iTRAQ-Based Proteomics Analysis of Serum Proteins in Wistar Rats Treated with Sodium Fluoride: Insight into the Potential Mechanism and Candidate Biomarkers of Fluorosis
Abstract
:1. Introduction
2. Results
2.1. Analytical Strategy for Rat Serum Proteome Identification under NaF Treatment
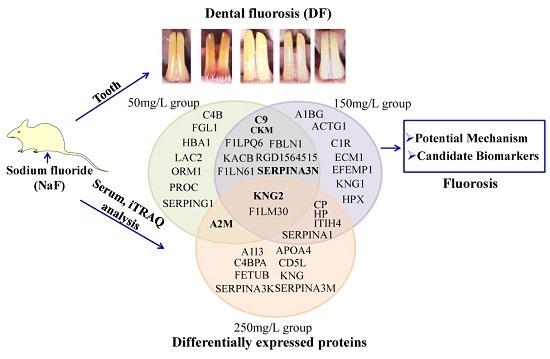
2.2. NaF Induced Dental Fluorosis in Wistar Rats
2.3. Serum Protein Expression Profiles of NaF Treated Rats
2.4. Functional Classification and Protein-Protein Interaction Analysis of Differential Expressed Proteins
2.5. Validation of Differential Expression of Proteins Found in iTRAQ via ELISA Assay
3. Discussion
4. Experimental Section
4.1. Animals and Treatment
4.2. Classification Principle for Rat’s Dental Fluorosis (DF)
4.3. Fluoride Ion Concentration Analysis
4.4. Depletion of High-Abundance Proteins
4.5. Protein Digestion and iTRAQ Labeling
4.6. LC-MS Analysis
4.7. Database Search and iTRAQ Data Analysis
4.8. Bioinformatics Analysis
4.9. ELISA Analysis for Protein Validation
4.10. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fuge, R. Sources of halogens in the environment, influences on human and animal health. Environ. Geochem. Health 1988, 10, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Chachra, D.; Turner, C.; Dunipace, A.; Grynpas, M.D. The effect of fluoride treatment on bone mineral in rabbits. Calcif. Tissue Int. 1999, 64, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Fejerskov, O.; Manji, F.; Baelum, V. The nature and mechanisms of dental fluorosis in man. J. Dent. Res. 1990, 69, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Grynpas, M.D. Fluoride effects on bone crystals. J. Bone Miner. Res. 1990, 5, S169–S175. [Google Scholar] [CrossRef] [PubMed]
- Kutlucan, A.; Koroglu, B.K.; Tamer, M.N.; Aydin, Y.; Baltaci, D.; Akdogan, M.; Ozturk, M.; Vural, H.; Ermis, F. The investigation of effects of fluorosis on thyroid volume in school-age children. Med. Glas. 2013, 10, 93–98. [Google Scholar]
- Mousny, M.; Omelon, S.; Wise, L.; Everett, E.T.; Dumitriu, M.; Holmyard, D.P.; Banse, X.; Devogelaer, J.P.; Grynpas, M.D. Fluoride effects on bone formation and mineralization are influenced by genetics. Bone 2008, 43, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Raina, S.K. A comment on effect of fluoride exposure on the intelligence of school children in Madhya Pradesh, India. J. Neurosci. Rural Pract. 2013, 4, 110–111. [Google Scholar] [CrossRef] [PubMed]
- Dharmaratne, R.W. Fluoride in drinking water and diet: The causative factor of chronic kidney diseases in the North Central Province of Sri Lanka. Environ. Health Prev. Med. 2015, 20, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Song, C.; Wu, H.; Chen, X.; Zhang, Y. Adverse effects of high concentrations of fluoride on characteristics of the ovary and mature oocyte of mouse. PLoS ONE 2015, 10, e0129594. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Pereira, H.A.B.; de Lima Leite, A.; Charone, S.; Lobo, J.G.; Cestari, T.M.; Peres-Buzalaf, C.; Buzalaf, M.A. Proteomic analysis of liver in rats chronically exposed to fluoride. PLoS ONE 2013, 8, e75343. [Google Scholar]
