 (46.4) | 1.0 | 0.9 [0.7–1.2] | 120 (44.6) | 0.6 | 0.9 [0.7–1.2] | 84 (49.1) | 0.5 | 1.1 [0.8–1.5] |
| Allele G | 1532 (73.1) | 637 (72.4) | - | 1.0 (Ref) | 399 (74.2) | - | 1.0 (Ref) | 238 (69.6) | - | 1.0 (Ref) |
| Allele C | 564 (26.9) | 243 (27.6) | 0.7 | 1.0 [0.8–1.2] | 139 (25.8) | 0.6 | 0.9 [0.7–1.1] | 104 (30.4) | 0.2 | 1.1 [0.9–1.5] |
| rs12975333 (Pre-miR-125a) | ||||||||||
| G/G | 1040 (99.2) | 436 (99.1) | - | 1.0 (ref) | 267 (99.3) | - | 1.0 (ref) | 169 (98.8) | - | 1.0 (ref) |
| G/T | 8 (0.8) | 4 (0.9) | 0.7 | 1.1 [0.3–3.9] | 2 (0.7) | 0.2 | 1.9 [0.5–6.5] | 2 (1.2) | 0.6 | 1.5 [0.3–7.3] |
| T/T | 0 | 0 | - | - | 0 | - | - | 0 | - | - |
| G/T + T/T | 8 (0.8) | 4 (0.9) | 0.7 | 1.1 [0.3–3.9] | 2 (0.7) | 0.2 | 1.9 [0.5–6.5] | 2 (1.2) | 0.6 | 1.5 [0.3–7.3] |
| Allele G | 2088 (99.6) | 876 (99.5) | - | 1.0 (ref) | 536 (99.6) | - | 1.0 (ref) | 340 (99.4) | - | 1.0 (ref) |
| Allele T | 8 (0.4) | 4 (0.5) | 0.7 | 1.1 [0.3–3.9] | 2 (0.4) | 0.2 | 1.9 [0.5–6.5] | 2 (0.6) | 0.6 | 1.5 [0.3–7.3] |
| rs2043556 (miR-605) | ||||||||||
| T/T | 376 (35.9) | 208 (47.3) | - | 1.0 (ref) | 128 (47.6) | - | 1.0 (ref) | 80 (46.8) | - | 1.0 (ref) |
| T/C | 571 (54.5) | 182 (41.3) | <10−4 | 0.5 [0.4–0.7] | 115 (42.7) | 0.0003 | 0.5 [0.4–0.7] | 67 (39.2) | 0.0009 | 0.5 [0.3–0.7] |
| C/C | 101 (9.6) | 50 (11.4) | 0.6 | 0.8 [0.6–1.3] | 26 (9.7) | 0.2 | 0.7 [0.5 –1.2] | 24 (14.0) | 0.6 | 1.1 [0.6–1.8] |
| T/C + C/C | 672 (64.1) | 232 (52.7) | <10−4 | 0.6 [0.4–0.7] | 141 (52.4) | 0.0006 | 0.6 [0.4–0.8] | 91 (53.2) | 0.02 | 0.6 [0.5–0.9] |
| Allele T | 1323 (63.1) | 598 (68.0) | - | 1.0 (ref) | 371 (69.0) | - | 1.0 (ref) | 227 (66.4) | - | 1.0 (ref) |
| Allele C | 773 (36.9) | 282 (32.0) | 0.01 | 0.8 [0.6–0.9] | 167 (31.0) | 0.01 | 0.7 [0.6–0.9] | 115 (33.6) | 0.4 | 0.9 [0.7–1.1] |
| rs4541843 (Pri-miR-182) | ||||||||||
| C/C | 386 (36.8) | 150 (34.1) | - | 1.0 (Ref) | 81 (30.1) | - | 1.0 (ref) | 69 (40.4) | - | 1.0 Ref |
| C/T | 473 (45.1) | 205 (46.6) | 0.4 | 1.1 [0.8–1.4] | 127 (47.2) | 0.1 | 1.2 [0.9–1.7] | 78 (45.6) | 0.6 | 0.9 [0.6–1.3] |
| T/T | 189 (18.1) | 85 (19.3) | 0.4 | 1.5 [0.8–1.5] | 61 (22.7) | 0.03 | 1.5 [1.0–2.2] | 24 (14.0) | 0.1 | 0.7 [0.4–1.1] |
| C/T + T/T | 662 (63.2) | 290 (65.9) | 0.3 | 1.1 [0.8–1.4] | 188 (69.9) | 0.04 | 1.3 [1.0–1.8] | 102 (59.6) | 0.3 | 0.8 [0.6–1.1] |
| Allele C | 1245 (59.4) | 505 (57.4) | - | 1.0 (Ref) | 289 (53.7) | - | 1.0 (ref) | 216 (63.2) | - | 1.0 (Ref) |
