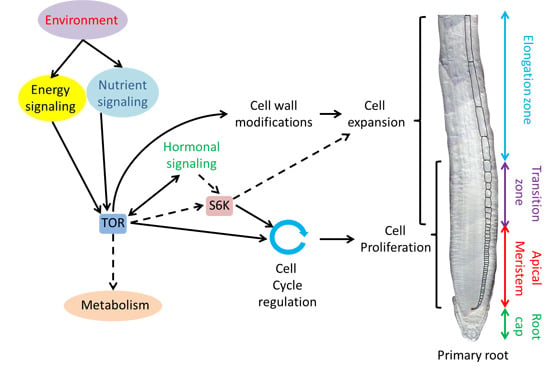
Spatial Regulation of Root Growth: Placing the Plant TOR Pathway in a Developmental Perspective
Abstract
:1. Introduction
2. Primary Root Morphology and Zonation Is the Result of Controlled Cell Proliferation, Differentiation and Expansion
2.1. Root Patterning and Zonation
2.1.1. Radial Patterning

2.1.2. Longitudinal Root Zonation

2.2. Cell Proliferation
2.2.1. Essential Cell Cycle Regulations in Plants
2.2.2. Quiescent Center and First Asymmetric Divisions

2.2.3. Apical Meristem
2.2.4. Basal Meristem
2.3. Cell Expansion, a Major Contributor to Root Growth
2.3.1. Post-Mitotic Cell Enlargement
2.3.2. Polarized Root Hair Growth
2.4. A Cross-Talk between Plant Hormones Controls the Balance between Cell Division and Differentiation
2.4.1. Establishment of a Hormone Gradient along the Root

2.4.2. Auxin and Cytokinin Regulate Proliferation and Elongation
3. The TOR Signaling Pathway, a Master Regulator of Root Growth Adaptation to Nutritional Conditions
3.1. The TOR Pathway a Conserved Major Regulator of Cell Growth in Eukaryotes
3.1.1. The TOR Pathway in Yeast and Animals
3.1.2. Conservation of the TOR Pathway in Plants
3.2. A Key Regulator of Plant Cell Growth
3.2.1. TOR Is a Global Regulator of Plant Growth
3.2.2. TOR Function in the Root Meristem

3.2.3. Regulation of Cell Expansion by TOR
3.2.4. S6K, an Important Element of the Plant TOR Pathway.
3.2.5. Regulation of Plant Metabolism by TOR
3.3. TOR as an Integrator of Environmental and Hormonal Signaling
3.3.1. Integration of Nutrient Status by TOR
3.3.2. Potential Connection with AMPK
3.3.3. TOR and Auxin

4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Montané, M.H.; Menand, B. ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 2013, 64, 4361–4374. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Venglat, P.; Qiu, S.; Feng, L.; Cao, Y.; Wang, E.; Xiang, D.; Wang, J.; Alexander, D.; Chalivendra, S.; et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 2012, 24, 4850–4874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Y.; McCormack, M.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 2013, 496, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Cederholm, H.M.; Iyer-Pascuzzi, A.S.; Benfey, P.N. Patterning the primary root in Arabidopsis. Wiley Interdiscip. Rev. 2012, 1, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Racolta, A.; Bryan, A.C.; Tax, F.E. The receptor-like kinases GSO1 and GSO2 together regulate root growth in Arabidopsis through control of cell division and cell fate specification. Dev. Dyn. 2014, 243, C1. [Google Scholar] [CrossRef] [PubMed]
- Lavenus, J.; Goh, T.; Roberts, I.; Guyomarc’h, S.; Lucas, M.; de Smet, I.; Fukaki, H.; Beeckman, T.; Bennett, M.; Laplaze, L. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 2013, 18, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Dolan, L.; Janmaat, K.; Willemsen, V.; Linstead, P.; Poethig, S.; Roberts, K.; Scheres, B. Cellular organisation of the Arabidopsis thaliana root. Development 1993, 119, 71–84. [Google Scholar] [PubMed]
- Wendrich, J.R.; Weijers, D. The Arabidopsis embryo as a miniature morphogenesis model. New Phytol. 2013, 199, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Olszewska, M.J. Autoradiographic and ultrastructural study of Cucurbita pepo root cells during their growth and differentiation. Histochemistry 1976, 49, 157–175. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, S.; Umeda, M. Cell-cycle control and plant development. In International Review of Cell and Molecular Biology; Academic Press: New Youk, NY, USA, 2011; Volume 291, pp. 227–261. [Google Scholar]
- Scofield, S.; Jones, A.; Murray, J.A. The plant cell cycle in context. J. Exp. Bot. 2014, 65, 2557–2562. [Google Scholar] [CrossRef] [PubMed]
- De Veylder, L.; Larkin, J.C.; Schnittger, A. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 2011, 16, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Magyar, Z.; Horvath, B.; Khan, S.; Mohammed, B.; Henriques, R.; de Veylder, L.; Bako, L.; Scheres, B.; Bogre, L. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 2012, 31, 1480–1493. [Google Scholar] [CrossRef] [PubMed]