- Zhou, B.H.; Zhao, J.; Liu, J.; Zhang, J.L.; Li, J.; Wang, H.W. Fluoride-induced oxidative stress is involved in the morphological damage and dysfunction of liver in female mice. Chemosphere 2015, 139, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Kurdi, M.S. Chronic fluorosis: The disease and its anaesthetic implications. Indian J. Anaesth. 2016, 60, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Asawa, K.; Singh, A.; Bhat, N.; Tak, M.; Shinde, K.; Jain, S. Association of temporomandibular joint signs & symptoms with dental fluorosis & skeletal manifestations in endemic fluoride areas of dungarpur district, Rajasthan, India. J. Clin. Diagn. Res. 2015, 9, ZC18–ZC21. [Google Scholar] [PubMed]
- Sebastian, S.T.; Soman, R.R.; Sunitha, S. Prevalence of dental fluorosis among primary school children in association with different water fluoride levels in Mysore district, Karnataka. Indian J. Dent. Res. 2016, 27, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Christie, D.P. The spectrum of radiographic bone changes in children with fluorosis. Radiology 1980, 136, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Kong, L.; Zhao, H.; Dong, R.; Zhou, J.; Lu, Y. Thoracic myelopathy caused by ossification of ligamentum flavum of which fluorosis as an etiology factor. J. Orthop. Surg. Res. 2006, 1. [Google Scholar] [CrossRef] [PubMed]
- Browne, D.; Whelton, H.; O’Mullane, D. Fluoride metabolism and fluorosis. J. Dent. 2005, 33, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhao, Y.; Liu, J.; Xirao, R.; Danzeng, S.; Daji, D.; Yan, Y. Brick tea fluoride as a main source of adult fluorosis. Food Chem. Toxicol. 2003, 41, 535–542. [Google Scholar] [CrossRef]
- Whyte, M.P.; Essmyer, K.; Gannon, F.H.; Reinus, W.R. Skeletal fluorosis and instant tea. Am. J. Med. 2005, 118, 78–82. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, B.S.; da Silva Cardoso, V.E.; Buzalaf, M.A. Fluoride ingestion from toothpaste and diet in 1- to 3-year-old Brazilian children. Community Dent. Oral Epidemiol. 2007, 35, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Denbesten, P.; Li, W. Chronic fluoride toxicity: Dental fluorosis. Monogr. Oral Sci. 2011, 22, 81–96. [Google Scholar] [PubMed]
- Sun, D.J.; Sun, Y.F. Report on epidemiology investigation of endemic fluorosis in China. Chin. J. Epidemiol. 2005, 20, 81–84. [Google Scholar]
- Lu, J.; Xu, Q.; Chen, H.; Li, J.; Chen, K. Comparative proteomics analysis of midgut samples from Takifugu rubripes exposed to excessive fluoride: Initial molecular response to fluorosis. Toxicol. Mech. Methods 2011, 21, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, C.A.; Leite, A.L.; Silva, T.L.; Santos, L.D.; Nogueira, F.C.; Oliveira, R.C.; Palma, M.S.; Domont, G.B.; Buzalaf, M.A. Proteomic analysis of kidney in rats chronically exposed to fluoride. Chem. Biol. Interact. 2009, 180, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Loo, J.A.; Wong, D.T. Human body fluid proteome analysis. Proteomics 2006, 6, 6326–6353. [Google Scholar] [CrossRef] [PubMed]
- Glen, A.; Evans, C.A.; Gan, C.S.; Cross, S.S.; Hamdy, F.C.; Gibbins, J.; Lippitt, J.; Eaton, C.L.; Noirel, J.; Wright, P.C.; et al. Eight-plex iTRAQ analysis of variant metastatic human prostate cancer cells identifies candidate biomarkers of progression: An exploratory study. Prostate 2010, 70, 1313–1332. [Google Scholar] [CrossRef] [PubMed]
