MicroRNA-34a: A Versatile Regulator of Myriads of Targets in Different Cancers
Abstract
:1. Introduction
2. Activation and Expression of miR-34a
3. miR-34a-Mediated Regulation of Different Targets
4. miR-34a Co-Operates with Multiple microRNAs to Target Different Genes
5. miR-34’s Role in Cancer Treatment
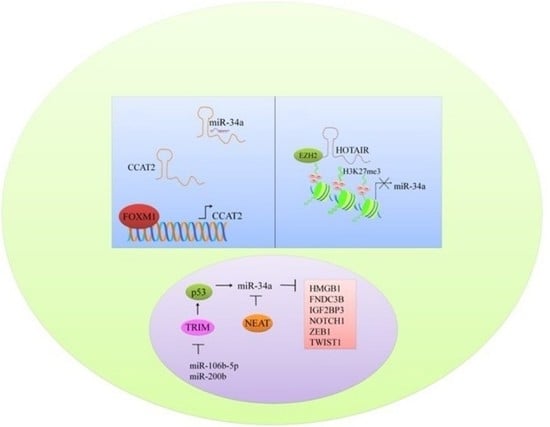
6. Multi-Layered Complexity of Competitive Endogenous RNAs Crosstalk and Competition with miR-34a
7. Nanotechnological Strategies to Deliver miR-34a
8. Natural Products-Mediated Regulation of miR-34a
9. Darker Side of miR-34a in Different Cancers
10. Clinical Trials Involving miR-34a
11. Conclusions
Author Contributions
Conflicts of Interest
Abbreviations
| ATM | Ataxia-telangiectasia Mutated Kinase |
| ATP | Adenosine-Triphosphate |
| ceRNA | Competitive endogenous RNA |
| Clp1 | Cleavage and Polyadenylation Factor I Subunit-1 |
| CSC | Cancer stem cell |
| DDX3X | DEAD-box RNA helicase |
| DHX9 | DEAH Box Polypeptide-9 |
| DLL1 | Delta-like ligand 1 |
| DMPM | Diffuse malignant peritoneal mesothelioma |
| dsRNA | Double-stranded RNA |
| DZnep | 3-Deazaneplanocin A |
| EMT | Epithelial to mesenchymal transition |
| ESCC | Esophageal squamous cell carcinoma |
| FNDC3B | Fibronectin Type III Domain Containing 3B |
| ghRNA | Guide hairpin RNA |
| HCC | Hepatocellular carcinoma |
| HHV-8 | Human herpesvirus 8 |
| HMGB1 | High Mobility Group Box-1 |
| HOTAIR | Hox transcript antisense intergenic RNA |
| IGF2BP3 | Insulin-like growth factor-2 mRNA-binding protein 3 |
| KSHV | Kaposi’s sarcoma-associated herpesvirus |
| LF-MFs | Low-frequency magnetic fields |
| Linc-ROR | Long intergenic non-protein coding RNA, regulator of reprogramming |
| LncARSR | lncRNA Activated in RCC with sunitinib resistance |
| LncRNAs | long non-coding RNAs |
| NEAT1 | Nuclear paraspeckle assembly transcript-1 |
| PDAC | Pancreatic ductal adenocarcinoma |
| PKR | Protein kinase (PKR) |
| PPC-1 | Primary prostate carcinoma-1 |
| RLR | RIG-I-like receptor |
| SIRT1 | Silent expression regulator-1 |
| SMAD3 | Mothers Against Decapentaplegic |
| ST3GAL5 | ST3 Beta-Galactoside Alpha-2,3-Sialyltransferase 5 |
| TKIs | Tyrosine-kinase-inhibitors |
| TLR | Toll-like receptor |
| TRIM8 | Tripartite motif containing protein-8 |
| TUG1 | Taurine upregulated 1 |
| VEGFA | Vascular endothelial growth factor-A |
References
- Ito, Y.; Inoue, A.; Seers, T.; Hato, Y.; Igarashi, A.; Toyama, T.; Taganov, K.D.; Boldin, M.P.; Asahara, H. Identification of targets of tumor suppressor microrna-34a using a reporter library system. Proc. Natl. Acad. Sci. USA 2017, 114, 3927–3932. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Peng, Y.; Lin, F.; Singh, R.K.; Mahato, R.I. Micellar delivery of mir-34a modulator rubone and paclitaxel in resistant prostate cancer. Cancer Res. 2017, 77, 3244–3254. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, C.; Liu, X.; Tang, D.G.; Wang, J. The microrna mir-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44HI stem-like NSCLC cells. PLoS ONE 2014, 9, e90022. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.D.; Parsons, C.; Slack, F.J. The tumor-suppressive and potential therapeutic functions of mir-34a in epithelial carcinomas. Expert Opin. Ther. Targets 2016, 20, 737–753. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Nakaoka, T.; Saito, H. MicroRNA-34a as a therapeutic agent against human cancer. J. Clin. Med. 2015, 4, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Li, L. Regulatory mechanisms and clinical perspectives of miR-34a in cancer. J. Cancer Res. Ther. 2014, 10, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Ren, Z.J.; Tang, J.H. MicroRNA-34a: A potential therapeutic target in human cancer. Cell Death Dis. 2014, 5, e1327. [Google Scholar] [CrossRef] [PubMed]