- Van Leene, J.; Boruc, J.; de Jaeger, G.; Russinova, E.; de Veylder, L. A kaleidoscopic view of the Arabidopsis core cell cycle interactome. Trends Plant Sci. 2011, 16, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, P.; Das Gupta, M.; Joseph, A.P.; Chatterjee, N.; Srinivasan, N.; Nath, U. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 2010, 22, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Berckmans, B.; de Veylder, L. Transcriptional control of the cell cycle. Curr. Opin. Plant Biol. 2009, 12, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Herve, C.; Dabos, P.; Bardet, C.; Jauneau, A.; Auriac, M.C.; Ramboer, A.; Lacout, F.; Tremousaygue, D. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 2009, 149, 1462–1477. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Potuschak, T.; Colon-Carmona, A.; Gutierrez, R.A.; Doerner, P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 2005, 102, 12978–12983. [Google Scholar] [CrossRef] [PubMed]
- Tremousaygue, D.; Garnier, L.; Bardet, C.; Dabos, P.; Herve, C.; Lescure, B. Internal telomeric repeats and “TCP domain” protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J. 2003, 33, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Van Leene, J.; Hollunder, J.; Eeckhout, D.; Persiau, G.; van de Slijke, E.; Stals, H.; van Isterdael, G.; Verkest, A.; Neirynck, S.; Buffel, Y.; et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 2010, 6, 397. [Google Scholar] [CrossRef] [PubMed]
- Cook, G.S.; Gronlund, A.L.; Siciliano, I.; Spadafora, N.; Amini, M.; Herbert, R.J.; Bitonti, M.B.; Graumann, K.; Francis, D.; Rogers, H.J. Plant WEE1 kinase is cell cycle regulated and removed at mitosis via the 26S proteasome machinery. J. Exp. Bot. 2013, 64, 2093–2106. [Google Scholar] [CrossRef] [PubMed]
- De Schutter, K.; Joubes, J.; Cools, T.; Verkest, A.; Corellou, F.; Babiychuk, E.; van der Schueren, E.; Beeckman, T.; Kushnir, S.; Inze, D.; et al. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 2007, 19, 211–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerinier, T.; Millan, L.; Crozet, P.; Oury, C.; Rey, F.; Valot, B.; Mathieu, C.; Vidal, J.; Hodges, M.; Thomas, M.; et al. Phosphorylation of p27(KIP1) homologs KRP6 and 7 by SNF1-related protein kinase-1 links plant energy homeostasis and cell proliferation. Plant J. 2013, 75, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Blomme, J.; Inze, D.; Gonzalez, N. The cell-cycle interactome: A source of growth regulators? J. Exp. Bot. 2014, 65, 2715–2730. [Google Scholar] [CrossRef] [PubMed]
- Dudits, D.; Abraham, E.; Miskolczi, P.; Ayaydin, F.; Bilgin, M.; Horvath, G.V. Cell-cycle control as a target for calcium, hormonal and developmental signals: The role of phosphorylation in the retinoblastoma-centred pathway. Ann. Bot. 2011, 107, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Wildwater, M.; Campilho, A.; Perez-Perez, J.M.; Heidstra, R.; Blilou, I.; Korthout, H.; Chatterjee, J.; Mariconti, L.; Gruissem, W.; Scheres, B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 2005, 123, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Weimer, A.K.; Nowack, M.K.; Bouyer, D.; Zhao, X.; Harashima, H.; Naseer, S.; de Winter, F.; Dissmeyer, N.; Geldner, N.; Schnittger, A. Retinoblastoma related1 regulates asymmetric cell divisions in Arabidopsis. Plant Cell 2012, 24, 4083–4095. [Google Scholar] [CrossRef] [PubMed]
- Forzani, C.; Aichinger, E.; Sornay, E.; Willemsen, V.; Laux, T.; Dewitte, W.; Murray, J.A. WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr. Biol. 2014, 24, 1939–1944. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Perilli, S.; Sabatini, S. Analysis of root meristem size development. Methods Mol. Biol. 2010, 655, 177–187. [Google Scholar] [PubMed]
- Gutierrez, C. The Arabidopsis cell division cycle. Arabidopsis Book 2009, 7, e0120. [Google Scholar] [CrossRef] [PubMed]
