miR-29a Participated in Nacre Formation and Immune Response by Targeting Y2R in Pinctada martensii
Abstract
:1. Introduction
2. Results
2.1. Sequence Verification of Mature Pm (Pinctada martensii)-miR-29a

2.2. Expression and Distribution of Pm-miR-29a in Different Tissues

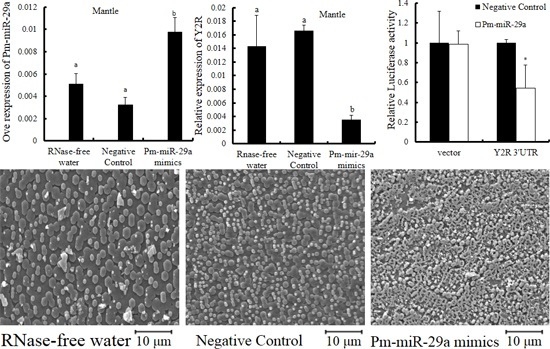
2.3. Functions of Pm-miR-29a in Nacreous Layer Formation

2.4. Target Prediction
2.5. Target Verification


3. Discussion
4. Experimental Section
4.1. Experimental Material
4.2. Small RNA Extraction and Template Preparation
| Primer Name | Primer Sequence | Function |
|---|---|---|
| 5′ adaptor | CGACUGGAGCACGAGGACACUGAAAA | miR-RACE |
| GSP1 | TTTTTTAAACTGATTTCAAATGGTGC | miR-RACE |
| mirRacer 5′ primer | CTGGAGCACGAGGACACTGA | miR-RACE |
| GSP2 | CTGAAAATAGCACCATTTGAAATCA | miR-RACE |
| mirRacer 3′primer | ATTCTAGAGGCCGAGGCGGCCGACATG | miR-RACE |
| RT-primer | ATTCTAGAGGCCGAGGCGGCCGACATGTTTTTTTTTTTTTTTTTTTTTTT | miR-RACE |
| Pm-miR-29a (F) | TAGCACCATTTGAAATCAGTTT | qRT-PCR |
| Pm-miR-29a (R) | TGCGTGTCGTGGAGTC | qRT-PCR |
| Pm-miR-29a (RT) | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAAACTGAT | qRT-PCR |
| U6 (F) | ATTGGAACGATACAGAGAAGATT | qRT-PCR |
| U6 (R) | ATTTGCGTGTCATCCTTGC | qRT-PCR |
| U6 (RT) | ATTTGCGTGTCATCCTTGC | qRT-PCR |
| GAPDH (F) | CACTCGCCAAGATAATCAACG | qRT-PCR |
| GAPDH (R) | CCATTCCTGTCAACTTCCCAT | qRT-PCR |
| Y2R-F1 | CGGACTAGTTCAAATATTCGATCGTGGGGAGCGTG | Vector Constructs |
| Y2R-R1 | CCAAGCTTCGTGATGAGCCGATGACCTCTCTTGA | Vector Constructs |
| FGF18-F | CGGACTAGTTTCGGCACAGACGGGTAACATTTCC | Vector Constructs |
| FGF18-R | CCCAAGCTTTACTGGCCATGGGATCCTCGGTGT | Vector Constructs |
| SK-F | CGGACTAGTGTGATGAAAAATGCAAATCAGGGTC | Vector Constructs |
| SK-R | CCCAAGCTTAAATGCCATGTCGGAATTCAGTATATAC | Vector Constructs |
| Y2R-F2 | TAGGGAGAACTTTAGCGGTCAA | qRT-PCR |
| Y2R-R2 | AAATCCAATCGCAATGAGACC | qRT-PCR |
| NF-κB-F | AGAAGAGACAGGCCAAAGAGCA | qRT-PCR |
| NF-κB-R | AGAGAGAACAGGCGTGAGAAGC | qRT-PCR |
| IL-17-F | AAGAAAACTTTGAACATGCCGTAC | qRT-PCR |
| IL-17-R | TAATCACATAATGCCAGGGACA | qRT-PCR |
4.3. miR-RACE
4.4. Quantitative Real-Time PCR (qRT-PCR)
4.5. Target Gene Prediction
4.6. Over-Expression of Pm-miR-29a in Vivo
4.7. Vector Construction
4.8. Cell Culture and Transfection
4.9. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs—Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–298. [Google Scholar] [CrossRef]
- Chang, T.C.; Mendell, J.T. MicroRNAs in vertebrate physiology and human disease. Annu. Rev. Genom. Hum. Genet. 2007, 8, 215–239. [Google Scholar] [CrossRef] [PubMed]
- Iuliano, R.; Vismara, M.F.M.; Dattilo, V.; Trapasso, F.; Baudi, F.; Perrotti, N. The role of microRNAs in cancer susceptibility. BioMed. Res. Int. 2013, 2013, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Krützfeldt, J.; Stoffel, M. MicroRNAs: A new class of regulatory genes affecting metabolism. Cell Metab. 2006, 4, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Marina, C.; Witold, F. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009, 21, 452–460. [Google Scholar]
- Kapinas, K.; Kessler, C.B.; Delany, A.M. miR-29 suppression of osteonectin in osteoblasts: Regulation during differentiation and by canonical wnt signaling. J. Cell. Biochem. 2009, 108, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Maurer, B.; Stanczyk, J.; Jungel, A.; Akhmetshina, A.; Trenkmann, M.; Brock, M.; Kowal-Bielecka, O.; Gay, R.E.; Michel, B.A.; Distler, J.H.; et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010, 62, 1733–1743. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hassan, M.Q.; Jafferji, M.; Aqeilan, R.I.; Garzon, R.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009, 284, 15676–15684. [Google Scholar] [CrossRef] [PubMed]