- Maroni, P.; Puglisi, R.; Mattia, G.; Care, A.; Matteucci, E.; Bendinelli, P.; Desiderio, M.A. In bone metastasis miR-34a-5p absence inversely correlates with met expression, while met oncogene is unaffected by miR-34a-5p in non-metastatic and metastatic breast carcinomas. Carcinogenesis 2017, 38, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.L.; Zhao, X.M.; Zhang, L.; Yang, P.; Fan, J.; Tang, Z.Y.; Zeng, Z.C. MicroRNA-34a expression levels in serum and intratumoral tissue can predict bone metastasis in patients with hepatocellular carcinoma. Oncotarget 2016, 7, 87246–87256. [Google Scholar] [CrossRef] [PubMed]
- Ghandadi, M.; Sahebkar, A. MicroRNA-34a and its target genes: Key factors in cancer multidrug resistance. Curr. Pharm. Des. 2016, 22, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Imani, S.; Zhang, X.; Hosseinifard, H.; Fu, S.; Fu, J. The diagnostic role of microRNA-34a in breast cancer: A systematic review and meta-analysis. Oncotarget 2017, 8, 23177–23187. [Google Scholar] [CrossRef] [PubMed]
- Rapti, S.M.; Kontos, C.K.; Christodoulou, S.; Papadopoulos, I.N.; Scorilas, A. MiR-34a overexpression predicts poor prognostic outcome in colorectal adenocarcinoma, independently of clinicopathological factors with established prognostic value. Clin. Biochem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.H.; Qin, Y.E.; Tang, W.F.; Tao, J.; Song, H.M.; Zuo, M. Mir-34a and mir-206 act as novel prognostic and therapy biomarkers in cervical cancer. Cancer Cell Int. 2017, 17, 63. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Fayyaz, S.; Shatynska-Mytsyk, I.; Javed, Z.; Jabeen, S.; Yaylim, I.; Gasparri, M.L.; Panici, P.B. Is miR-34a a well-equipped swordsman to conquer temple of molecular oncology? Chem. Biol. Drug Des. 2016, 87, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.G. MiR-34—A microRNA replacement therapy is headed to the clinic. Front. Genet. 2012, 3, 120. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Knight, R.A. MiR-34: From bench to bedside. Oncotarget 2014, 5, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.Y.; Ferrajoli, A.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. Microrna therapeutics in cancer—An emerging concept. EBioMedicine 2016, 12, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Salzman, D.W.; Nakamura, K.; Nallur, S.; Dookwah, M.T.; Metheetrairut, C.; Slack, F.J.; Weidhaas, J.B. MiR-34 activity is modulated through 5′-end phosphorylation in response to DNA damage. Nat. Commun. 2016, 7, 10954. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Mao, Y.; Zhao, Y.; He, Y. Ddx3x promotes the biogenesis of a subset of mirnas and the potential roles they played in cancer development. Sci. Rep. 2016, 6, 32739. [Google Scholar] [CrossRef] [PubMed]
- Kawai, S.; Amano, A. BRCA1 regulates microRNA biogenesis via the drosha microprocessor complex. J. Cell. Biol. 2012, 197, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Xiao, Z.; Tong, J.H.; To, K.F.; Fang, X.; Cheng, A.S.; Chen, Y. EZH2 coupled with hotair to silence microRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma. Int. J. Cancer 2017, 140, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Bai, G.; Li, Y.; Feng, Y.; Wang, L. A positive feedback loop of long noncoding RNA ccat2 and foxm1 promotes hepatocellular carcinoma growth. Am. J. Cancer Res. 2017, 7, 1423–1434. [Google Scholar] [PubMed]
