Decreased Expression of Alpha-L-Fucosidase Gene FUCA1 in Human Colorectal Tumors
Abstract
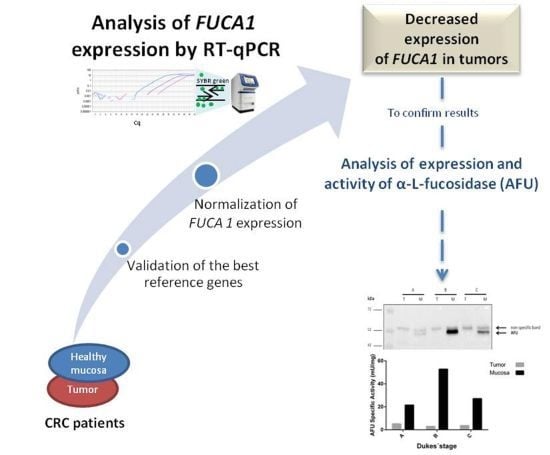
:1. Introduction
2. Results and Discussion
2.1. Specificity of Primers and PCR Amplification Efficiency
2.2. Expression Stability of Candidate Reference Genes in Colorectal Tissues
2.3. Expression of the Target Gene FUCA1 in Tumor and Normal Mucosa from CRC Patients
2.4. Validation of the RT-qPCR Data: Expression of Alpha-l-Fucosidase Protein
3. Experimental Section
3.1. Patients
3.2. RNA Extraction
3.3. cDNA Synthesis
3.4. Primers for the Target Gene FUCA1 and Candidate Reference Genes
3.5. Quantitative Real-Time PCR (qPCR)
3.6. Western Blot
3.7. Alpha-l-Fucosidase Activity and Protein Determination in Tumor and Normal Colorectal Mucosa
3.8. Statistical Analyses
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Cunningham, D.; Atkin, W.; Lenz, H.J.; Lynch, H.T.; Minsky, B.; Nordlinger, B.; Starling, N. Colorectal cancer. Lancet 2010, 375, 1030–1047. [Google Scholar]
- Hakomori, S. Glycosylation defining cancer malignancy: New wine in an old bottle. Proc. Natl. Acad. Sci. USA 2002, 99, 10231–10233. [Google Scholar]
- Meany, D.L.; Chan, D.W. Aberrant glycosylation associated with enzymes as cancer biomarkers. Clin. Proteomics 2011, 8, 7. [Google Scholar]
- Drake, P.M.; Cho, W.; Li, B.; Prakobphol, A.; Johansen, E.; Anderson, N.L.; Regnier, F.E.; Gibson, B.W.; Fisher, S.J. Sweetening the pot: Adding glycosylation to the biomarker discovery equation. Clin. Chem 2010, 56, 223–236. [Google Scholar]
- Mechref, Y.; Hu, Y.; Garcia, A.; Zhou, S.; Desantos-Garcia, J.L.; Hussein, A. Defining putative glycan cancer biomarkers by MS. Bioanalysis 2012, 4, 2457–2469. [Google Scholar]
- Fernández-Rodríguez, J.; Feijoo-Carnero, C.; Merino-Trigo, A.; Páez de la Cadena, M.; Rodríguez-Berrocal, F.J.; de Carlos, A.; Butrón, M.; Martínez-Zorzano, V.S. Immunohistochemical analysis of sialic acid and fucose composition in human colorectal adenocarcinoma. Tumor Biol 2000, 21, 153–164. [Google Scholar]
- Misonou, Y.; Shida, K.; Korekane, H.; Seki, Y.; Noura, S.; Ohue, M.; Miyamoto, Y. Comprehensive clinico-glycomic study of 16 colorectal cancer specimens: Elucidation of aberrant glycosylation and its mechanistic causes in colorectal cancer cells. J. Proteome Res 2009, 8, 2990–3005. [Google Scholar]
- Balog, C.I.; Stavenhagen, K.; Fung, W.L.; Koeleman, C.A.; McDonnell, L.A.; Verhoeven, A.; Mesker, W.E.; Tollenaar, R.A.; Deelder, A.M.; Wuhrer, M. N-glycosylation of colorectal cancer tissues: A liquid chromatography and mass spectrometry-based investigation. Mol. Cell. Proteomics 2012, 11, 571–585. [Google Scholar]
- Fernández-Rodríguez, J.; Páez de la Cadena, M.; Martínez-Zorzano, V.S.; Rodríguez-Berrocal, F.J. Fucose levels in sera and in tumours of colorectal adenocarcinoma patients. Cancer Lett 1997, 121, 147–153. [Google Scholar]
- Muinelo-Romay, L.; Gil-Martín, E.; Fernández-Briera, A. α(1,2)fucosylation in human colorectal carcinoma. Oncol. Lett 2010, 1, 361–366. [Google Scholar]
- Johnson, S.W.; Alhadeff, J.A. Mammalian alpha-l-fucosidases. Comp. Biochem. Physiol. B 1991, 99, 479–488. [Google Scholar]