- Colon-Carmona, A.; You, R.; Haimovitch-Gal, T.; Doerner, P. Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999, 20, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Sasabe, M.; Boudolf, V.; de Veylder, L.; Inze, D.; Genschik, P.; Machida, Y. Phosphorylation of a mitotic kinesin-like protein and a MAPKKK by cyclin-dependent kinases (CDKs) is involved in the transition to cytokinesis in plants. Proc. Natl. Acad. Sci. USA 2011, 108, 17844–17849. [Google Scholar] [CrossRef] [PubMed]
- Sasabe, M.; Machida, Y. Regulation of organization and function of microtubules by the mitogen-activated protein kinase cascade during plant cytokinesis. Cytoskeleton 2012, 69, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Vanstraelen, M.; Baloban, M.; Da Ines, O.; Cultrone, A.; Lammens, T.; Boudolf, V.; Brown, S.C.; de Veylder, L.; Mergaert, P.; Kondorosi, E. APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2009, 106, 11806–11811. [Google Scholar] [CrossRef] [PubMed]
- Menges, M.; de Jager, S.M.; Gruissem, W.; Murray, J.A. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005, 41, 546–566. [Google Scholar] [CrossRef] [PubMed]
- Churchman, M.L.; Brown, M.L.; Kato, N.; Kirik, V.; Hulskamp, M.; Inze, D.; de Veylder, L.; Walker, J.D.; Zheng, Z.; Oppenheimer, D.G.; et al. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 2006, 18, 3145–3157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, B.; Nieuwland, J.; Murray, J.A. The Arabidopsis CDK inhibitor ICK3/KRP5 is rate limiting for primary root growth and promotes growth through cell elongation and endoreduplication. J. Exp. Bot. 2013, 64, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Boudolf, V.; Lammens, T.; Boruc, J.; Van Leene, J.; van den Daele, H.; Maes, S.; van Isterdael, G.; Russinova, E.; Kondorosi, E.; Witters, E.; et al. CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 2009, 150, 1482–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.O.; Davidson, J.M.; Duronio, R.J. Endoreplication: Polyploidy with purpose. Genes Dev. 2009, 23, 2461–2477. [Google Scholar] [CrossRef] [PubMed]
- Jovtchev, G.; Schubert, V.; Meister, A.; Barow, M.; Schubert, I. Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 2006, 114, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Sliwinska, E.; Mathur, J.; Bewley, J.D. On the relationship between endoreduplication and collet hair initiation and tip growth, as determined using six Arabidopsis thaliana root-hair mutants. J. Exp. Bot. 2015. [Google Scholar] [CrossRef] [PubMed]
- Forouzesh, E.; Goel, A.; Mackenzie, S.A.; Turner, J.A. In vivo extraction of Arabidopsis cell turgor pressure using nanoindentation in conjunction with finite element modeling. Plant J. 2013, 73, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.H.; Holman, T.J.; Sorensen, I.; Cancho-Sanchez, E.; Wells, D.M.; Swarup, R.; Knox, J.P.; Willats, W.G.; Ubeda-Tomas, S.; Holdsworth, M.; et al. Multi-omics analysis identifies genes mediating the extension of cell walls in the Arabidopsis thaliana root elongation zone. Front. Cell Dev. Biol. 2015, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Rayle, D.L.; Cleland, R.E. The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992, 99, 1271–1274. [Google Scholar] [CrossRef] [PubMed]
- McQueen-Mason, S.; Cosgrove, D.J. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc. Natl. Acad. Sci. USA 1994, 91, 6574–6578. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Bashline, L.; Lei, L.; Gu, Y. Cellulose synthesis and its regulation. Arabidopsis Book 2014, 12, e0169. [Google Scholar] [CrossRef] [PubMed]