- Li, J.; Wang, K.; Chen, X.; Meng, H.; Song, M.; Wang, Y.; Xu, X.; Bai, Y. Transcriptional activation of microRNA-34a by NF-kappa B in human esophageal cancer cells. BMC Mol. Biol. 2012, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ren, W.; Chen, K. MiR-34a promotes apoptosis and inhibits autophagy by targeting HMGB1 in acute myeloid leukemia cells. Cell. Physiol. Biochem. 2017, 41, 1981–1992. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Song, X.; Zhu, J.; Li, M.; Ji, Y.; Wu, F.; Chen, Y.; Cui, X.; Hu, J.; Wang, L.; et al. Tumor suppressor microRNA-34a inhibits cell migration and invasion by targeting mmp-2/mmp-9/fndc3b in esophageal squamous cell carcinoma. Int. J. Oncol. 2017, 51, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Huang, T.; Siu, H.L.; Wong, C.C.; Dong, Y.; Wu, F.; Zhang, B.; Wu, W.K.; Cheng, A.S.; Yu, J.; et al. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol. Cancer 2017, 16, 77. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Liu, Y.; Wei, H.Y.; Lv, K.Z.; Fu, P.F. Large intergenic non-coding rna-ror reverses gemcitabine-induced autophagy and apoptosis in breast cancer cells. Oncotarget 2016, 7, 59604–59617. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chen, N.; Gong, Y.; Xiao, R.; Wang, W.; Pan, Z. The long non-coding RNA neat1 enhances epithelial-to-mesenchymal transition and chemoresistance via the miR-34a/c-met axis in renal cell carcinoma. Oncotarget 2017. [Google Scholar] [CrossRef]
- Mastropasqua, F.; Marzano, F.; Valletti, A.; Aiello, I.; Di Tullio, G.; Morgano, A.; Liuni, S.; Ranieri, E.; Guerrini, L.; Gasparre, G.; et al. TRIM8 restores p53 tumour suppressor function by blunting n-myc activity in chemo-resistant tumours. Mol. Cancer 2017, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Fujino, T.; Yokosuka, A.; Higurashi, H.; Yokokawa, R.; Sakurai, R.; Harashima, W.; Miki, Y.; Fujiwara, Y.; Mimaki, Y.; Hayakawa, M. Au-1 from agavaceae plants causes transient increase in p21/cip1 expression in renal adenocarcinoma achn cells in an mir-34-dependent manner. J. Nat. Med. 2017, 71, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, X.; Zhang, Y.; Hu, Y.; Shen, X.; Zhu, W. Long non-coding RNA tug1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p. Oncotarget 2017, 8, 31386–31394. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A. Non-coding rnas: A tale of junk turning into treasure. Non Coding RNA Res. 2016, 1, 2. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Akiyama, Y.; Yuasa, Y. Multiple-to-multiple relationships between micrornas and target genes in gastric cancer. PLoS ONE 2013, 8, e62589. [Google Scholar] [CrossRef] [PubMed]
- Hart, M.; Rheinheimer, S.; Leidinger, P.; Backes, C.; Menegatti, J.; Fehlmann, T.; Grasser, F.; Keller, A.; Meese, E. Identification of miR-34a-target interactions by a combined network based and experimental approach. Oncotarget 2016, 7, 34288–34299. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.H.; Chen, Z.N.; Li, L.; Chen, J.; Mo, X.W.; Qin, Y.Z.; Wei, W.E.; Qin, H.Q.; Lin, Y.; Chen, J.S. E2f-1 promotes DAPK2-induced anti-tumor immunity of gastric cancer cells by targeting miR-34a. Tumour Biol. 2016, 37, 15925–15936. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Lin, H.; Li, G.; Sun, Y.; Shen, J.; Xu, J.; Lin, C.; Yeh, S.; Cai, X.; Chang, C. Cisplatin enhances nk cells immunotherapy efficacy to suppress HCC progression via altering the androgen receptor (ar)-ulbp2 signals. Cancer Lett. 2016, 373, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Dong, K.; Lin, F.; Long, M.; Ouyang, Y.; Wei, J.; Chen, X.; Weng, Y.; He, T.; et al. Tumor suppressor miR-34a targets pd-l1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell. Signal 2015, 27, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Yao, W.; Xiao, W.; Li, H.; Xu, H.; Lang, B. MicroRNA-34a functions as an anti-metastatic microRNA and suppresses angiogenesis in bladder cancer by directly targeting cd44. J. Exp. Clin. Cancer Res. 2014, 33, 779. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, Y.; Wang, J. Overexpression of microRNA-34a-5p inhibits proliferation and promotes apoptosis of human cervical cancer cells by downregulation of Bcl-2. Oncol. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, M.; Geng, F. Omega-3 polyunsaturated fatty acids eicosapentaenoic acid and docosahexaenoic acid enhance dexamethasone sensitivity in multiple myeloma cells by the p53/miR-34a/bcl-2 axis. Biochemistry 2017, 82, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Niu, Y.; Li, D.; Liu, Z. Lncrna C2DAT1 promotes cell proliferation, migration, and invasion by targeting miR-34a-5p in osteosarcoma cells. Oncol. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhou, H.; Miao, Y.; Li, N.; Zhao, L.; Jia, L. Mirna expression profiles reveal the involvement of mir-26a, mir-548l and mir-34a in hepatocellular carcinoma progression through regulation of st3gal5. Lab. Investig. 2017, 97, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Gao, S.; Song, X.; Dong, W.; Zhou, H.; Zhao, L.; Jia, L. Comprehensive N-glycan profiles of hepatocellular carcinoma reveal association of fucosylation with tumor progression and regulation of fut8 by micrornas. Oncotarget 2016, 7, 61199–61214. [Google Scholar] [CrossRef] [PubMed]
- Jian, C.; Tu, M.J.; Ho, P.Y.; Duan, Z.; Zhang, Q.; Qiu, J.X.; DeVere White, R.W.; Wun, T.; Lara, P.N.; Lam, K.S.; et al. Co-targeting of DNA, rna, and protein molecules provides optimal outcomes for treating osteosarcoma and pulmonary metastasis in spontaneous and experimental metastasis mouse models. Oncotarget 2017, 8, 30742–30755. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Shui, L.; Lou, G.; Ye, B.; Zhu, W.; Wang, J.; Wu, S.; Xu, X.; Mao, L.; Xu, W.; et al. 0404 inhibits hepatocellular carcinoma through a p53/miR-34a/sirt1 positive feedback loop. Sci. Rep. 2017, 7, 4396. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, F.J.; Zhang, H.G.; Xu, X.Z.; Jia, R.C.; Yao, L.; Qiao, P.F. MiR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-beta/smad4 pathway. World J. Gastroenterol. 2017, 23, 1816–1827. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Ding, L.; Xu, Q.; Shi, G.; Li, X.; Li, X.; Ji, J.; Zhang, D.; Wang, Y.; Wang, T.; et al. Lf-mf inhibits iron metabolism and suppresses lung cancer through activation of p53-miR-34a-e2f1/e2f3 pathway. Sci. Rep. 2017, 7, 749. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Guerrero, A.; Kelnar, K.; Peltier, H.J.; Bader, A.G. Synergy between next generation egfr tyrosine kinase inhibitors and mir-34a in the inhibition of non-small cell lung cancer. Lung Cancer 2017, 108, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Pyzer, A.R.; Stroopinsky, D.; Rosenblatt, J.; Anastasiadou, E.; Rajabi, H.; Washington, A.; Tagde, A.; Chu, J.H.; Coll, M.; Jiao, A.L.; et al. Muc1 inhibition leads to decrease in pd-l1 levels via upregulation of mirnas. Leukemia 2017. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; He, X.; Li, M.; Shi, F.; Wu, D.; Pan, M.; Guo, M.; Zhang, R.; Luo, S.; Gu, N.; et al. MiRNA-34a overexpression inhibits multiple myeloma cancer stem cell growth in mice by suppressing tgif2. Am. J. Transl. Res. 2016, 8, 5433–5443. [Google Scholar] [PubMed]