- Roberto, V.P.; Tiago, D.M.; Silva, I.A.; Cancela, M.L. miR-29a is an enhancer of mineral deposition in bone-derived systems. Arch. Biochem. Biophys. 2014, 564, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Zheng, Z.; Du, X.; Wang, Q.; Huang, R.; Deng, Y.; Shi, S.; Zhao, X. Identification and characterization of microRNAs in pearl oyster Pinctada martensii by solexa deep sequencing. Mar. Biotechnol. 2014, 16, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, M.; Yu, H.; Han, J.; Song, C.; Ma, R.; Fang, J. Computational identification of microRNAs in peach expressed sequence tags and validation of their precise sequences by miR-RACE. Mol. Biol. Rep. 2012, 39, 1975–1987. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Kawashima, T.; Koyanagi, R.; Gyoja, F.; Tanaka, M.; Ikuta, T.; Shoguchi, E.; Fujiwara, M.; Shinzato, C.; Hisata, K.; et al. Draft genome of the pearl oyster Pinctada fucata: A platform for understanding bivalve biology. DNA Res. 2012, 19, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Zheng, Z.; Tian, R.; Du, X.; Wang, Q.; Huang, R. MicroRNA, pm-miR-2305, participates in nacre formation by targeting pearlin in pearl oyster Pinctada martensii. Int. J. Mol. Sci. 2015, 16, 21442–21453. [Google Scholar] [CrossRef] [PubMed]
- Eric, N.J.; Delanyc, A.M.; Lakshmi, S. Post-transcriptional regulation in osteoblasts using localized delivery of miR-29a inhibitor from nanofibers to enhance extracellular matrix deposition. Acta Biomater. 2014, 10, 3571–3580. [Google Scholar]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Kriegel, A.J.; Liu, Y.; Fang, Y.; Ding, X.; Liang, M. The miR-29 family: Genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol. Genom. 2012, 44, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Mitic, K.; Stanojevic, S.; Kustrimovic, N.; Vujic, V.; Dimitrijevic, M. Neuropeptide y modulates functions of inflammatory cells in the rat: Distinct role for Y1, Y2 and Y5 receptors. Peptides 2011, 32, 1626–1633. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.; Xapelli, S.; Santos, T.; Silva, A.P.; Cristovao, A.; Cortes, L.; Malva, J.O. Neuropeptide Y modulation of interleukin-1β (IL-1β)-induced nitric oxide production in microglia. J. Biol. Chem. 2010, 285, 41921–41934. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Z.; Huang, X.D.; Li, Q.; He, M.X. Interleukin-17 in pearl oyster (Pinctada fucata): Molecular cloning and functional characterization. Fish Shellfish Immunol. 2013, 34, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Xu, S.; Liu, X.; Zhang, Q.; Xu, X.; Liu, M.; Hua, M.; Li, N.; Yao, H.; Cao, X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat. Immunol. 2011, 12, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, Z.; Tu, J.; Zhu, W.; Ge, J.; Zheng, X.; Yang, L.; Pan, X.; Yan, H.; Zhu, J. MicroRNA-29a regulates pro-inflammatory cytokine secretion and scavenger receptor expression by targeting LPL in oxLDL-stimulated dendritic cells. FEBS Lett. 2011, 585, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bai, Y.; Han, J.; Chen, M.; Kayesh, E.; Jiang, W.; Fang, J. Bioinformatics prediction of miRNAs in the Prunus persica genome with validation of their precise sequences by miR-RACE. J. Plant Physiol. 2013, 170, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Fang, J.; Li, X.; Liu, H.; Chao, C. Identification and characterization of 27 conserved microRNAs in citrus. Planta 2009, 230, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Jiao, Y.; Deng, Y.; Du, X.; Huang, R.; Wang, Q.; Chen, W. Tissue inhibitor of metalloproteinase gene from pearl oyster Pinctada martensii participates in nacre formation. Biochem. Biophys. Res. Commun. 2014, 450, 300–305. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, R.; Zheng, Z.; Huang, R.; Jiao, Y.; Du, X. miR-29a Participated in Nacre Formation and Immune Response by Targeting Y2R in Pinctada martensii. Int. J. Mol. Sci. 2015, 16, 29436-29445. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161226182
Tian R, Zheng Z, Huang R, Jiao Y, Du X. miR-29a Participated in Nacre Formation and Immune Response by Targeting Y2R in Pinctada martensii. International Journal of Molecular Sciences. 2015; 16(12):29436-29445. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161226182
Chicago/Turabian StyleTian, Rongrong, Zhe Zheng, Ronglian Huang, Yu Jiao, and Xiaodong Du. 2015. "miR-29a Participated in Nacre Formation and Immune Response by Targeting Y2R in Pinctada martensii" International Journal of Molecular Sciences 16, no. 12: 29436-29445. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161226182