Heat-Responsive Photosynthetic and Signaling Pathways in Plants: Insight from Proteomics
Abstract
:1. Introduction
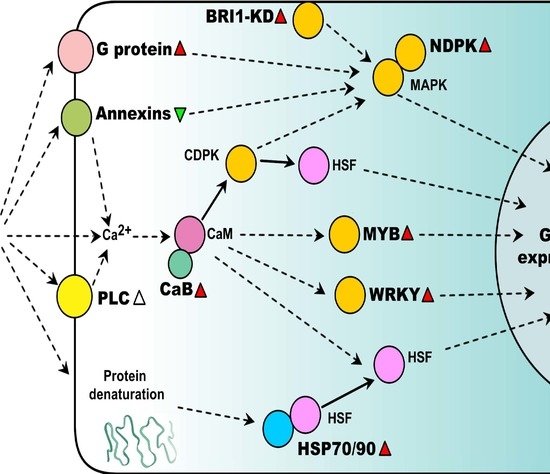
2. Heat-Responsive Signaling Transduction
2.1. Ca2+ Signaling Pathways
2.2. G Protein-Mediated Signaling
2.3. Kinase Signaling Pathways
2.4. Heat-Responsive Transcription Factors
3. Chlorophyll Synthesis Is Disturbed by Heat Stress
4. Photosystem (PS) II and PS I Are Disrupted under Heat Stress
5. CO2 Fixation Is Inhibited Under Heat Stress
6. Photorespiration Is Enhanced to Cope with Heat Stress
7. Energy Supply Is Essential for Heat Tolerance
8. Conclusions and Perspectives
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 2DE | Two-dimensional electrophoretic |
| BRI1-KD | Brassinosteroid-insensitive I-kinase domain |
| C4-PPDK | C4-specific pyruvate orthophosphate dikinase |
| CA | Carbonic anhydrase |
| CDPK | Calcium-dependent protein kinases |
| Chl | Chlorophyll |
| DIGE | Two-dimensional fluorescence difference in gel |
| ESI-Q-TOF | Electrospray ionization quadrupole time-of-flight |
| fstH | Filamentation temperature-sensitive H |
| FBPA | Fructose-bisphosphate aldolase |
| G protein | GTP-binding protein |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| GDC | Glycine decarboxylase |
| GLDC | Glycine dehydrogenases |
| GO | Glycolate oxidase |
| HSF | Heat shock factor |
| HSPs | Heat shock proteins |
| iTRAQ | Isobaric tags for relative and absolute quantitation |
| LHC | Light-harvesting chlorophyll a/b binding protein |
| MALDI | Matrix-assisted laser desorption/ionization |
| MAPK | Mitogen-activated protein kinase |
| MAPK | Mitogen-activated protein kinase |
| NDPK | Nucleoside diphosphate kinase |
| OEC | Oxygen-evolving complex |
| PEPCK | Phosphoenolpyruvate carboxykinase |
| PLC | Phospholipase C |
| POR | Protochlorophyllide reductase |
| PS | Photosystem |
| ROS | Reactive oxygen species |
| RCA | Rubisco activase |
| RuBisCO | Ribulose-1,5-bisphosphate carboxylase/oxygenase |
| RuBP | Ribulose bisphosphate |
| SBPase | Sedoheptulose-1,7-bisphosphatase |
| TK | Chloroplast transketolase |
| TOF | Time-of-flight |
| TOF/TOF | Tandem time-of-flight |
| WOC | Water oxidizing complex |
References
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Long, S.P.; Ort, D.R. More than taking the heat: Crops and global change. Curr. Opin. Plant Biol. 2010, 13, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Lobell, D.B.; Burke, M.; Tebaldi, C.; Mastrandrea, M.D.; Falcon, W.P.; Naylor, R.L. Prioritizing climate change adaptation needs for food security in 2030. Science 2008, 319, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.D.; New, M.; Parker, D.E.; Martin, S.; Rigor, I.G. Surface air temperature and its changes over the past 150 years. Rev. Geophys. 1999, 37, 173–199. [Google Scholar] [CrossRef]
- Mathur, S.; Agrawal, D.; Jajoo, A. Photosynthesis: Response to high temperature stress. J. Photochem. Photobiol. B. 2014, 137, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Ashraf, M. Some important physiological selection criteria for salt tolerance in plants. Flora 2004, 199, 361–376. [Google Scholar] [CrossRef]
- Nouri, M.-Z.; Moumeni, A.; Komatsu, S. Abiotic stresses: Insight into gene regulation and protein expression in photosynthetic pathways of plants. Int. J. Mol. Sci. 2015, 16, 20392–20416. [Google Scholar] [CrossRef] [PubMed]
- Gururani, M.A.; Venkatesh, J.; Tran, L.-S.P. Regulation of Photosynthesis during Abiotic Stress-Induced Photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef] [PubMed]
- Qu, A.-L.; Ding, Y.-F.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Johnova, P.; Skalak, J.; Saiz-Fernandez, I.; Brzobohaty, B. Plant responses to ambient temperature fluctuations and water-limiting conditions: A proteome-wide perspective. BBA-Proteins Proteom. 2016, 1864, 916–931. [Google Scholar] [CrossRef] [PubMed]
