Interaction of DNA with Simple and Mixed Ligand Copper(II) Complexes of 1,10-Phenanthrolines as Studied by DNA-Fiber EPR Spectroscopy
Abstract
:1. Introduction
2. DNA-Fiber EPR Spectra of Cu(II) Complexes


3. Interaction of Cu(II) Complexes of Phen and Its Derivatives with DNA
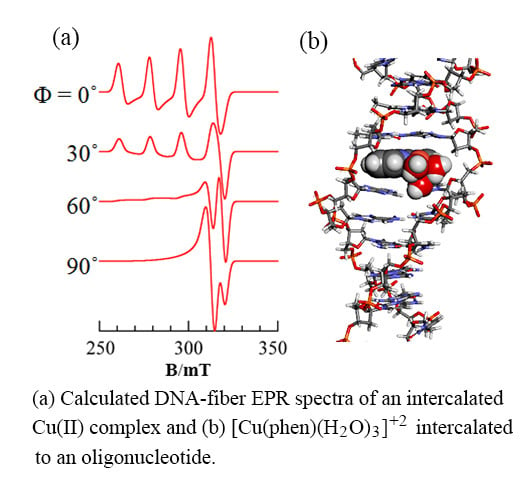
3.1. 1:1 Complexes of Cu(II) with Phen and Its Methyl Derivatives




- (1).
- It is not clear if mono-phen Cu(II) complex intercalates as [Cu(phen)(H2O)2]2+ after removing the apically coordinated water molecule from [Cu(phen)(H2O)3]2+ or as [Cu(phen)(H2O)4]2+ keeping the two apically coordinated water molecules. At pH 7.0, the presence of [Cu(phen)(H2O)2(OH)]+, [Cu(5,6-dmp)(H2O)2(OH)]+, or [Cu(2,9-dmp)(H2O)2(OH)]+ should also be considered. The positive charge of the complexes decreases by dissociation of a proton from the coordinating water molecule, which does not favor binding to a negatively charged oligo-nucleotide. However, the removal of an apically coordinated water molecule results in the formation of flat coordination plane, which is favorable for intercalative binding.
- (2).
- It would be interesting to investigate whether one of the coordinating water molecules in the complex is substituted with phosphate oxygen or N7 of the guanine residue to produce a covalent interaction.
- (3).
- It is also important to investigate how the binding affinity changes when effecting changes in the base sequence of nucleotides.
3.2. Ternary Complexes with Amino Acids, [Cu(phen)(AA)]n+(n = 1 or 2)


3.3. Cu(II) Complexes of 1,10-Phenanthroline-derived Alkyl Amine


3.4. [Cu(phen)2(H2O)]2+

3.5. Ternary Copper(II)–Phen–Edda Complex [Cu(phen)(edda)]



3.6. Ternary Cu(II) Complexes of Cationic Schiff Bases and N-Heteroaromatic Diimines

| Complex | Frozen Solution (−150 °C) | B-Form DNA Fiber (r.t.) * | ||
|---|---|---|---|---|
| g|| | A||/(10−4 cm−1) | g|| | A||/(10−4 cm−1) | |
| 1 | 2.26 | 165 | 2.23 | 184 |
| 2 | 2.26 | 167 | 2.23 | 178 |
| 3 | 2.26 | 170 | 2.23 | 178 |


3.7. Binary Complex of Cu(II) with 2,2′-Bipyridine (bpy)

4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sigel, A.; Siegel, H. (Eds.) Metal Ions in Biological Systems; Marcel Dekker Inc.: New York, NY, USA, 1996; Volumes 32, 33.
- Sigel, A.; Siegel, H. (Eds.) Metal Ions in Biological Systems; Marcel Dekker Inc.: New York, NY, USA, 2004; Volume 42.