- Intra, J.; Perotti, M.E.; Pavesi, G.; Horner, D. Comparative and phylogenetic analysis of alpha-l-fucosidase genes. Gene 2007, 392, 34–46. [Google Scholar]
- Lin, S.P.; Chang, J.H.; Páez de la Cadena, M.; Chang, T.F.; Lee-Chen, G.J. Mutation identification and characterization of a Taiwanese patient with fucosidosis. J. Hum. Genet 2007, 52, 553–556. [Google Scholar]
- Leray, G.; Deugnier, Y.; Jouanolle, A.M.; Lehry, D.; Bretagne, J.F.; Campion, J.P.; Brissot, P.; Le Treut, A. Biochemical aspects of alpha-l-fucosidase in hepatocellular carcinoma. Hepatology 1989, 9, 249–252. [Google Scholar]
- Hutchinson, W.L.; Johnson, P.J.; Du, M.Q.; Williams, R. Serum and tissue alpha-l-fucosidase activity in the pre-clinical and clinical stages of hepatocellular carcinoma. Clin. Sci 1991, 81, 177–182. [Google Scholar]
- Vesce, F.; Biondi, C. Alpha-l-fucosidase activity in endometrial, cervical and ovarian cancer. Eur. J. Gynaecol. Oncol 1983, 4, 135–138. [Google Scholar]
- Wang, J.W.; Ambros, R.A.; Weber, P.B.; Rosano, T.G. Fucosyltransferase and alpha-l-fucosidase activities and fucose levels in normal and malignant endometrial tissue. Cancer Res 1995, 55, 3654–3658. [Google Scholar]
- Gil-Martín, E.; Gil-Seijo, S.; Nieto-Novoa, C.; Fernández-Briera, A. Elevation of acid glycosidase activities in thyroid and gastric tumors. Int. J. Biochem. Cell Biol 1996, 28, 651–657. [Google Scholar]
- Páez de la Cadena, M.; Fernández-Rodríguez, J.; de Carlos, A.; Martínez-Zorzano, V.S.; Gil-Martín, E.; Rodríguez-Berrocal, F.J. Low levels of alpha-l-fucosidase activity in colorectal cancer are due to decreased amounts of the enzymatic protein and are related with Dukes’ stage. Int. J. Oncol 1996, 9, 747–754. [Google Scholar]
- Fernández-Rodríguez, J.; Ayude, D.; Páez de la Cadena, M.; Martínez-Zorzano, V.S.; de Carlos, A.; Caride-Castro, A.; de Castro, G.; Rodríguez-Berrocal, F.J. Alpha-l-fucosidase enzyme in the prediction of colorectal cancer patients at high risk of tumor recurrence. Cancer Detect. Prev 2000, 24, 143–149. [Google Scholar]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Aspects Med 2006, 27, 95–125. [Google Scholar]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; Strategies and considerations. Genes Immun 2005, 6, 279–284. [Google Scholar]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem 2005, 344, 141–143. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 2001, 25, 402–408. [Google Scholar]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalisation of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004, 64, 5245–5250. [Google Scholar]
- Matthijsen, R.A.; Derikx, J.P.M.; Kuipers, D.; van Dam, R.M.; Dejong, C.H.C.; Buurman, W.A. Enterocyte shedding and epithelial lining repair following ischemia of the human small intestine attenuate inflammation. PLoS One 2009, 4, e7045. [Google Scholar]
- Fu, J.; Bian, L.; Zhao, L.; Dong, Z.; Gao, X.; Luan, H.; Sun, Y.; Song, H. Identification of genes for normalization of quantitative real-time PCR data in ovarian tissues. Acta Biochim. Biophys. Sin 2010, 42, 568–574. [Google Scholar]
- Kulkarni, B.; Mohammed, I.; Hopkinson, A.; Dua, H.S. Validation of endogenous control genes for gene expression studies on human ocular surface epithelium. PLoS One 2011, 6, e22301. [Google Scholar]