- Grover, A.; Mittal, D.; Negi, M.; Lavania, D. Generating high temperature tolerant transgenic plants: Achievements and challenges. Plant Sci. 2013, 205, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Liu, C.F.; Chen, X.B. Proteomics of rice in response to heat stress and advances in genetic engineering for heat tolerance in rice. Plant Cell Rep. 2011, 30, 2155–2165. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.R.; Xu, C.P. Identification and characterization of proteins associated with plant tolerance to heat stress. J. Integr. Plant Biol. 2008, 50, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Abreu, I.A.; Farinha, A.P.; Negrao, S.; Goncalves, N.; Fonseca, C.; Rodrigues, M.; Batista, R.; Saibo, N.J.M.; Oliveira, M.M. Coping with abiotic stress: Proteome changes for crop improvement. J. Proteom. 2013, 93, 145–168. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Chen, S. Understanding information processes at the proteomics level. In Springer Handbook of Bio-/Neuroinformatics; Kasabov, N., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 57–72. [Google Scholar]
- Shakeel, S.N.; Aman, S.; Haq, N.N.; Heckathorn, S.A.; Luthe, D. Proteomic and transcriptomic analyses of Agave americana in response to heat stress. Plant Mol. Biol. Rep. 2013, 31, 840–851. [Google Scholar] [CrossRef]
- Xu, C.P.; Huang, B.R. Root proteomic responses to heat stress in two Agrostis grass species contrasting in heat tolerance. J. Exp. Bot. 2008, 59, 4183–4194. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.P.; Huang, B.R. Differential proteomic response to heat stress in thermal Agrostis scabra and heat-sensitive Agrostis stolonifera. Physiol. Plant. 2010, 139, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Rocco, M.; Arena, S.; Renzone, G.; Scippa, G.S.; Lomaglio, T.; Verrillo, F.; Scaloni, A.; Marra, M. Proteomic analysis of temperature stress-responsive proteins in Arabidopsis thaliana rosette leaves. Mol. Biosyst. 2013, 9, 1257–1267. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Chen, Q.Z.; Kong, L.Q.; Xia, F.S.; Yan, H.F.; Zhu, Y.Q.; Mao, P.S. Proteomic and physiological analysis of the response of oat (Avena sativa) seeds to heat stress under different moisture conditions. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.H.; Lin, K.H.; Chen, S.C.; Shen, Y.H.; Lo, H.F. Proteomic analysis of broccoli (Brassica oleracea) under high temperature and waterlogging stresses. Bot. Stud. 2015, 56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.H.; Li, G.W.; Huang, W.; Bi, T.; Chen, G.Y.; Tang, Z.C.; Su, W.A.; Sun, W.N. Proteomic study of Carissa spinarum in response to combined heat and drought stress. Proteomics 2010, 10, 3117–3129. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, N.; Donnart, T.; Nouri, M.Z.; Komatsu, S. Tissue-specific defense and thermo-adaptive mechanisms of soybean seedlings under heat stress revealed by proteomic approach. J. Proteome Res. 2010, 9, 4189–4204. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Eldakak, M.; Paudel, B.; Kim, D.-W.; Hemmati, H.; Basu, C.; Rohila, J.S. Leaf proteome analysis reveals prospective drought and heat stress response mechanisms in soybean. BioMed Res. Int. 2016, 2016, 6021047. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, H.; Song, L.; Shu, Y.; Gu, W. Comparative proteomics analysis reveals the mechanism of pre-harvest seed deterioration of soybean under high temperature and humidity stress. J. Proteom. 2012, 75, 2109–2127. [Google Scholar] [CrossRef] [PubMed]
- Ashoub, A.; Baeumlisberger, M.; Neupaertl, M.; Karas, M.; Bruggemann, W. Characterization of common and distinctive adjustments of wild barley leaf proteome under drought acclimation, heat stress and their combination. Plant Mol. Biol. Rep. 2015, 87, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Rollins, J.A.; Habte, E.; Templer, S.E.; Colby, T.; Schmidt, J.; von Korff, M. Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeum vulgare L.). J. Exp. Bot. 2013, 64, 3201–3212. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wei, Z.; Qiao, Z.; Wu, Z.; Cheng, L.; Wang, Y. Proteomics analysis of alfalfa response to heat stress. PLoS ONE 2013, 8, e82725. [Google Scholar] [CrossRef] [PubMed]