- Boersma, A.J.; Megens, R.P.; Feringa, B.L.; Roelfes, G. DNA-based asymmetric catalysis. Chem. Soc. Rev. 2010, 39, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- Roelfes, G.; Boersma, A.J.; Feringa, B.L. Highly enantioselective DNA-based catalysis. Chem. Commun. 2006, 6, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Sugiyama, H. DNA as a chiral scaffold for asymmetric synthesis. Molecules 2012, 17, 12792–12803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Draksharapu, A.; Boersma, A.J.; Leising, M.; Meetsma, A; Browne, W.R.; Roelfes, G. Binding of Cu(II) polypyridyl complexes to DNA and consequences for DNA-based asymmetric catalysis. Dalton Trans. 2015, 44, 3647–3655. [Google Scholar] [CrossRef] [PubMed]
- Sigman, D.S.; Graham, D.R.; D’Aurora, V.; Stern, A.M. Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J. Biol. Chem. 1979, 254, 12269–12272. [Google Scholar] [PubMed]
- Sigman, D.S.; Mazumder, A.; Perrin, D.M. Chemical nucleases. Chem. Rev. 1993, 93, 2295–2316. [Google Scholar] [CrossRef]
- Sigman, D.S.; Landgraf, R.; Perrin, D.M.; Pearson, L. Nucleic acid chemistry of the cuprous complexes of 1,10-phenanthroline and derivatives. In Metal Ions in Biological Systems; Sigel, A., Sigel, H., Eds.; Dekker: New York, NY, USA, 1996; Volume 33, pp. 485–513. [Google Scholar]
- Downey, K.M.; Que, B.G.; So, A.G. Degradation of DNA by 1,10-phenanthroline. Biochem. Biophys. Res. Commun. 1980, 93, 264–270. [Google Scholar] [CrossRef]
- Veal, J.M.; Rill, R.L. Sequence specificity of DNA cleavage by bis(1,10)-phenanthroline Cu(I): Effects of single base pair transitions on the cleavage of preferred pyrimidine-purine-pyrimidine triplets. Biochemistry 1989, 28, 3243–3250. [Google Scholar] [CrossRef] [PubMed]
- Veal, J.M.; Rill, R.L. Noncovalent DNA binding of bis(1,10-phenanthroline)Cu(I) and related compounds. Biochemistry 1991, 30, 1132–1140. [Google Scholar] [CrossRef] [PubMed]
- Veal, J.M.; Merchant, K.; Rill, R.L. The influence of reducing agent and 1,10-phenanthroline concentration on DNA cleavage by phenanthroline + copper. Nucleic Acids Res. 1991, 19, 3383–3388. [Google Scholar] [CrossRef] [PubMed]
- Ranford, J.D.; Sadler, P.J.; Tocher, D.A. Cytotoxicity and antiviral activity of transition-metal salicylato complexes and crystal structure of bis(diisopropylsalicylato)(1,10-phenanthroline)Cu(II). J. Chem. Soc. Dalton Trans. 1993, 33, 3393–3399. [Google Scholar] [CrossRef]
- Patra, A.K.; Nethaji, M.; Chakravarty, A.R. Red-light photosensitized cleavage of DNA by (l-lysine)(phenanthroline base)copper(II) complexes. Dalton Trans. 2005, 6, 2798–2804. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.K.; Nethaji, M.; Chakravarty, A.R. Synthesis, crystal structure, DNA binding and photo-induced DNA cleavage activity of (S-methyl-l-cysteine)-Cu(II) complexes of heterocyclic bases. J. Inorg. Biochem. 2007, 101, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.K.; Bhowmick, T.; Roy, S.; Ramakumar, S.; Chakravarty, A.R. Cu(II) Complexes of l-arginine as netropsin mimics showing DNA cleavage activity in red light. Inorg. Chem. 2009, 48, 2932–2943. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Saha, S.; Majumdar, R.; Dighe, R.R; Chakravarty, A.R. DNA photocleavage and anticancer activity of terpyridine Cu(II) complexes having phenanthroline bases. Polyhedron 2010, 29, 2787–2794. [Google Scholar] [CrossRef]