- Glen, A.; Gan, C.S.; Hamdy, F.C.; Eaton, C.L.; Cross, S.S.; Catto, J.W.; Wright, P.C.; Rehman, I. iTRAQ-facilitated proteomic analysis of human prostate cancer cells identifies proteins associated with progression. J. Proteome Res. 2008, 7, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteom. 2004, 3, 1154–1169. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T. Evolution of the lectin-complement pathway and its role in innate immunity. Nat. Rev. Immunol. 2002, 2, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Schwaeble, W.; Dahl, M.R.; Thiel, S.; Stover, C.; Jensenius, J.C. The mannan-binding lectin-associated serine proteases (MASPs) and MAp19: Four components of the lectin pathway activation complex encoded by two genes. Immunobiology 2002, 205, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M.; Fujita, T. The lectin pathway. Res. Immunol. 1996, 147, 115–118. [Google Scholar] [CrossRef]
- Wang, Y.; Bjes, E.S.; Esser, A.F. Molecular aspects of complement-mediated bacterial killing. Periplasmic conversion of C9 from a protoxin to a toxin. J. Biol. Chem. 2000, 275, 4687–4692. [Google Scholar] [CrossRef] [PubMed]
- Podack, E.R.; Müller-Eberhard, H.J.; Horst, H.; Hoppe, W. Membrane attach complex of complement (MAC): Three-dimensional analysis of MAC-phospholipid vesicle recombinants. J. Immunol. 1982, 128, 2353–2357. [Google Scholar] [PubMed]
- Müller, F.; Renne, T. Novel roles for factor XII-driven plasma contact activation system. Curr. Opin. Hematol. 2008, 15, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Markiewski, M.M.; Nilsson, B.; Ekdahl, K.N.; Mollnes, T.E.; Lambris, J.D. Complement and coagulation: Strangers or partners in crime? Trends Immunol. 2007, 28, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Lund, K.; Refsnes, M.; Ramis, I.; Dunster, C.; Boe, J.; Schwarze, P.E.; Skovlund, E.; Kelly, F.J.; Kongerud, J. Human exposure to hydrogen fluoride induces acute neutrophilic, eicosanoid, and antioxidant changes in nasal lavage fluid. Inhal. Toxicol. 2002, 14, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Aydin, G.; Çiçek, E.; Akdoğan, M.; Gökalp, O. Histopathological and biochemical changes in lung tissues of rats following administration of fluoride over several generations. J. Appl. Toxicol. 2003, 23, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Schwarze, P.E.; Låg, M.; Becher, R.; Thrane, E.V.; Samuelsen, J.T.; Hetland, R.B.; Refsnes, M. Role of signal transduction pathways in lung inflammatory responses. Toxicol. Lett. 2000, 112, 165–170. [Google Scholar] [CrossRef]
- Feinman, R.D. The proteinase-binding reaction of α 2M. Ann. N. Y. Acad. Sci. 1994, 737, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Langston, C.S. The aberrant central venous catheter and its complications. Radiology 1971, 100, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Park, K.H.; Ryu, E.; Tosakulwong, N.; Wu, Y.; Edwards, A.O. Common variation in the SERPING1 gene is not associated with age-related macular degeneration in two independent groups of subjects. Mol. Vis. 2009, 15, 200–207. [Google Scholar] [PubMed]
- Matsushita, M.; Endo, Y.; Fujita, T. Cutting edge: Complement-activating complex of ficolin and mannose-binding lectin-associated serine protease. J. Immunol. 2000, 164, 2281–2284. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.E.; Mejia, P.; Lu, F. Biological activities of C1 inhibitor. Mol. Immunol. 2008, 45, 4057–4063. [Google Scholar] [CrossRef] [PubMed]
- Olsen, G.N.; Harris, J.O.; Castle, J.R.; Waldman, R.H.; Karmgard, H.J. α-1-antitrypsin content in the serum, alveolar macrophages, and alveolar lavage fluid of smoking and nonsmoking normal subjects. J. Clin. Investig. 1975, 55, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Travis, J.; Salvesen, G.S. Human plasma proteinase inhibitors. Annu. Rev. Biochem. 1983, 52, 655–709. [Google Scholar] [CrossRef] [PubMed]