| Allele T | 851 (40.6) | 375 (42.6) | 0.3 | 1.0 [0.9–1.2] | 249 (46.3) | 0.01 | 1.2 [1.0–1.5] | 126 (36.8) | 0.2 | 0.8 [0.6–1.0] |
| Families with 2 BC and/or OC Cases (n = 148) | Families with ≥3 BC and/or OC Cases (n = 121) | ||||||
|---|---|---|---|---|---|---|---|
| Genotype or Allele | Controls (n = 1048) (%) | BC Cases (%) | p-Value a | OR [95% CI] | BC Cases (%) | p-Value a | OR [95% CI] |
| rs2043556 (miR-605) | |||||||
| T/T | 376 (35.9) | 67 (45.3) | - | 1.0 (Ref) | 61 (50.4) | - | 1.0 (Ref) |
| T/C | 571 (54.5) | 68 (45.9) | 0.08 | 0.7 [0.5–1.0] | 47 (38.9) | 0.003 | 0.5 [0.4–0.8] |
| C/C | 101 (9.6) | 13 (8.8) | 0.3 | 0.7 [0.4–1.4] | 13 (10.7) | 0.5 | 0.7 [0.4–1.5] |
| T/C + C/C | 672 (64.1) | 81 (54.7) | 0.06 | 0.7 [0.5–1.0] | 60 (49.6) | 0.006 | 0.5 [0.4–0.9] |
| Allele T | 1323 (63.1) | 202 (68.2) | - | 1.0 (Ref) | 169 (69.8) | - | 1.0 (Ref) |
| Allele C | 773 (36.9) | 94 (31.8) | 0.1 | 0.8 [0.6–1.0] | 73 (30.2) | 0.06 | 0.7 [0.5–1.0] |
| rs4541843 (Pri-miR-182) | |||||||
| C/C | 386 (36.8) | 45 (30.4) | - | 1.0 (Ref) | 36 (29.8) | - | 1.0 (Ref) |
| C/T | 473 (45.1) | 70 (47.3) | 0.2 | 1.2 [0.8–1.8] | 57 (47.1) | 0.2 | 1.2 [0.8–1.9] |
| T/T | 189 (18.1) | 33 (22.3) | 0.1 | 1.4 [0.8–2.4] | 28 (23.1) | 0.09 | 1.5 [0.9–2.6] |
| C/T + T/T | 662 (63.2) | 103 (69.6) | 0.1 | 1.3 [0.9–1.9] | 85 (70.2) | 0.1 | 1.3 [0.9–2.0] |
| Allele C | 1245 (59.4) | 160 (54.1) | - | 1.0 (Ref) | 129 (53.3) | - | 1.0 (Ref) |
| Allele T | 851 (40.6) | 136 (45.9) | 0.09 | 1.2 [0.9–1.5] | 113 (46.7) | 0.07 | 1.2 [0.9–1.6] |
| Number of Risk Alleles (a) | Controls (n = 1048) (%) | All BC Cases (n = 440) | Families with ≥2 BC and/or OC cases (n = 269) | Families with a Single Case, Diagnosis at ≤50 Years of Age (n = 171) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BC Cases (%) | OR [95% CI] | p-Value (b) | BC Cases (%) | OR [95% CI] | p-Value (b) | BC Cases (%) | OR [95% CI] | p-Value (b) | ||
| 0 risk alleles | 153 (14.6) | 49 (11.1) | 1.0 (Ref) | - | 22 (8.2) | 1.0 (Ref) | - | 27 (15.8) | 1.0 (Ref) | - |
| 1 risk allele | 381 (36.4) | 128 (29.1) | 1.0 [0.7–1.5] | 0.8 | 73 (27.1) | 1.3 [0.7–2.2] | 0.3 | 55 (32.2) | 0.8 [0.4–1.3] | 0.4 |
| 2 risk alleles | 336 (32.1) | 168 (38.2) | 1.5 [1.0–2.2] | 0.01 | 105 (39) | 2.1 [1.3–3.5] | 0.001 | 63 (36.9) | 1.0 [0.6–1.7] | 0.9 |