- Selvakumar, B.; Rajendiran, V.; Maheswari, P.U.; Stoeckli-Evans, H.; Palaniandavar, M. Structures, spectra, and DNA-binding properties of mixed ligand Cu(II) complexes of iminodiacetic acid: The novel role of diimine co-ligands on DNA conformation and hydrolytic and oxidative double strand DNA cleavage. J. Inorg. Biochem. 2006, 100, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Rajendiran, V.; Karthik, R.; Palaniandavar, M.; Stoeckli-Evans, H.; Periasamy, V.S.; Akbarsha, M.A.; Srinag, B.S.; Krishnamurthy, H. Mixed-ligand Cu(II)-phenolate complexes: effect of coligand on enhanced DNA and protein binding, DNA cleavage, and anticancer activity. Inorg. Chem. 2007, 46, 8208–8221. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, S.; Palaniandavar, M. Spectroscopic and voltammetric studies on copper complexes of 2,9-dimethyl-1,10-phenanthrolines bound to calf thymus DNA. Inorg. Chem. 2007, 37, 693–700. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Rajendiran, V.; Palaniandavar, M.; Periasamy, V.S.; Srinag, B.S.; Krishnamurthy, H.; Akbarsha, M.A. Induction of cell death by ternary Cu(II) complexes of l-tyrosine and diimines: Role of coligands on DNA binding and cleavage and anticancer activity. Inorg. Chem. 2009, 48, 1309–1322. [Google Scholar] [CrossRef] [PubMed]
- Alemón-Medina, R.; Breña-Valle, M.; Muñoz-Sánchez, J.L.; Gracia-Mora, M.I.; Ruiz-Azuara, L. Induction of oxidative damage by copper-based antineoplastic drugs (Casiopeı´nas®). Cancer Chemother. Pharmacol. 2007, 60, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Ambundo, E.A.; Deydier, M.-V.; Grall, A.J.; Aguera-Vega, N.; Dresel, L.T.; Cooper, T.H.; Heeg, M.J.; Ochrymowycz, L.A.; Rorabacher, D.B. Influence of coordination geometry upon Cu(II/I) redox potentials. Physical parameters for twelve Cu tripodal ligand complexes. Inorg. Chem. 1999, 38, 4233–4242. [Google Scholar] [CrossRef]
- Ng, C.H.; Kong, K.C.; Von, S.T.; Balra, P.; Jensen, P.; Thirthagir, E.; Hamada, H.; Chikira, M. Synthesis, characterization, DNA-binding study and anticancer properties of ternary metal(II) complexes of edda and an intercalating ligand. Dalton Trans. 2008, 4, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Chikira, M. DNA-fiber EPR spectroscopy as a tool to study DNA–metal complex interactions: DNA binding of hydrated Cu(II) ions and Cu(II) complexes of amino acids and peptides. J. Inorg. Biochem. 2008, 102, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Chikira, M.; Tomizawa, Y.; Fukita, T.; Sugisaki, D.; Sugawara, N.; Yamazaki, T.; Sasano, A.; Shindo, H.; Palaniandavar, M.; Antholine, W.E. DNA-fiber EPR study of the orientation of Cu(II) complexes of 1,10-phenanthroline and its derivatives bound to DNA: Mono(phenanthroline)-Cu(II) and its ternary complexes with amino acids. J. Inorg. Biochem. 2002, 89, 163–173. [Google Scholar] [CrossRef]
- Hirohama, T.; Kuranuki, Y.; Ebina, E.; Sugizaki, T.; Arii, H.; Chikira, M.; Selvi, P.T.; Palaniandavar, M. Cu(II) complexes of 1,10-phenanthroline-derived ligands: Studies on DNA binding properties and nuclease activity. J. Inorg. Biochem. 2005, 99, 1205–1219. [Google Scholar] [CrossRef] [PubMed]
- Atherton, N.M. Electron Spin Resonance, Theory and Applications; John Wiley & Sons Inc.: New York, NY, USA, 1973. [Google Scholar]
- Wilson, R.; Kivelson, D. ESR Linewidths in Solution. IV. Experimental studies of anisotropic and spin - Rotational effects in Cu complexes. J. Chem. Phys. 1966, 44, 4445–4452. [Google Scholar] [CrossRef]