- Gadek, J.E.; Fells, G.A.; Zimmerman, R.L.; Rennard, S.I.; Crystal, R.G. Antielastases of the human alveolar structures. Implications for the protease-antiprotease theory of emphysema. J. Clin. Investig. 1981, 68, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Duranton, J.; Adam, C.; Bieth, J.G. Kinetic mechanism of the inhibition of cathepsin G by α1-antichymotrypsin and α1-proteinase inhibitor. Biochemistry 1998, 37, 11239–11245. [Google Scholar] [CrossRef] [PubMed]
- Joe, B.; Nagaraju, A.; Gowda, L.R.; Basrur, V.; Lokesh, B.R. Mass-spectrometric identification of T-Kininogen I/thiostatin as an acute-phase inflammatory protein suppressed by curcumin and capsaicin. PLoS ONE 2014, 9, e107565. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Gigli, I.; Nussenzweig, V. Human C4-binding protein. II. Role in proteolysis of C4b by C3b-inactivator. J. Exp. Med. 1978, 148, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Hillarp, A.; Dahlback, B. Novel subunit in C4b-binding protein required for protein S binding. J. Biol. Chem. 1988, 263, 12759–12764. [Google Scholar] [PubMed]
- Hillarp, A.; Dahlback, B. Cloning of cDNA coding for the β chain of human complement component C4b-binding protein: Sequence homology with the α chain. Proc. Natl. Acad. Sci. USA 1990, 87, 1183–1187. [Google Scholar] [CrossRef] [PubMed]
- Tschopp, J. Circular polymerization of the membranolytic ninth component of complement. Dependence on metal ions. J. Biol. Chem. 1984, 259, 10569–10573. [Google Scholar] [PubMed]
- Liszewski, M.K.; Atkinson, J.P. Regulatory proteins of complement. In The Human Complement System in Health and Disease; Volanakis, J.E., Frank, M.M., Eds.; Marcel Dekker: New York, NY, USA, 1998; pp. 149–165. [Google Scholar]
- Wang, J.; Lu, D.; Zheng, Z.A.; Chen, K.Z. Study on toxicity of sodium fluoride. J. Hyg. Res. 1995, 19, 39. [Google Scholar]
- Wang, Z.H.; Yu, Y.W.; Zhao, Y.H.; Qiao, W.B.; Zhang, X.D.; Xing, Y.R. Daily quantity of drinking water and stool of male SD rat. Chin. J. Lab. Anim. Sci. 2004, 14, 228. [Google Scholar]






| Protein Name | Gene Name | Swissport Accession No. | Protein Score | Coverage (%) | No. of Peptides | 50 mg/L NaF vs. Control (Ratio) | 150 mg/L NaF vs. Control (Ratio) | 250 mg/L NaF vs. Control (Ratio) |
|---|---|---|---|---|---|---|---|---|
| α-1-inhibitor 3 | A1I3 | P14046 | 135.27 | 73.3 | 107 | 0.80 | 1.11 | 1.52 (↑) * |
| Actin, cytoplasmic 2 | ACTG1 | P63259 | 18.87 | 52.8 | 11 | 0.92 | 2.60 (↑) * | 0.84 |
| α-1-acid glycoprotein | ORM1 | P02764 | 11.39 | 39.5 | 8 | 2.60 (↑) * | 0.65 | 0.64 |
| α-1-antiproteinase | SERPINA1 | P17475 | 40.96 | 61.3 | 33 | 0.85 | 0.73 (↓) * | 0.78 (↓) * |
| α-1B-glycoprotein | A1BG | Q9EPH1 | 36.84 | 52.2 | 27 | 0.81 | 0.66 (↓) * | 1.24 |
| α-2-macroglobulin | A2M | P06238 | 67.4 | 43.7 | 43 | 3.01 (↑) * | 0.90 | 0.73 (↓) * |
| Apolipoprotein A-IV | APOA4 | P02651 | 46.78 | 83.9 | 40 | 0.99 | 0.93 | 1.28 (↑) * |
| C4b-binding protein α chain | C4BPA | Q5M891 | 18.71 | 28.3 | 10 | 1.23 | 0.9 | 0.80 (↓) * |
| C9 protein | C9 | Q5BKC4 | 41 | 55.2 | 29 | 1.34 (↑) * | 0.78 (↓) * | 0.82 |
| CD5 antigen-like | CD5L | Q4KM75 | 20 | 42.8 | 10 | 0.93 | 0.83 | 0.71 (↓) * |
| Ceruloplasmin | CP | G3V7K3 | 84.04 | 63.3 | 57 | 1.07 | 0.65 (↓) * | 0.73 (↓) * |
| Complement component 4, gene 2 | C4B | Q6MG90 | 15.46 | 40 | 46 | 1.45 (↑) * | 1.18 | 1.23 |
| Creatine kinase M-type | CKM | P00564 | 11.05 | 31.5 | 6 | 0.73 (↓) * | 1.30 (↑) * | 1.11 |