- Sharmin, S.A.; Alam, I.; Rahman, M.A.; Kim, K.H.; Kim, Y.G.; Lee, B.H. Mapping the leaf proteome of Miscanthus sinensis and its application to the identification of heat-responsive proteins. Planta 2013, 238, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Scafaro, A.P.; Haynes, P.A.; Atwell, B.J. Physiological and molecular changes in Oryza meridionalis Ng., a heat-tolerant species of wild rice. J. Exp. Bot. 2010, 61, 191–202. [Google Scholar]
- Lee, D.; Ahsan, N.; Lee, S.; Kang, K.Y.; Bahk, J.D.; Lee, I.; Lee, B. A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 2007, 7, 3369–3383. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Chen, H.; Li, X.; Yang, M.; Liu, G.; Shen, S. A comparative proteomic analysis of rice seedlings under various high-temperature stresses. Biochim. Biophys. Acta 2009, 1794, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Krishnan, P.; Mishra, V.; Kumar, R.; Ramakrishnan, B.; Singh, N.K. Proteomic changes in rice leaves grown under open field high temperature stress conditions. Mol. Biol. Rep. 2015, 42, 1545–1558. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.J.; Muthurajan, R.; Rahman, H.; Selvam, J.; Peng, S.B.; Zou, Y.B.; Jagadish, K.S.V. Source-sink dynamics and proteomic reprogramming under elevated night temperature and their impact on rice yield and grain quality. New Phytol. 2013, 197, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Gammulla, C.G.; Pascovici, D.; Atwell, B.J.; Haynes, P.A. Differential metabolic response of cultured rice (Oryza sativa) cells exposed to high- and low-temperature stress. Proteomics 2010, 10, 3001–3019. [Google Scholar] [CrossRef] [PubMed]
- Timabud, T.; Yin, X.; Pongdontri, P.; Komatsu, S. Gel-free/label-free proteomic analysis of developing rice grains under heat stress. J. Proteom. 2016, 133, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, W.; Zhang, B.; Zhou, J.; Wang, Y.; Yang, Q.; Ke, Y.; He, H. Phosphoproteins regulated by heat stress in rice leaves. Proteome Sci. 2011, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.H.; Zhu, G.S.; Guo, Q.S.; Zhu, Z.B.; Wang, C.L.; Liu, Z.Y. A comparative proteomic analysis of Pinellia ternata leaves exposed to heat stress. Int. J. Mol. Sci. 2013, 14, 20614–20634. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, J.; Liu, Q.; Ben, C.; Todd, C.D.; Shi, J.; Yang, Y.; Hu, X. Comparative proteomic analysis of the thermotolerant plant Portulaca oleracea acclimation to combined high temperature and humidity stress. J. Proteome Res. 2012, 11, 3605–3623. [Google Scholar] [CrossRef] [PubMed]
- Lara, M.V.; Borsani, J.; Budde, C.O.; Lauxmann, M.A.; Lombardo, V.A.; Murray, R.; Andreo, C.S.; Drincovich, M.F. Biochemical and proteomic analysis of Dixiland peach fruit (Prunus persica) upon heat treatment. J. Exp. Bot. 2009, 60, 4315–4333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Xu, L.; Zhu, X.W.; Gong, Y.Q.; Xiang, F.; Sun, X.C.; Liu, L.W. Proteomic analysis of heat stress response in leaves of radish (Raphanus sativus L.). Plant Mol. Biol. Rep. 2013, 31, 195–203. [Google Scholar] [CrossRef]
- Majoul-Haddad, T.; Bancel, E.; Martre, P.; Triboi, E.; Branlard, G. Effect of short heat shocks applied during grain development on wheat (Triticum aestivum L.) grain proteome. J. Cereal Sci. 2013, 57, 486–495. [Google Scholar] [CrossRef]
- Majoul, T.; Bancel, E.; Triboi, E.; Hamida, J.B.; Branlard, G. Proteomic analysis of the effect of heat stress on hexaploid wheat grain: Characterization of heat-responsive proteins from total endosperm. Proteomics 2003, 3, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Laino, P.; Shelton, D.; Finnie, C.; De Leonardis, A.M.; Mastrangelo, A.M.; Svensson, B.; Lafiandra, D.; Masci, S. Comparative proteome analysis of metabolic proteins from seeds of durum wheat (cv. Svevo) subjected to heat stress. Proteomics 2010, 10, 2359–2368. [Google Scholar] [PubMed]