- Barragan, V.; Leidi, E.O.; Andres, Z.; Rubio, L.; De Luca, A.; Fernandez, J.A.; Cubero, B.; Pardo, J.M. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 2012, 24, 1127–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippar, K.; Ivashikina, N.; Ache, P.; Christian, M.; Luthen, H.; Palme, K.; Hedrich, R. Auxin activates KAT1 and KAT2, two K+-channel genes expressed in seedlings of Arabidopsis thaliana. Plant J. 2004, 37, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Cramer, G.R.; Lynch, J.; Lauchli, A.; Epstein, E. Influx of Na, K, and Ca into roots of salt-stressed cotton seedlings: Effects of supplemental Ca. Plant Physiol. 1987, 83, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Bichet, A.; Desnos, T.; Turner, S.; Gradjean, O.; Höfte, H. BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 2001, 25, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Burk, D.H.; Liu, B.; Zhong, R.; Morrison, W.H.; Ye, Z.H. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 2001, 13, 807–827. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Ioki, M.; Braybrook, S.; Li, S.; Ye, Z.H.; Julie Lee, Y.R.; Hotta, T.; Chang, A.; Tian, J.; Wang, G.; et al. Kinesin-4 Functions in vesicular transport on cortical microtubules and regulates cell wall mechanics during cell elongation in plants. Mol. Plant 2015, 8, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- De Cnodder, T.; Vissenberg, K.; van der Straeten, D.; Verbelen, J.P. Regulation of cell length in the Arabidopsis thaliana root by the ethylene precursor 1-aminocyclopropane-1-carboxylic acid: A matter of apoplastic reactions. New Phytol. 2005, 168, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Grierson, C.; Nielsen, E.; Ketelaarc, T.; Schiefelbein, J. Root hairs. Arabidopsis Book 2014, 12, e0172. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.D.; Yi, K.; Breuninger, H.; Catarino, B.; Menand, B.; Dolan, L. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl. Acad. Sci. USA 2013, 110, 9571–9576. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef] [PubMed]
- De Vos, D.; Vissenberg, K.; Broeckhove, J.; Beemster, G.T. Putting theory to the test: Which regulatory mechanisms can drive realistic growth of a root? PLoS Comput. Biol. 2014, 10, e1003910. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Dai, X.; de-Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Dello Ioio, R.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A genetic framework for the control of cell division and differentiation in the root meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Galweiler, L.; Guan, C.; Muller, A.; Wisman, E.; Mendgen, K.; Yephremov, A.; Palme, K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 1998, 282, 2226–2230. [Google Scholar] [PubMed]
- Grieneisen, V.A.; Xu, J.; Marée, A.F.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007, 449, 1008–1013. [Google Scholar] [CrossRef] [PubMed]
- Band, L.R.; Wells, D.M.; Fozard, J.A.; Ghetiu, T.; French, A.P.; Pound, M.P.; Wilson, M.H.; Yu, L.; Li, W.; Hijazi, H.I.; et al. Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 2014, 26, 862–875. [Google Scholar] [CrossRef] [PubMed]
- Moubayidin, L.; Perilli, S.; dello Ioio, R.; di Mambro, R.; Costantino, P.; Sabatini, S. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 2010, 20, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Mockaitis, K.; Estelle, M. Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 2008, 24, 55–80. [Google Scholar] [CrossRef] [PubMed]
- Mahonen, A.P.; ten Tusscher, K.; Siligato, R.; Smetana, O.; Diaz-Trivino, S.; Salojarvi, J.; Wachsman, G.; Prasad, K.; Heidstra, R.; Scheres, B. PLETHORA gradient formation mechanism separates auxin responses. Nature 2014, 515, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wei, L.; Xu, J.; Zhai, Q.; Jiang, H.; Chen, R.; Chen, Q.; Sun, J.; Chu, J.; Zhu, L.; et al. Arabidopsis Tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 2010, 22, 3692–3709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Swarup, R.; Bennett, M.; Schaller, G.E.; Kieber, J.J. Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr. Biol. 2013, 23, 1979–1989. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Ramirez, A.; Diaz-Trivino, S.; Blilou, I.; Grieneisen, V.A.; Sozzani, R.; Zamioudis, C.; Miskolczi, P.; Nieuwland, J.; Benjamins, R.; Dhonukshe, P.; et al. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 2012, 150, 1002–1015. [Google Scholar] [CrossRef] [PubMed]