- Chikira, M.; Suda, S.; Nakabayashi, T.; Fujiwara, Y.; Ejiri, T.; Yoshikawa, M.; Kobayashi, N.; Shindo, H. Electron spin resonance study of the binding structures of cationic water-soluble metalloporphyrins on highly oriented deoxyribonucleic acid fibres. J. Chem. Soc. Dalton Trans. 1995, 8, 1325–1331. [Google Scholar] [CrossRef]
- Shindo, H.; Woolen, J.B.; Pheiffer, B.S.; Zimmerman, B. Nonuniform backbone conformation of deoxyribonucleic acid indicated by phosphorus-31 nuclear magnetic resonance chemical shift anisotropy. Biochemistry 1980, 19, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Chikira, M.; Iiyama, T.; Sakamoto, K.; Antholine, W.E.; Petering, D.H. Orientation of iron bleomycin and porphyrin complexes on DNA fibers. Inorg. Chem. 2000, 39, 1779–1786. [Google Scholar] [CrossRef] [PubMed]
- Maki, A.H.; McGarvey, B.R. Electron spin resonance in transition metal chelates. I. Cu(II) bis-acetylacetonate. J. Chem. Phys. 1958, 29, 31–34. [Google Scholar] [CrossRef]
- Maki, A.H.; McGarvey, B.R. Electron spin resonance in transition metal chelates. II. Cu(II) bis-salicylaldehyde-imine. J. Chem. Phys. 1958, 29, 35–38. [Google Scholar] [CrossRef]
- Gustafson, R.L.; Martell, A.E. Hydrolytic tendencies of metal chelate compounds. V. Hydrolysis and dimerization of Cu(II) chelates of 1,2-diamines. J. Am. Chem. Soc. 1959, 81, 525–529. [Google Scholar] [CrossRef]
- Perrin, D.; Sharma, V. Complex formation by hydrolysed Cu(II) ions in aqueous solution. J. Inorg. Nucl. Chem. 1966, 28, 1271–1278. [Google Scholar] [CrossRef]
- Lu, L.-P.; Zhu, M.-L.; Yang, P. Crystal structure and nuclease activity of mono(1,10-phenanthroline) copper complex. J. Inorg. Biochem. 2003, 95, 31–36. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09 Revision D.01; Release Notes; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. AMBER 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.-Y. R.E.D. Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011, 39, W511–W517. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, S.; Palaniandavar, M. Spectral and Electrochemical Behavior of Cu(II)-phenanthrolines bound to calf thymus DNA. [(5,6-dimethyl-OP)2Cu]2+ (5,6-dimethyl-OP) 5,6-dimethyl-1,10-phenanthroline) induces a conformational transition from B to Z DNA. Inorg. Chem. 1998, 37, 3927–3934. [Google Scholar] [CrossRef] [PubMed]
- Preston, H.S.; Kennard, C.H.L. Stereochemistry of rigid chelate–metal complexes. Part III. Crystal structure of dichloroaquo-(2,9-dimethyl-1,10-phenanthroline)-Cu(II). J. Chem. Soc. A 1969, 2955–2958. [Google Scholar] [CrossRef]
- Addison, A.W.; Rao, T.N.; Reedijk, J.; van Rijn, J.; Verschoor, G.C. Synthesis, structure, and spectroscopic properties of Cu(II) compounds containing nitrogen-sulphur donor ligands; the crystal and molecular structure of aqua[l,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]Cu(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 1349–1356. [Google Scholar] [CrossRef]
- Seng, H.-L.; Wang, W.-S.; Kong, S.-M.; Alan Ong, H.-K.; Win, Y.-F.; Abd Rahman, R.N.Z.R.; Chikira, M.; Leong, W.-K.; Ahmad, M.-B.; Khoo, A.; et al. Biological and cytoselective anticancer properties of Cu(II)-polypyridyl complexes modulated by auxillary methylated glycine ligand. Biometals 2012, 25, 1061–1081. [Google Scholar] [CrossRef] [PubMed]
- Malik, G.S.; Singh, S.P.; Tandon, J.P. Mixed ligand complexes involving ligands of biological importance [Cu(II)-1,10-phenanthroline or 2,2′-bipyridyl-α-amino acids]. Monatsch. Chem. 1979, 110, 149–155. [Google Scholar]