- Yang, F.; Jorgensen, A.D.; Li, H.; Sondergaard, I.; Finnie, C.; Svensson, B.; Jiang, D.; Wollenweber, B.; Jacobsen, S. Implications of high-temperature events and water deficits on protein profiles in wheat (Triticum aestivum L. cv. Vinjett) grain. Proteomics 2011, 11, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dinler, B.S.; Vignjevic, M.; Jacobsen, S.; Wollenweber, B. Physiological and proteome studies of responses to heat stress during grain filling in contrasting wheat cultivars. Plant Sci. 2015, 230, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-T.; Ma, L.; Duan, W.; Wang, B.-C.; Li, J.-H.; Xu, H.-G.; Yan, X.-Q.; Yan, B.-F.; Li, S.-H.; Wang, L.-J. Differential proteomic analysis of grapevine leaves by iTRAQ reveals responses to heat stress and subsequent recovery. BMC Plant Biol. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wu, L.; Zhao, F.; Zhang, D.; Li, N.; Zhu, G.; Li, C.; Wang, W. Phosphoproteomic analysis of the response of maize leaves to drought, heat and their combination stress. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Finka, A.; Goloubinoff, P. How do plants feel the heat. Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Goraya, G.K.; Kaur, B.; Asthir, B.; Bala, S.; Kaur, G.; Farooq, M. Rapid injuries of high temperature in plants. J. Plant Biol. 2017, 60, 298–305. [Google Scholar] [CrossRef]
- Graff, J.M.; Young, T.N.; Johnson, J.D.; Blackshear, P.J. Phosphorylation-regulated calmodulin binding to a prominent cellular substrate for protein kinase C. J. Biol. Chem. 1989, 264, 21818–21823. [Google Scholar] [PubMed]
- Jami, S.K.; Clark, G.B.; Ayele, B.T.; Ashe, P.; Kirti, P.B. Genome-wide comparative analysis of annexin superfamily in plants. PLoS ONE 2012, 7, e47801. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.M. Annexin-mediated calcium signalling in plants. Plants 2014, 3, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Rohila, J.S.; Chen, M.; Chen, S.; Chen, J.; Cerny, R.L.; Dardick, C.; Canlas, P.E.; Xu, X.; Gribskov, M.; Kanrar, S. Protein–protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J. 2006, 46, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.Y.; Cheong, N.E.; Lee, D.C.; Je, D.Y.; Bahk, J.D.; Cho, M.J.; Lee, S.Y. Cloning and sequencing analysis of a full-length cDNA encoding a G protein α subunit, SGA1, from soybean. Plant Physiol. 1995, 108, 1315–1316. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Signaling through G protein coupled receptors. Plant Signal. Behav. 2009, 4, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zeng, X.; Ding, X.; Li, S.; Yu, C.; Zhu, Y. Ectopic expression of a vesicle trafficking gene, OsRab7, from Oryza sativa, confers tolerance to several abiotic stresses in Escherichia coli. Afr. J. Biotechnol. 2011, 10, 6941–6946. [Google Scholar]
- Yadav, D.K.; Islam, S.M.S.; Tuteja, N. Rice heterotrimeric G-protein gamma subunits (RGG1 and RGG2) are differentially regulated under abiotic stress. Plant Signal. Behav. 2012, 7, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Wu, Y.; Venkataraman, G.; Sopory, S.K.; Tuteja, N. Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): Role in salinity and heat stress and cross-talk with phospholipase C. Plant J. 2007, 51, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.H.; Lee, S.Y.; Park, J.H.; Lee, Y.; Jung, H.S.; Chi, Y.H.; Jung, Y.J.; Chae, H.B.; Shin, M.R.; Kim, W.Y.; et al. Stress-driven structural and functional switching of Ypt1p from a GTPase to a molecular chaperone mediates thermo tolerance in Saccharomyces cerevisiae. FASEB J. 2015, 29, 4424–4434. [Google Scholar] [PubMed]