- Wei, R.; Stewart, E.A.; Amoaku, W.M. Suitability of endogenous reference genes for gene expression studies with human intraocular endothelial cells. BMC Res. Notes 2013, 6, 46. [Google Scholar]
- Valente, V.; Teixeira, S.A.; Neder, L.; Okamoto, O.K.; Oba-Shinjo, S.M.; Marie, S.K.; Scrideli, C.A.; Paçó-Larson, M.L.; Carlotti, C.G., Jr. Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol. Biol. 2009, 10, 17–27. [Google Scholar]
- Fu, L.Y.; Jia, H.L.; Dong, Q.Z.; Wu, J.C.; Zhao, Y.; Zhou, H.J.M.; Ren, N.; Ye, Q.H.; Qin, L.X. Suitable reference genes for real-time PCR in human HBV-related hepatocelular carcinoma with different clinical prognoses. BMC Cancer 2009, 9, 49. [Google Scholar]
- Kerr, C.A.; Dunne, R.; Hines, B.M.; Zucker, M.; Cosgrove, L.; Ruszkiewicz, A.; Lockett, T.; Head, R. Measuring the combinatorial expression of solute transporters and metalloproteinases transcripts in colorectal cancer. BMC Res. Notes 2009, 2, 164. [Google Scholar]
- Sørby, L.A.; Andersen, S.N.; Bukholm, I.R.; Jacobsen, M.B. Evaluation of suitable reference genes for normalization of real-time reverse transcription PCR analysis in colon cancer. J. Exp. Clin. Cancer Res 2010, 29, 144–152. [Google Scholar]
- Hellemans, J.; Mortier, G.; de Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 2007, 8, R19. [Google Scholar]
- De Bustos, C.; Ramos, E.; Young, J.M.; Tran, R.K.; Menzel, U.; Langford, C.F.; Eichler, E.E.; Hsu, L.; Henikoff, S.; Dumanski, J.P.; et al. Tissue-specific variation in DNA methylation levels along human chromosome 1. Epigenetics Chromatin 2009, 2, 7. [Google Scholar]
- Kawamura, Y.I.; Toyota, M.; Kawashima, R.; Hagiwara, T.; Suzuki, H.; Imai, K.; Shinomura, Y.; Tokino, T.; Kannagi, R.; Dohi, T. DNA hypermethylation contributes to incomplete synthesis of carbohydrate determinants in gastrointestinal cancer. Gastroenterology 2008, 135, 142–151. [Google Scholar]
- Horvat, T.; Zoldoš, V.; Lauc, G. Evolutional and clinical implications of the epigenetic regulation of protein glycosylation. Clin. Epigenetics 2011, 2, 425–432. [Google Scholar]
- Moriwaki, K.; Narisada, M.; Imai, T.; Shinzaki, S.; Miyoshi, E. The effect of epigenetic regulation of fucosylation on TRAIL-induced apoptosis. Glycoconj. J 2010, 27, 649–659. [Google Scholar]
- Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-time PCR, Methods and Applications; Meuer, S., Wittwer, C., Nakagawara, K., Eds.; Springer Press: Heidelberg, Germany, 2001; pp. 21–34. [Google Scholar]

| Gen | Amplification efficiency (%) | R2 | Slope | Cq median | Cq range |
|---|---|---|---|---|---|
| B2M | 101.00 | 0.989 | −3.2992 | 25.54 | 19.23–33.80 |
| GAPDH | 93.40 | 0.997 | −3.4913 | 25.68 | 18.28–33.75 |
| HPRT1 | 90.61 | 0.993 | −3.5696 | 31.25 | 23.09–36.28 |
| PPIA | 104.37 | 0.999 | −3.2214 | 23.32 | 17.82–31.01 |
| RPLP0 | 92.95 | 0.994 | −3.5034 | 23.65 | 18.33–29.64 |
| FUCA1 | 106.24 | 0.997 | −3.1810 | 27.96 | 23.08–36.12 |
| Stability ranking | GeNorm | NormFinder |
|---|---|---|
| Best pair | RPLP0/HPRT1 | RPLP0/HPRT1 |
| Ranking | Gen | Gen |
| 1 | RPLP0 | HPRT1 |
| 2 | HPRT1 | RPLP0 |