| EGF-containing fibulin-like Extracellular matrix protein 1 | EFEMP1 | Q6AXN2 | 5.56 | 22.1 | 3 | 1.17 | 1.42 (↑) * | 1.07 |
| Extracellular matrix protein 1 | ECM1 | Q62894 | 12 | 33.6 | 6 | 1.19 | 1.30 (↑) * | 1.17 |
| Fetub protein | FETUB | Q6IRS6 | 33.22 | 66.7 | 22 | 0.93 | 0.92 | 1.34 (↑) * |
| Fibrinogen-like protein 1 | FGL1 | Q5M8C6 | 3.51 | 17.8 | 3 | 1.50 (↑) * | 1.08 | 1.02 |
| Fibulin 1 (Predicted) | FBLN1 | D3ZQ25 | 10.57 | 30.2 | 6 | 1.40 (↑) * | 1.40 (↑) * | 1.09 |
| Haptoglobin | HP | P06866 | 32.67 | 63.7 | 24 | 1.26 | 0.44 (↓) * | 0.55 (↓) * |
| Hemoglobin subunit α-1/2 | HBA1 | P01946 | 11.09 | 57 | 8 | 0.78 (↓) * | 0.89 | 0.95 |
| Hemopexin | HPX | P20059 | 52.31 | 68.7 | 37 | 1.09 | 0.60 (↓) * | 0.72 |
| Igκ chain C region, B allele | KACB | P01835 | 16.1 | 95.3 | 15 | 0.65 (↓) * | 0.63 (↓) * | 0.86 |
| Igλ-2 chain C region | LAC2 | P20767 | 12 | 87.5 | 10 | 0.77 (↓) * | 0.77 | 0.79 |
| Inter α-trypsin inhibitor, heavy chain 4 | ITIH4 | Q5EBC0 | 72.55 | 60.1 | 46 | 1.00 | 0.75 (↓) * | 0.72 (↓) * |
| Kininogen-1 | KNG | P08934 | 23.51 | 43.2 | 20 | 1.06 | 1.04 | 1.27 (↑) * |
| Plasma protease C1 inhibitor | SERPING1 | Q6P734 | 21.73 | 39.3 | 14 | 1.28 (↑) * | 0.94 | 1.01 |
| Protein C | PROC | Q68FY8 | 4.17 | 22.1 | 2 | 1.33 (↑) * | 1.35 | 1.10 |
| Protein C1r (Fragment) | C1R | D4A1T6 | 8.03 | 19.4 | 4 | 1.15 | 1.40 (↑) * | 1.25 |
| Protein RGD1564515 (Fragment) | RGD1564515 | F1LYQ4 | 11.49 | 49.6 | 7 | 0.66 (↓) * | 0.66 (↓) * | 1.13 |
| Serine protease inhibitor A3K | SERPINA3K | P05545 | 30.76 | 62.5 | 30 | 0.84 | 0.89 | 1.53 (↑) * |
| Serine protease inhibitor A3M (Fragment) | SERPINA3M | F1LR92 | 18.03 | 44.3 | 14 | 1.10 | 1.06 | 1.45 (↑) |
| Serine protease inhibitor A3N | SERPINA3N | P09006 | 29.27 | 62.4 | 26 | 1.37 (↑) * | 0.67 (↓) * | 0.90 |
| T-kininogen 1 | KNG1 | P01048 | 8.01 | 63.3 | 33 | 1.38 | 0.45 (↓) * | 0.64 |
| T-kininogen 2 | KNG2 | P08932 | 45.83 | 65.4 | 34 | 1.95 (↑) * | 0.56 (↓) * | 0.68 (↓) * |
| Uncharacterized protein | Ighm | F1LN61 | 35.49 | 71.1 | 41 | 0.69 (↓) * | 0.60 (↓) * | 0.81 |
| Uncharacterized protein | Ighm | F1LM30 | 25.71 | 50 | 20 | 0.67 (↓) * | 0.47 (↓) * | 0.49 (↓) * |
| Uncharacterized protein (Fragment) | Ighm | F1LPQ6 | 18.67 | 63.4 | 17 | 0.75 (↓) * | 0.49 (↓) * | 0.84 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Zeng, B.; Zhang, H.; Chen, C.; Wu, Y.; Wang, N.; Wu, Y.; Shen, L. iTRAQ-Based Proteomics Analysis of Serum Proteins in Wistar Rats Treated with Sodium Fluoride: Insight into the Potential Mechanism and Candidate Biomarkers of Fluorosis. Int. J. Mol. Sci. 2016, 17, 1644. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17101644
Wei Y, Zeng B, Zhang H, Chen C, Wu Y, Wang N, Wu Y, Shen L. iTRAQ-Based Proteomics Analysis of Serum Proteins in Wistar Rats Treated with Sodium Fluoride: Insight into the Potential Mechanism and Candidate Biomarkers of Fluorosis. International Journal of Molecular Sciences. 2016; 17(10):1644. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17101644
Chicago/Turabian StyleWei, Yan, Beibei Zeng, Hua Zhang, Cheng Chen, Yanli Wu, Nanlan Wang, Yanqiu Wu, and Liming Shen. 2016. "iTRAQ-Based Proteomics Analysis of Serum Proteins in Wistar Rats Treated with Sodium Fluoride: Insight into the Potential Mechanism and Candidate Biomarkers of Fluorosis" International Journal of Molecular Sciences 17, no. 10: 1644. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17101644