- Moon, H.; Lee, B.; Choi, G.; Shin, D.; Prasad, D.T.; Lee, O.; Kwak, S.-S.; Kim, D.H.; Nam, J.; Bahk, J.; et al. NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc. Natl. Acad. Sci. USA 2003, 100, 358–363. [Google Scholar] [PubMed]
- Dooki, A.D.; Mayer-Posner, F.J.; Askari, H.; Zaiee, A.A.; Salekdeh, G.H. Proteomic responses of rice young panicles to salinity. Proteomics 2006, 6, 6498–6507. [Google Scholar] [CrossRef] [PubMed]
- Maleki, M.; Naghavi, M.R.; Alizadeh, H.; Poostini, K.; Mishani, C.A. Comparison of protein changes in the leaves of two bread wheat cultivars with different sensitivity under salt stress. Annu. Res. Rev. Biol. 2014, 4, 1784–1797. [Google Scholar] [CrossRef]
- Singh, A.; Bhatnagar, N.; Pandey, A.; Pandey, G.K. Plant phospholipase C family: Regulation and functional role in lipid signaling. Cell Calcium 2015, 58, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, F.; Yan, P.; Jing, W.; Zhang, C.; Kudla, J.; Zhang, W. A phosphoinositide-specific phospholipase C pathway elicits stress-induced Ca2+ signals and confers salt tolerance to rice. New Phytol. 2017, 214, 1172–1187. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-R.; Yang, A.-F.; Yue, G.-D.; Gao, Q.; Yin, H.-Y.; Zhang, J.-R. Enhanced expression of phospholipase C 1 (ZmPLC1) improves drought tolerance in transgenic maize. Planta 2008, 227, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Krckova, Z.; Brouzdova, J.; Danek, M.; Kocourkova, D.; Rainteau, D.; Ruelland, E.; Valentova, O.; Pejchar, P.; Martinec, J. Arabidopsis non-specific phospholipase C1: Characterization and its involvement in response to heat stress. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Samad, A.F.A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.A.; Zainal, Z.; Ismail, I. MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- El-kereamy, A.; Bi, Y.-M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Efeoglu, B.; Ekmekci, Y.; Cicek, N. Physiological responses of three maize cultivars to drought stress and recovery. S. Afr. J. Bot. 2009, 75, 34–42. [Google Scholar] [CrossRef]
- Farhad, M.; Babak, A.M.; Reza, Z.M.; Hassan, R.M.; Afshin, T. Response of proline, soluble sugars, photosynthetic pigments and antioxidant enzymes in potato (Solanum tuberosum L.) to different irrigation regimes in greenhouse condition. Aust. J. Crop Sci. 2011, 5, 55–60. [Google Scholar]
- Mohanty, S.; Grimm, B.; Tripathy, B.C. Light and dark modulation of chlorophyll biosynthetic genes in response to temperature. Planta 2006, 224, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Selstam, E.; Schelin, J.; Brain, T.; Williams, W.P. The effects of low pH on the properties of protochlorophyllide oxidoreductase and the organization of prolamellar bodies of maize (Zea mays). FEBS J. 2002, 269, 2336–2346. [Google Scholar] [CrossRef]
- Berry Bjorkman, O. Photosynthetic response and adaptation to temperature in higher plants. Annu. Rev. Plant Biol. 2003, 31, 491–543. [Google Scholar] [CrossRef]
- Srivastava, A.; Guisse, B.; Greppin, H.; Strasser, R.J. Regulation of antenna structure and electron transport in Photosystem II of Pisum sativum under elevated temperature probed by the fast polyphasic chlorophyll a fluorescence transient: OKJIP. Biochim. Biophys. Acta 1997, 1320, 95–106. [Google Scholar] [CrossRef]
- Takahashi, S.; Bauwe, H.; Badger, M.R. Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol. 2007, 144, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Havaux, M. Rapid photosynthetic adaptation to heat stress triggered in potato leaves by moderately elevated temperatures. Plant Cell Environ. 1993, 16, 461–467. [Google Scholar] [CrossRef]
- Chen, L.; Jia, H.; Du, L.; Tian, Q.; Gao, Y.; Liu, Y. Release of the oxygen-evolving complex subunits from photosystem II membranes in phosphorylation condition under light stress. Chin. J. Chem. 2011, 29, 2631–2636. [Google Scholar] [CrossRef]