- Doerner, P.C.J. How are plant growth regulators involved in cell cycle control? Plant Horm. Res. 2000, 1–27. [Google Scholar]
- Tromas, A.B.N.; Muller, P.; Khodus, T.; Paponov, I.A.; Palme, K.; Ljung, K.; Lee, J.Y.; Benfey, P.; Murray, J.A.; Scheres, B.; et al. The AUXIN BINDING PROTEIN 1 is required for differential auxin responses mediating root growth. PLoS ONE 2009, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Roudier, F.; Fedorova, E.; Lebris, M.; Lecomte, P.; Gyorgyey, J.; Vaubert, D.; Horvath, G.; Abad, P.; Kondorosi, A.; Kondorosi, E. The Medicago species A2-type cyclin is auxin regulated and involved in meristem formation but dispensable for endoreduplication-associated developmental programs. Plant Physiol. 2003, 131, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Fujiwara, S.; Miura, K.; Stacey, N.; Yoshimura, M.; Schneider, K.; Adachi, S.; Minamisawa, K.; Umeda, M.; Sugimoto, K. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 2009, 21, 2284–2297. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Shimizu, K.; Ishida, T.; Sugimoto, K.; Umeda, M. Differential regulation of B2-type CDK accumulation in Arabidopsis roots. Plant Cell Rep. 2014, 33, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Kajihara, T.; Okamura, C.; Kim, Y.; Katagiri, Y.; Okushima, Y.; Matsunaga, S.; Hwang, I.; Umeda, M. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr. Biol. 2013, 23, 1812–1817. [Google Scholar] [CrossRef] [PubMed]
- Rück, A.; Palms, K.; Venis, M.A.; Napier, R.M.; Felle, H. Patch-clamp analysis establishes a role for an auxin binding protein in the auxin stimulation of plasma membrane current in Zea mays protoplasts. Plant J. 1993, 4, 41–46. [Google Scholar] [CrossRef]
- Thiel, G.; Blatt, M.R.; Fricker, M.D.; White, I.R.; Millner, P. Modulation of K+ channels in Vicia stomatal guard cells by peptide homologs to the auxin-binding protein C terminus. Proc. Natl. Acad. Sci. USA 1993, 90, 11493–11497. [Google Scholar] [CrossRef] [PubMed]
- Blancaflor, E.B.; Hasenstein, K.H. Growth and microtubule orientation of Zea mays roots subjected to osmotic stress. Int. J. Plant. Sci. 1995, 156, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Scheres, B. Cell polarity: ROPing the ends together. Curr. Opin. Plant Biol. 2005, 8, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Grandont, L.; Li, H.; Hauschild, R.; Paque, S.; Abuzeineh, A.; Rakusova, H.; Benkova, E.; Perrot-Rechenmann, C.; Friml, J. Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature 2014, 516, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jurgens, G.; Alonso, J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Ubeda-Tomas, S.; Beemster, G.T.; Bennett, M.J. Hormonal regulation of root growth: Integrating local activities into global behaviour. Trends Plant Sci. 2012, 17, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arabidopsis Book 2014, 12, e0168. [Google Scholar] [CrossRef] [PubMed]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, D.; Sonenberg, N. The Akt of translational control. Oncogene 2005, 24, 7426–7434. [Google Scholar] [CrossRef] [PubMed]
- Zinzalla, V.; Stracka, D.; Oppliger, W.; Hall, M.N. Activation of mTORC2 by association with the ribosome. Cell 2011, 144, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Serfontein, J.; Nisbet, R.E.; Howe, C.J.; de Vries, P.J. Evolution of the TSC1/TSC2-TOR signaling pathway. Sci. Signal. 2010, 3, ra49. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, T.J.; Zwartkruis, F.J.; Bos, J.L.; Snel, B. Evolution of the TOR pathway. J. Mol. Evol. 2011, 73, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Crespo, J.L.; Diaz-Troya, S.; Florencio, F.J. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 2005, 139, 1736–1749. [Google Scholar] [CrossRef] [PubMed]
