ADAR1 and MicroRNA; A Hidden Crosstalk in Cancer
Abstract
:1. The Role of RNA Editing in Eukaryotes
2. ADAR and A-to-I Editing
2.1. Target Sites of A-to-I Editing and ADAR Binding
2.2. Structure, Expression, and Localization of ADARs
2.3. ADAR1
2.4. ADAR2
2.5. ADAR3
3. Overwhelming Evidence of RNA Editing by ADARs in Cancer
4. MicroRNA Biogenesis and Influence of RNA Editing
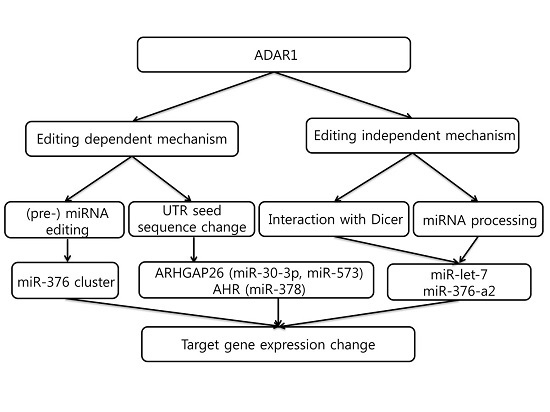
5. Editing-Dependent ADAR Effects in Cancer
6. Editing-Independent Effects of ADAR
7. MicroRNAs That Regulate ADAR1
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Baltimore, D. Our genome unveiled. Nature 2001, 409, 814–816. [Google Scholar] [CrossRef] [PubMed]
- Keegan, L.P.; Gallo, A.; O’Connell, M.A. The many roles of an RNA editor. Nat. Rev. Genet. 2001, 2, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Gott, J.M.; Emeson, R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000, 34, 499–531. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef] [PubMed]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.D.; Kim, T.T.; Walsh, T.; Kobayashi, Y.; Matise, T.C.; Buyske, S.; Gabriel, A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004, 14, 1719–1725. [Google Scholar] [CrossRef] [PubMed]
- Ramaswami, G.; Li, J.B. RADAR: A rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2013. [Google Scholar] [CrossRef] [PubMed]
- Kiran, A.; Baranov, P.V. DARNED: A DAtabase of RNa EDiting in humans. Bioinformatics 2010, 26, 1772–1776. [Google Scholar] [CrossRef] [PubMed]
- Athanasiadis, A.; Rich, A.; Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004, 2, e391. [Google Scholar] [CrossRef] [PubMed]
- Morse, D.P.; Aruscavage, P.J.; Bass, B.L. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. USA 2002, 99, 7906–7911. [Google Scholar] [CrossRef] [PubMed]
- Bahn, J.H.; Ahn, J.; Lin, X.; Zhang, Q.; Lee, J.-H.; Civelek, M.; Xiao, X. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 2015, 6, 6355. [Google Scholar] [CrossRef] [PubMed]
- Macbeth, M.R.; Schubert, H.L.; VanDemark, A.P.; Lingam, A.T.; Hill, C.P.; Bass, B.L. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 2005, 309, 1534–1539. [Google Scholar] [CrossRef] [PubMed]
- Ryter, J.M.; Schultz, S.C. Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998, 17, 7505–7513. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K.; Yoo, C.; Kim, U.; Murray, J.; Estes, P.; Cash, F.; Liebhaber, S. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991, 10, 3523. [Google Scholar] [PubMed]
- Lehmann, K.A.; Bass, B.L. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 2000, 39, 12875–12884. [Google Scholar] [CrossRef] [PubMed]
- Vitali, P.; Basyuk, E.; Le Meur, E.; Bertrand, E.; Muscatelli, F.; Cavaillé, J.; Huttenhofer, A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005, 169, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Miyakoda, M.; Yang, W.; Khillan, J.; Stachura, D.L.; Weiss, M.J.; Nishikura, K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004, 279, 4952–4961. [Google Scholar] [CrossRef] [PubMed]
- Hartner, J.C.; Schmittwolf, C.; Kispert, A.; Müller, A.M.; Higuchi, M.; Seeburg, P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004, 279, 4894–4902. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, M.; Maas, S.; Single, F.N.; Hartner, J.; Rozov, A.; Burnashev, N.; Feldmeyer, D.; Sprengel, R.; Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000, 406, 78–81. [Google Scholar] [PubMed]