| 3 | PPIA | GAPDH |
| 4 | B2M | PPIA |
| 5 | GAPDH | B2M |
| Dukes’ stage | N | Tumor | Normal mucosa | pa | ||
|---|---|---|---|---|---|---|
| Median | Range | Median | Range | |||
| Stages A + B + C | 31 | 0.00452 | 0.00017–0.10387 | 0.01184 | 0.00009–0.20345 | <0.01 |
| Stage A | 8 | 0.00650 | 0.00017–0.00867 | 0.02324 | 0.00009–0.09498 | <0.05 |
| Stage B | 11 | 0.00452 | 0.00048–0.10387 | 0.04132 | 0.00101–0.20345 | <0.05 |
| Stage C | 12 | 0.00315 | 0.00035–0.01510 | 0.00688 | 0.00016–0.15438 | NS * |
| Features of tumors | N | Median | Range | pa | |
|---|---|---|---|---|---|
| Tumor differentiation | Well | 4 | 0.00373 | 0.00017–0.00750 | 0.192 |
| Moderate | 21 | 0.00416 | 0.00035–0.09684 | ||
| Poor | 6 | 0.00788 | 0.00386–0.10387 | ||
| Tumor location | Right colon | 12 | 0.00508 | 0.00173–0.01510 | 0.388 |
| Left colon | 14 | 0.00575 | 0.00017–0.10387 | ||
| Rectum | 5 | 0.00060 | 0.00035–0.09684 | ||
| Clinical-pathological variable | Number of patients | |
|---|---|---|
| Gender | Male | 21 |
| Female | 10 | |
| Age | Range | 56–89 |
| Mean ± SD | 70 ± 9.41 | |
| Tumor Dukes’ stage | A | 8 |
| B | 11 | |
| C | 12 | |
| Tumor differentiation | Well | 4 |
| Moderate | 21 | |
| Poor | 6 | |
| Tumor location | Right colon | 12 |
| Left colon | 14 | |
| Rectum | 5 | |
| Gen | Protein name | GenBank ID | Amplicon size (bp) | Primers sequences (5′to 3′) |
|---|---|---|---|---|
| B2M | beta-2-microglobulin | NM_004048 | 228 | F:TTTCATCCATCCGACATTGA R:CCTCCATGATGCTGCTTACA |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | NM_002046 | 238 | F:GAGTCAACGGATTTGGTCGT R:TTGATTTTGGAGGGATCTCG |
| HPRT1 | hypoxanthine-guanine phosphoribosyltransferase | NM_000194 | 248 | F:CCCCACGAAGTGTTGGATA R:AGCAGATGGCCACAGAACT |
| PPIA | peptidyl-prolyl cis-trans isomerase A | NM_021130 | 188 | F:CAAGAAGATCACCATTGCT R:AGGGAACAAGGAAAACAT |
| RPLP0 | 60S acidic ribosomal protein P0 | NM_001002 | 140 | F:GCAATGTTGCCAGTGTCTG R:GCCTTGACCTTTTCAGCAA |
| FUCA1 | tissue alpha-L-fucosidase | NM_000147 | 190 | F:AGTCACCCTGTTGCCTATGG R:TTTGGCGCTTTTAGATTGCT |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Otero-Estévez, O.; Martínez-Fernández, M.; Vázquez-Iglesias, L.; Páez de la Cadena, M.; Rodríguez-Berrocal, F.J.; Martínez-Zorzano, V.S. Decreased Expression of Alpha-L-Fucosidase Gene FUCA1 in Human Colorectal Tumors. Int. J. Mol. Sci. 2013, 14, 16986-16998. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms140816986
Otero-Estévez O, Martínez-Fernández M, Vázquez-Iglesias L, Páez de la Cadena M, Rodríguez-Berrocal FJ, Martínez-Zorzano VS. Decreased Expression of Alpha-L-Fucosidase Gene FUCA1 in Human Colorectal Tumors. International Journal of Molecular Sciences. 2013; 14(8):16986-16998. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms140816986
Chicago/Turabian StyleOtero-Estévez, Olalla, Mónica Martínez-Fernández, Lorena Vázquez-Iglesias, María Páez de la Cadena, Francisco J. Rodríguez-Berrocal, and Vicenta S. Martínez-Zorzano. 2013. "Decreased Expression of Alpha-L-Fucosidase Gene FUCA1 in Human Colorectal Tumors" International Journal of Molecular Sciences 14, no. 8: 16986-16998. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms140816986