- El Bezawy, R.; de Cesare, M.; Pennati, M.; Deraco, M.; Gandellini, P.; Zuco, V.; Zaffaroni, N. Antitumor activity of miR-34a in peritoneal mesothelioma relies on c-met and axl inhibition: Persistent activation of erk and akt signaling as a possible cytoprotective mechanism. J. Hematol. Oncol. 2017, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Imani, S.; Wei, C.; Cheng, J.; Khan, M.A.; Fu, S.; Yang, L.; Tania, M.; Zhang, X.; Xiao, X.; Zhang, X.; et al. MicroRNA-34a targets epithelial to mesenchymal transition-inducing transcription factors (emt-tfs) and inhibits breast cancer cell migration and invasion. Oncotarget 2017, 8, 21362–21379. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Maitah, M.Y.; Ginnebaugh, K.R.; Li, Y.; Bao, B.; Gadgeel, S.M.; Sarkar, F.H. Inhibition of hedgehog signaling sensitizes nsclc cells to standard therapies through modulation of emt-regulating mirnas. J. Hematol. Oncol. 2013, 6, 77. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Yang, M.; Jiang, R.; An, N.; Wang, X.; Liu, B. Long non-coding rna hotair regulates the proliferation, self-renewal capacity, tumor formation and migration of the cancer stem-like cell (csc) subpopulation enriched from breast cancer cells. PLoS ONE 2017, 12, e0170860. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Itano, K.; Harada, Y.; Asada, K.; Oikawa, K.; Kashiwazako, M.; Okuyama, H.; Kumagai, K.; Takanashi, M.; Sudo, K.; et al. Development of novel small hairpin rnas that do not require processing by dicer or ago2. Mol. Ther. 2016, 24, 1278–1289. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of cerna crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous microrna sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Ding, J.; Chen, C.; Wu, Z.J.; Liu, B.; Gao, Y.; Chen, W.; Liu, F.; Sun, W.; Li, X.F.; et al. Exosome-transmitted lncarsr promotes sunitinib resistance in renal cancer by acting as a competing endogenous rna. Cancer Cell 2016, 29, 653–668. [Google Scholar] [CrossRef] [PubMed]
- Fanini, F.; Fabbri, M. Cancer-derived exosomic micrornas shape the immune system within the tumor microenvironment: State of the art. Semin. Cell. Dev. Biol. 2017, 67, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, K.; Yamamoto, Y.; Matsuoka, R.; Ochiya, T. Maintaining good mirnas in the body keeps the doctor away? Perspectives on the relationship between food-derived natural products and micrornas in relation to exosomes/extracellular vesicles. Mol. Nutr. Food Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, M.; Lee, W.; Santarelli, L.; Neuzil, J. Exosome-derived micrornas in cancer metabolism: Possible implications in cancer diagnostics and therapy. Exp. Mol. Med. 2017, 49, e285. [Google Scholar] [CrossRef] [PubMed]
- Urgard, E.; Brjalin, A.; Langel, U.; Pooga, M.; Rebane, A.; Annilo, T. Comparison of peptide- and lipid-based delivery of miR-34a-5p mimic into ppc-1 cells. Nucleic Acid Ther. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.; Liang, X.; Han, H.; Wang, H.; Yang, Y.; Li, Q. An ATP-responsive codelivery system of doxorubicin and miR-34a to synergistically inhibit cell proliferation and migration. Mol. Pharm. 2017, 14, 2323–2332. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, Y.; Liang, X.; Huang, Y.; Li, Q. Chondroitin sulfate-functionalized polyamidoamine as a tumor-targeted carrier for miR-34a delivery. Acta Biomater. 2017, 57, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Ginnebaugh, K.R.; Li, Y.; Padhye, S.B.; Sarkar, F.H. Molecular targets of naturopathy in cancer research: Bridge to modern medicine. Nutrients 2015, 7, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Li, Y.; Bao, B.; Kong, D.; Sarkar, F.H. Epigenetic regulation of miRNA-cancer stem cells nexus by nutraceuticals. Mol. Nutr. Food Res. 2014, 58, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Zhao, F.; Wang, H.; Cai, S. MiR-34a-5p promotes multi-chemoresistance of osteosarcoma through down-regulation of the dll1 gene. Sci. Rep. 2017, 7, 44218. [Google Scholar] [CrossRef] [PubMed]