| 3 risk alleles | 153 (14.6) | 79 (18) | 1.6 [1.0–2.4] | 0.02 | 57 (21.2) | 2.5 [1.5–4.4] | 0.0006 | 22 (12.9) | 0.8 [0.4–1.4] | 0.5 |
| 4 risk alleles | 25 (2.4) | 16 (36) | 1.9 [0.9–3.8] | 0.08 | 12 (4.5) | 3.2 [1.4–7.2] | 0.006 | 4 (2.3) | 0.8 [0.2–2.6] | 1.0 |
| p-trend (c) | 0.0005 | <10−4 | 0.9755 | |||||||
| Global p (d) | 0.005 | 0.0001 | 0.6970 | |||||||
| Number of Risk Alleles (a) | Controls (n = 1048) (%) | Families with Two BC and/or OC Cases (n = 148) | Families with ≥3 BC and/or OC Cases (n = 121) | ||||
|---|---|---|---|---|---|---|---|
| BC Cases (%) | OR [95% CI] | p-Value (b) | BC Cases (%) | OR [95% CI] | p-Value (b) | ||
| 0 risk alleles | 153 (14.6) | 10 (6.8) | 1.0 (Ref) | - | 12 (8.1) | 1.0 (Ref) | - |
| 1 risk allele | 381 (36.4) | 29 (19.6) | 1.1 [0.5–2.4] | 0.8 | 44 (29.7) | 1.4 [0.7–2.8] | 0.2 |
| 2 risk alleles | 336 (32.1) | 47 (31.8) | 2.1 [1.0–4.3] | 0.03 | 58 (39.2) | 2.2 [1.1–4.2] | 0.01 |
| 3 risk alleles | 153 (14.6) | 30 (20.3) | 3.0 [1.4–6.3] | 0.003 | 27 (18.2) | 2.2 [1.0–4.6] | 0.02 |
| 4 risk alleles | 25 (2.4) | 5 (3.4) | 2.9 [0.9–9.3] | 0.06 | 7 (4.7) | 3.4 [1.2–9.5] | 0.02 |
| p-trend(c) | 0.0001 | 0.001 | |||||
| Global p (d) | 0.001 | 0.02 | |||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales, S.; De Mayo, T.; Gulppi, F.A.; Gonzalez-Hormazabal, P.; Carrasco, V.; Reyes, J.M.; Gómez, F.; Waugh, E.; Jara, L. Genetic Variants in pre-miR-146a, pre-miR-499, pre-miR-125a, pre-miR-605, and pri-miR-182 Are Associated with Breast Cancer Susceptibility in a South American Population. Genes 2018, 9, 427. https://0-doi-org.brum.beds.ac.uk/10.3390/genes9090427
Morales S, De Mayo T, Gulppi FA, Gonzalez-Hormazabal P, Carrasco V, Reyes JM, Gómez F, Waugh E, Jara L. Genetic Variants in pre-miR-146a, pre-miR-499, pre-miR-125a, pre-miR-605, and pri-miR-182 Are Associated with Breast Cancer Susceptibility in a South American Population. Genes. 2018; 9(9):427. https://0-doi-org.brum.beds.ac.uk/10.3390/genes9090427
Chicago/Turabian StyleMorales, Sebastián, Tomas De Mayo, Felipe Andrés Gulppi, Patricio Gonzalez-Hormazabal, Valentina Carrasco, José Miguel Reyes, Fernando Gómez, Enrique Waugh, and Lilian Jara. 2018. "Genetic Variants in pre-miR-146a, pre-miR-499, pre-miR-125a, pre-miR-605, and pri-miR-182 Are Associated with Breast Cancer Susceptibility in a South American Population" Genes 9, no. 9: 427. https://0-doi-org.brum.beds.ac.uk/10.3390/genes9090427