- Yang, Y.; Pogni, R.; Basosi, R. Mixed-ligand complexes of Cu(II)-1,10-o-phenanthroline and its analogues characterized by computer-aided electron spin resonance spectroscopy. J. Chem. Soc. Faraday Trans. 1 1989, 85, 3995–4009. [Google Scholar] [CrossRef]
- Wang, Z.-M.; Lin, H.-K.; Zhou, Z.-F.; Xu, M.; Liu, T.-F.; Zhu, S.-R.; Chen, Y.-T. Cu(II) complexes with N,N′-dialkyl-1,10-phenanthroline–2,9-dimethanamine: synthesis, characterization, DNA-binding thermodynamical and kinetic studies. Bioorg. Med. Chem. 2001, 9, 2849–2855. [Google Scholar] [CrossRef]
- Harada, W.; Nojima, T.; Shibayama, A.; Ueda, H.; Shindo, H.; Chikira, M. How amino acids control the binding of Cu(II) ions to DNA: I—The role of the hydroxyl group of serine and threonine in fixing the orientation of the complexes. J. Inorg. Biochem. 1996, 64, 273–285. [Google Scholar] [CrossRef]
- Murphy, G.; Nagle, P.; Murphy, B.; Hathaway, B. Crystal structures, electronic properties and structural pathways of four [Cu(phen)2Cl][Y] complexes (Y = BF4−·0.5H2O, PF6−, CF3SO3−·H2O or BPh4−). J. Chem. Soc. Dalton Trans. 1997, 2645–2652. [Google Scholar] [CrossRef]
- Murphy, G.; O’Sullivan, C.; Murphy, B.; Hathaway, B. Comparative crystallography 5. Crystal structures, electronic properties, and structural pathways of five [Cu(phen)2Br][Y] complexes, Y = [Br]−1 H2O, [ClO4]−, [NO3]− H2O, [PF6]−, and [BPh4]−. Inorg. Chem. 1998, 37, 240–248. [Google Scholar] [CrossRef]
- Sigman, D.S.; Spassky, A.; Rimsky, S.; Buc, H. Conformational analysis of lac promoters using the nuclease activity of 1,10-phenanthroline-copper ion. Biopolymers 1985, 24, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Drew, H.R. Structural specificities of five commonly used DNA nucleases. J. Mol. Biol. 1984, 176, 535–557. [Google Scholar] [CrossRef]
- Pope, L.E.; Sigman, D.S. Secondary structure specificity of the nuclease activity of the 1,10-phenanthroline-copper complex. Proc. Natl. Acad. Sci. USA 1984, 81, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, M.D.; Yoon, C.; Goyne, T.; Thederahn, T.B.; Sigman, D.S. Nuclease activity of 1,10-phenanthroline-copper ion: Reaction with CGCGAATTCGCG and its complexes with netropsin and EcoRI. Biochemistry 1986, 25, 7401–7408. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, F.; Rimsky, S.; Spassky, A. DNA-stacking interactions determine the sequence specificity of the deoxyribonuclease activity of 1,10-phenanthroline-copper ion. J. Mol. Biol. 1996, 260, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Robertazzi, A.; Vargiu, A.V.; Magistrato, A.; Ruggerone, P.; Carloni, P.; Hoog, P.; Reedijk, J. Copper-1,10-phenanthroline complexes binding to DNA: Structural predictions from molecular simulations. J. Phys. Chem. B 2009, 113, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Von, S.-T.; Seng, H.L.; Lee, H.-B.; Ng, S.W.; Chikira, M.; Kitamura, Y.; Ng, C.-H. DNA molecular recognition and cellular selectivity of anticancer metal(II) complexes of EDDA and phenanthroline: Multiple targets. J. Biol. Inorg. Chem. 2012, 17, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Iwasaki, Y.; Kimura, M.; Yamada, A.; Muto, Y.; Tateishi, M.; Arii, H.; Kitamura, Y.; Chikira, M. Conformational change of ternary Cu(II) complexes of cationic Schiff-bases and N-heteroaromatic amines induced by intercalative binding to DNA. Inorg. Chem. Commun. 2011, 14, 1461–1464. [Google Scholar] [CrossRef]
- Kivelson, N.; Neiman, R. ESR line shapes in glasses of copper complexes. J. Chem. Phys. 1961, 35, 149–155. [Google Scholar] [CrossRef]