- Krause, C.J.; Popp, O.; Thirunarayanan, N.; Dittmar, G.; Lipp, M.; Muller, G. MicroRNA-34a promotes genomic instability by a broad suppression of genome maintenance mechanisms downstream of the oncogene KSHV-VGPCR. Oncotarget 2016, 7, 10414–10432. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, P.; Li, Y.; Liu, G.; Zhou, B.; Zhan, L.; Zhou, Z.; Sun, X. The quantitative analysis by stem-loop real-time PCR revealed the microRNA-34a, microrna-155 and microRNA-200c overexpression in human colorectal cancer. Med. Oncol. 2012, 29, 3113–3118. [Google Scholar] [CrossRef] [PubMed]
- Dutta, K.K.; Zhong, Y.; Liu, Y.T.; Yamada, T.; Akatsuka, S.; Hu, Q.; Yoshihara, M.; Ohara, H.; Takehashi, M.; Shinohara, T.; et al. Association of microrna-34a overexpression with proliferation is cell type-dependent. Cancer Sci. 2007, 98, 1845–1852. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase i study of mrx34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]


| Target | Target Type | Cancer | Reference |
|---|---|---|---|
| FNDC3B (Fibronectin Type III Domain Containing 3B) | Gene | Esophageal | [25] |
| HMGB1 (High Mobility Group Box-1) | Gene | Acute Myeloid Leukemia | [24] |
| IGF2BP3 (Insulin-like growth factor-2 mRNA-binding protein 3) | Gene | Gastric | [26] |
| Linc-ROR (long intergenic non-protein coding RNA, regulator of reprogramming) | lncRNA | Breast | [27] |
| MMP2 (matrix metalloproteinase-2) | Gene | Esophageal | [25] |
| NEAT1 (Nuclear paraspeckle assembly transcript-1) | lncRNA | Renal | [28] |
| N-MYC (basic helix-loop-helix protein 37) | Gene | Renal/colorectal | [29] |
| SIRT1 (silent expression regulator-1) | Gene | Renal | [30] |
| TUG1 (Taurine upregulated 1) | lncRNA | Endometrial | [31] |
| Target | Mechanism | Biological Function | References |
|---|---|---|---|
| DAPK2 (Death-associated protein kinase 2), Sp1 (specificity protein 1) | miR-34a inhibits apoptosis of dendritic cells through repression of DAPK2/Sp1 pathway | Immunotherapy | [35] |
| AR (Androgen Receptor) | Negative regulation of AR by miR-34a increases ULBP2 expression and enhances NK cell activity | Immunotherapy | [36] |
| PD-L1 (Programmed death-ligand 1) | miR-34a can modulate PD-L1, an important mediator of immune response | Immunotherapy | [37] |
| VEGFA (Vascular endothelial growth factor A) | LncRNA-TUG1 functions as a miR-34a sponge in hepatoblastoma cells, and TUG1-miR-34a-VEGFA axis affects angiogenesis | Angiogenesis | [31] |
| CD44 | Reduced blood vessel density observed in tumors treated with miR-34a | Angiogenesis | [38] |
| Bcl-2 | miR-34a promotes apoptosis | Apotosis | [39,40] |
| Sirtuin-1 (SIRT-1) | SIRT-1 is negatively regulated by miR-34a | Apoptosis | [41] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farooqi, A.A.; Tabassum, S.; Ahmad, A. MicroRNA-34a: A Versatile Regulator of Myriads of Targets in Different Cancers. Int. J. Mol. Sci. 2017, 18, 2089. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102089
Farooqi AA, Tabassum S, Ahmad A. MicroRNA-34a: A Versatile Regulator of Myriads of Targets in Different Cancers. International Journal of Molecular Sciences. 2017; 18(10):2089. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102089
Chicago/Turabian StyleFarooqi, Ammad Ahmad, Sobia Tabassum, and Aamir Ahmad. 2017. "MicroRNA-34a: A Versatile Regulator of Myriads of Targets in Different Cancers" International Journal of Molecular Sciences 18, no. 10: 2089. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102089