- Tsuruoka, N.; Arima, M.; Yoshida, N.; Okada, S.; Sakamoto, A.; Hatano, M.; Satake, H.; Arguni, E.; Wang, J.Y.; Yang, J.H.; et al. ADAR1 protein induces adenosine-targeted DNA mutations in senescent Bcl6 gene-deficient cells. J. Biol. Chem. 2013, 288, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Singh, M.M.; Na, E.; Agassandian, K.; Zimmerman, M.B.; Johnson, A.K. Altered ADAR 2 equilibrium and 5HT(2C) R editing in the prefrontal cortex of ADAR 2 transgenic mice. Genes Brain Behav. 2011, 10, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Zimmerman, M.B.; Beltz, T.G.; Johnson, A.K. Affect-related behaviors in mice misexpressing the RNA editing enzyme ADAR2. Physiol. Behav. 2009, 97, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.B.; Samuel, C.E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell. Biol. 1995, 15, 5376–5388. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.B.; Thomis, D.C.; Hans, S.L.; Samuel, C.E. Mechanism of interferon action: Double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 1995, 210, 508–511. [Google Scholar] [CrossRef] [PubMed]
- Aicardi, J.; Goutieres, F. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann. Neurol. 1984, 15, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Liddicoat, B.J.; Piskol, R.; Chalk, A.M.; Ramaswami, G.; Higuchi, M.; Hartner, J.C.; Li, J.B.; Seeburg, P.H.; Walkley, C.R. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 2015, 349, 1115–1120. [Google Scholar] [CrossRef] [PubMed]
- Mannion, N.M.; Greenwood, S.M.; Young, R.; Cox, S.; Brindle, J.; Read, D.; Nellåker, C.; Vesely, C.; Ponting, C.P.; McLaughlin, P.J. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014, 9, 1482–1494. [Google Scholar] [CrossRef] [PubMed]
- Toth, A.M.; Li, Z.; Cattaneo, R.; Samuel, C.E. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J. Biol. Chem. 2009, 284, 29350–29356. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wolff, K.C.; Samuel, C.E. RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology 2010, 396, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, H.; Nilsson, J.; Damgaard, C.K.; Egebjerg, J.; Kjems, J. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol. Cell. Biol. 2001, 21, 7862–7871. [Google Scholar] [CrossRef] [PubMed]
- Fritz, J.; Strehblow, A.; Taschner, A.; Schopoff, S.; Pasierbek, P.; Jantsch, M.F. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol. Cell. Biol. 2009, 29, 1487–1497. [Google Scholar] [CrossRef] [PubMed]
- Mittaz, L.; Scott, H.S.; Rossier, C.; Seeburg, P.H.; Higuchi, M.; Antonarakis, S.E. Cloning of a human RNA editing deaminase (ADARB1) of glutamate receptors that maps to chromosome 21q22. 3. Genomics 1997, 41, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.; Gommans, W.M. Novel exon of mammalian ADAR2 extends open reading frame. PLoS ONE 2009, 4, e4225. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, M.; Single, F.N.; Köhler, M.; Sommer, B.; Sprengel, R.; Seeburg, P.H. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron-exon structure determines position and efficiency. Cell 1993, 75, 1361–1370. [Google Scholar] [CrossRef]
- Mittaz, L.; Antonarakis, S.E.; Higuchi, M.; Scott, H.S. Localization of a novel human RNA-editing deaminase (hRED2 or ADARB2) to chromosome 10p15. Hum. Genet. 1997, 100, 398–400. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-X.; Cho, D.; Wang, Q.; Lai, F.; Carter, K.C.; Nishikura, K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single-and double-stranded RNA binding domains. RNA 2000, 6, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Melcher, T.; Maas, S.; Herb, A.; Sprengel, R.; Higuchi, M.; Seeburg, P.H. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem. 1996, 271, 31795–31798. [Google Scholar] [CrossRef] [PubMed]