- Cusumano, M.; Giannetto, A. The Interaction of mixed-ligand square-planar complexes with calf thymus DNA. J. Inorg. Biochem. 1997, 65, 137–144. [Google Scholar] [CrossRef]
- Masuda, H.; Yamauchi, O. A structural basis for nucleic base-metallointercalator lnteractions: Crystal structure of [Pt(2,2′-bipyridine)(ethylenediamine)]·AMP·10H2O (AMP = adenosine 5′-monophosphate). Inorg. Chim. Acta 1987, 136, L29–L31. [Google Scholar] [CrossRef]
- Yamauchi, O.; Odani, A.; Shimata, R.; Kosaka, Y. Spectroscopic evidence for stacking and electrostatic interactions between nucleoside 5′-monophosphates and a platinum DNA intercalator, (2,2′-bipyridine)(ethylenediamine)platinum(II) in dilute aqueous solution. Inorg. Chem. 1986, 25, 3337–3339. [Google Scholar] [CrossRef]
- Odani, A.; Shimata, R.; Masuda, H.; Yamauchi, O. Platinum DNA intercalator-mononucleotide adduct formation. Cooperativity between aromatic ring stacking and electrostatic interactions. Inorg. Chem. 1991, 30, 2133–2138. [Google Scholar] [CrossRef]
- Goto, M.; Matsumoto, T.; Sumimoto, M.; Kurosaki, H. Intramolecular stacking of two aromatic rings in the platinum(II) coordination sphere: Preparation, crystal structures, and 1H-NMR spectra of bipyridine(N-arylmethyl-1,2-ethanediamine)-platinum(II) nitrate. Bull. Chem. Soc. Jpn. 2000, 73, 97–105. [Google Scholar] [CrossRef]
- Goto, M.; Sumimoto, M.; Matsumoto, T.; Iwasaki, M.; Tanaka, H.; Kurosaki, Y. 1H-NMR spectra of ternary platinum(II) complexes with N-ethyl- or N-benzyl-1,2-ethanediamine and 2,2′-bipyridine or 1,10-phenanthroline: Intramolecular aromatic-aromatic interaction in coordination sphere, and it’s solvent and temperature effects. Bull. Chem. Soc. Jpn. 2000, 73, 1589–1598. [Google Scholar] [CrossRef]
- Lueth, M.S.; Kapinos, L.E.; Song, B.; Sigel, H.; Lippert, B. Extent of intramolecular stacking interactions in the mixed-ligand complexes formed in aqueous solution by Cu(II), 2,2′-bipyridine or 1,10-phenanthroline and 2′-deoxyguanosine 5′-monophosphate. J. Chem. Soc. Dalton Trans. 1999, 3, 357–365. [Google Scholar] [CrossRef]
- Gilbert, B.C.; Silvester, S.; Walton, P.H.; Whitewood, A.C. DNA damage via intercalation of copper complexes and activation by ascorbate and peroxides: Direct EPR evidence for hydroxyl radical formation and reaction. J. Chem. Soc. Perkin Trans. 1999, II 9, 1891–1895. [Google Scholar] [CrossRef]
- Götz, A.W.; Clark, M.A.; Walker, R.C. An extensible interface for ab initio QM/MM molecular dynamics simulations with Amber. J. Comput. Chem. 2014, 35, 95–108. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chikira, M.; Ng, C.H.; Palaniandavar, M. Interaction of DNA with Simple and Mixed Ligand Copper(II) Complexes of 1,10-Phenanthrolines as Studied by DNA-Fiber EPR Spectroscopy. Int. J. Mol. Sci. 2015, 16, 22754-22780. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160922754
Chikira M, Ng CH, Palaniandavar M. Interaction of DNA with Simple and Mixed Ligand Copper(II) Complexes of 1,10-Phenanthrolines as Studied by DNA-Fiber EPR Spectroscopy. International Journal of Molecular Sciences. 2015; 16(9):22754-22780. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160922754
Chicago/Turabian StyleChikira, Makoto, Chew Hee Ng, and Mallayan Palaniandavar. 2015. "Interaction of DNA with Simple and Mixed Ligand Copper(II) Complexes of 1,10-Phenanthrolines as Studied by DNA-Fiber EPR Spectroscopy" International Journal of Molecular Sciences 16, no. 9: 22754-22780. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160922754