- Imamura, S.; Ishiwata, A.; Watanabe, S.; Yoshikawa, H.; Tanaka, K. Expression of budding yeast FKBP12 confers rapamycin susceptibility to the unicellular red alga Cyanidioschyzon merolae. Biochem. Biophys. Res. Commun. 2013, 439, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, M.M.; Kim, S.; Delauney, A.J.; Verma, D.P. Arabidopsis target of rapamycin interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 2006, 18, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Sormani, R.; Yao, L.; Menand, B.; Ennar, N.; Lecampion, C.; Meyer, C.; Robaglia, C. Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 2007, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Menand, B.; Desnos, T.; Nussaume, L.; Berger, F.; Bouchez, D.; Meyer, C.; Robaglia, C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 2002, 99, 6422–6427. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Qiu, S.; Venglat, P.; Xiang, D.; Feng, L.; Selvaraj, G.; Datla, R. Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 2011, 155, 1367–1382. [Google Scholar] [CrossRef] [PubMed]
- Henriques, R.; Bogre, L.; Horvath, B.; Magyar, Z. Balancing act: matching growth with environment by the TOR signalling pathway. J. Exp. Bot. 2014, 65, 2691–2701. [Google Scholar] [CrossRef] [PubMed]
- Deprost, D.; Yao, L.; Sormani, R.; Moreau, M.; Leterreux, G.; Nicolai, M.; Bedu, M.; Robaglia, C.; Meyer, C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007, 8, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Sheen, J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 2012, 287, 2836–2842. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.H.; Veit, B.; Hanson, M.R. The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 2005, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Deprost, D.; Truong, H.N.; Robaglia, C.; Meyer, C. An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem. Biophys. Res. Commun. 2005, 326, 844–850. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.; Azzopardi, M.; Clement, G.; Dobrenel, T.; Marchive, C.; Renne, C.; Martin-Magniette, M.L.; Taconnat, L.; Renou, J.P.; Robaglia, C.; et al. Mutations in the Arabidopsis homolog of LST8/GbetaL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 2012, 24, 463–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, C.S.; Ahn, H.K.; Pai, H.S. Overexpression of the PP2A regulatory subunit Tap46 leads to enhanced plant growth through stimulation of the TOR signalling pathway. J. Exp. Bot. 2015, 66, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Robaglia, C.; Menand, B.; Lei, Y.; Sormani, R.; Nicolai, M.; Gery, C.; Teoule, E.; Deprost, D.; Meyer, C. Plant growth: The translational connection. Biochem. Soc. Trans. 2004, 32, 581–584. [Google Scholar] [CrossRef] [PubMed]
- Leiber, R.M.; John, F.; Verhertbruggen, Y.; Diet, A.; Knox, J.P.; Ringli, C. The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 2010, 22, 1898–1908. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bassham, D.C. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 2010, 5, e11883. [Google Scholar] [CrossRef] [PubMed]
- Schepetilnikov, M.; Dimitrova, M.; Mancera-Martinez, E.; Geldreich, A.; Keller, M.; Ryabova, L.A. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013, 32, 1087–1102. [Google Scholar] [CrossRef] [PubMed]
- Schepetilnikov, M.; Kobayashi, K.; Geldreich, A.; Caranta, C.; Robaglia, C.; Keller, M.; Ryabova, L.A. Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J. 2011, 30, 1343–1356. [Google Scholar] [CrossRef] [PubMed]
- Turck, F.; Kozma, S.C.; Thomas, G.; Nagy, F. A heat-sensitive Arabidopsis thaliana kinase substitutes for human p70s6k function in vivo. Mol. Cell. Biol. 1998, 18, 2038–2044. [Google Scholar] [PubMed]