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Paz-Yaacov, N.; Bazak, L.; Buchumenski, I.; Porath, H.T.; Danan-Gotthold, M.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015, 13, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Diao, L.; Yu, S.; Xu, X.; Li, J.; Zhang, R.; Yang, Y.; Werner, H.M.; Eterovic, A.K.; Yuan, Y. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell 2015, 28, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.H.M.; Lin, C.H.; Qi, L.; Fei, J.; Li, Y.; Yong, K.J.; Liu, M.; Song, Y.; Chow, R.K.K.; Ng, V.H.E. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut 2013, 63, 832–843. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.H.; Qamra, A.; Tan, K.T.; Guo, J.; Yang, H.; Qi, L.; Lin, J.S.; Ng, V.H.; Song, Y.; Hong, H.; et al. ADAR-Mediated RNA Editing Predicts Progression and Prognosis of Gastric Cancer. Gastroenterology 2016, 151, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.-R.; Qiao, J.-J.; Chan, T.H.M.; Zhu, Y.-H.; Li, F.-F.; Liu, H.; Fei, J.; Li, Y.; Guan, X.-Y.; Chen, L. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014, 74, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Nemlich, Y.; Greenberg, E.; Ortenberg, R.; Besser, M.J.; Barshack, I.; Jacob-Hirsch, J.; Jacoby, E.; Eyal, E.; Rivkin, L.; Prieto, V.G. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J. Clin. Investig. 2013, 123, 2703–2718. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Grosshans, H. Regulation of MicroRNAs; Springer Science & Business Media: New York, NY, USA, 2010. [Google Scholar]
- Cho, C.J.; Myung, S.-J.; Chang, S. The Role of ADAR1 in Gastric Tumorigenesis. Unpublished work. 2017. [Google Scholar]
- Choudhury, Y.; Tay, F.C.; Lam, D.H.; Sandanaraj, E.; Tang, C.; Ang, B.-T.; Wang, S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig. 2012, 122, 4059–4076. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hui, H.; Guo, Z.; Zhang, W.; Hu, Y.; He, T.; Tai, Y.; Peng, P.; Wang, L. ADAR1 regulates ARHGAP26 gene expression through RNA editing by disrupting miR-30b-3p and miR-573 binding. RNA 2013, 19, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Fukami, T.; Gotoh, S.; Takamiya, M.; Aoki, Y.; Nakajima, M. RNA Editing Modulates Human Hepatic Aryl Hydrocarbon Receptor Expression by Creating MicroRNA Recognition Sequence. J. Biol. Chem. 2016, 291, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Heale, B.S.; Keegan, L.P.; McGurk, L.; Michlewski, G.; Brindle, J.; Stanton, C.M.; Caceres, J.F.; O'connell, M.A. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009, 28, 3145–3156. [Google Scholar] [CrossRef] [PubMed]
- Ota, H.; Sakurai, M.; Gupta, R.; Valente, L.; Wulff, B.-E.; Ariyoshi, K.; Iizasa, H.; Davuluri, R.V.; Nishikura, K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013, 153, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]




| Change in MicroRNA Expression and Editing Level upon ADAR1 Knockdown | No. |
|---|---|
| microRNA expression level increased >50% | 17 |
| microRNA expression level decreased >50% | 51 |
| Editing of pre-microRNA | 4 |
| Editing of microRNA seed sequence | 1 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, C.J.; Myung, S.-J.; Chang, S. ADAR1 and MicroRNA; A Hidden Crosstalk in Cancer. Int. J. Mol. Sci. 2017, 18, 799. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18040799
Cho CJ, Myung S-J, Chang S. ADAR1 and MicroRNA; A Hidden Crosstalk in Cancer. International Journal of Molecular Sciences. 2017; 18(4):799. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18040799
Chicago/Turabian StyleCho, Charles J., Seung-Jae Myung, and Suhwan Chang. 2017. "ADAR1 and MicroRNA; A Hidden Crosstalk in Cancer" International Journal of Molecular Sciences 18, no. 4: 799. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18040799