- Turck, F.; Zilbermann, F.; Kozma, S.C.; Thomas, G.; Nagy, F. Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol. 2004, 134, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.J.; Werner-Fraczek, J.; Chang, I.F.; Bailey-Serres, J. Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol. 2003, 132, 2086–2097. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, S.I.; Zvelebil, M.J.; Shuttleworth, S.J.; Hancox, T.; Saghir, N.; Timms, J.F.; Waterfield, M.D. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem. J. 2007, 404, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Creff, A.; Sormani, R.; Desnos, T. The two Arabidopsis RPS6 genes, encoding for cytoplasmic ribosomal proteins S6, are functionally equivalent. Plant Mol. Biol. 2010, 73, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, B.; Ekim, B.; Fingar, D.C. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 2012, 441, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Henriques, R.; Magyar, Z.; Monardes, A.; Khan, S.; Zalejski, C.; Orellana, J.; Szabados, L.; de la Torre, C.; Koncz, C.; Bogre, L. Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J. 2010, 29, 2979–2993. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef] [PubMed]
- Caldana, C.; Li, Y.; Leisse, A.; Zhang, Y.; Bartholomaeus, L.; Fernie, A.R.; Willmitzer, L.; Giavalisco, P. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 2013, 73, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Kim, S.; Shin, Y.J.; Hur, Y.S.; Kim, W.Y.; Lee, M.S.; Cheon, C.I.; Verma, D.P. Ribosomal protein S6, a target of rapamycin, is involved in the regulation of rRNA genes by possible epigenetic changes in Arabidopsis. J. Biol. Chem. 2014, 289, 3901–3912. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardie, D.G. AMP-activated protein kinase: A cellular energy sensor with a key role in metabolic disorders and in cancer. Biochem. Soc. Trans. 2011, 39, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Polge, C.; Thomas, M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007, 12, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Robaglia, C.; Thomas, M.; Meyer, C. Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 2012, 15, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Emanuelle, S.; Hossain, M.I.; Moller, I.E.; Pedersen, H.L.; van de Meene, A.M.; Doblin, M.S.; Koay, A.; Oakhill, J.S.; Scott, J.W.; Willats, W.G.; et al. SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 2015, 82, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Ogawa-Ohnishi, M.; Mori, A.; Matsubayashi, Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 2010, 329, 1065–1067. [Google Scholar] [CrossRef] [PubMed]
- Menand, B. Reverse Genetics Study of the Gene Coding for the TARGET OF RAPAMYCIN protein in Arabidopsis thaliana (AtTOR), the Homolog of a Kinase Controling Growth in Eucaryotes. Ph.D. Thesis, Louis Pasteur University, Strasbourg, France, 2002. [Google Scholar]
- Rademacher, E.H.; Moller, B.; Lokerse, A.S.; Llavata-Peris, C.I.; van den Berg, W.; Weijers, D. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011, 68, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Betz, C.; Hall, M.N. Where is mTOR and what is it doing there? J. Cell Biol. 2013, 203, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.P.; Ludwig, C.; Zampieri, M.; Weisser, H.; Aebersold, R.; Sauer, U. Dynamic phosphoproteomics reveals TORC1-dependent regulation of yeast nucleotide and amino acid biosynthesis. Sci. Signal. 2015, 8, rs4. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrada, A.; Montané, M.-H.; Robaglia, C.; Menand, B. Spatial Regulation of Root Growth: Placing the Plant TOR Pathway in a Developmental Perspective. Int. J. Mol. Sci. 2015, 16, 19671-19697. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160819671
Barrada A, Montané M-H, Robaglia C, Menand B. Spatial Regulation of Root Growth: Placing the Plant TOR Pathway in a Developmental Perspective. International Journal of Molecular Sciences. 2015; 16(8):19671-19697. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160819671
Chicago/Turabian StyleBarrada, Adam, Marie-Hélène Montané, Christophe Robaglia, and Benoît Menand. 2015. "Spatial Regulation of Root Growth: Placing the Plant TOR Pathway in a Developmental Perspective" International Journal of Molecular Sciences 16, no. 8: 19671-19697. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160819671