- Che, Y.; Fu, A.; Hou, X.; Mcdonald, K.L.; Buchanan, B.B.; Huang, W.; Luan, S. C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 16247–16252. [Google Scholar] [CrossRef] [PubMed]
- Schuhmann, H.; Adamska, I. Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiol. Plant. 2012, 145, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Burke, J.J.; Velten, J.; Xin, Z. FtsH11 protease plays a critical role in Arabidopsis thermotolerance. Plant J. 2006, 48, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Sonoike, K. Photoinhibition of photosystem I. Physiol. Plant. 2011, 142, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Majeau, N.; Arnoldo, M.; Coleman, J.R. Modification of carbonic anhydrase activity by antisense and over-expression constructs in transgenic tobacco. Plant Mol. Biol. 1994, 25, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Tianpei, X.; Mao, Z.; Zhu, Y.; Li, S. Expression of rice mature carbonic anhydrase gene increase E. coli tolerance to heat stress. Appl. Biochem. Biotechnol. 2015, 176, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Raines, C.A. Increasing photosynthetic carbon assimilation in C3 plants to improve crop yield: Current and future strategies. Plant Physiol. 2011, 155, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Craftsbrandner, S.J.; Salvucci, M.E. Sensitivity of photosynthesis in a C4 plant, maize, to heat stress. Plant Physiol. 2002, 129, 1773–1780. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, M.E.; Craftsbrandner, S.J. Relationship between the heat tolerance of photosynthesis and the thermal stability of Rubisco activase in plants from contrasting thermal environments. Plant Physiol. 2004, 134, 1460–1470. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, X.; Zhou, Z.; Feng, X.; Yang, W.X.; Jiang, D. Two Rubisco activase isoforms may play different roles in photosynthetic heat acclimation in the rice plant. Physiol. Plant. 2010, 139, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Kurek, I.; Chang, T.K.; Bertain, S.; Madrigal, A.; Liu, L.; Lassner, M.; Zhu, G. Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell 2007, 19, 3230–3241. [Google Scholar] [CrossRef] [PubMed]
- Rokka, A.; Zhang, L.; Aro, E. Rubisco activase: An enzyme with a temperature-dependent dual function? Plant J. 2001, 25, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.P.; Willingham, N.M.; Lloyd, J.C.; Raines, C.A. Reduced sedoheptulose-1,7-bisphosphatase levels in transgenic tobacco lead to decreased photosynthetic capacity and altered carbohydrate accumulation. Planta 1998, 204, 27–36. [Google Scholar] [CrossRef]
- Feng, L.; Wang, K.; Li, Y.; Tan, Y.; Kong, J.; Li, H.; Li, Y.; Zhu, Y. Overexpression of SBPase enhances photosynthesis against high temperature stress in transgenic rice plants. Plant Cell Rep. 2007, 26, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Guo, A.; Jin, X.; Yang, Q.; Wang, D.; Sun, Y.; Huang, Q.; Wang, L.; Peng, C.; Wang, X. The beta subunit of glyceraldehyde 3-phosphate dehydrogenase is an important factor for maintaining photosynthesis and plant development under salt stress—Based on an integrative analysis of the structural, physiological and proteomic changes in chloroplasts in Thellungiella halophila. Plant Sci. 2015, 236, 223–238. [Google Scholar] [PubMed]
- Ishimaru, K.; Ichikawa, H.; Matsuoka, M.; Ohsugi, R. Analysis of a C4 maize pyruvate, orthophosphate dikinase expressed in C3 transgenic Arabidopsis plants. Plant Sci. 1997, 129, 57–64. [Google Scholar] [CrossRef]
- Walker, R.P.; Paoletti, A.; Leegood, R.C.; Famiani, F. Phosphorylation of phosphoenolpyruvate carboxykinase (PEPCK) and phosphoenolpyruvate carboxylase (PEPC) in the flesh of fruits. Plant Physiol. Biochem. 2016, 108, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Monreal, J.; Feria, A.B.; Vinardell, J.M.; Vidal, J.; Echevarria, C.; Garciamaurino, S. ABA modulates the degradation of phosphoenolpyruvate carboxylase kinase in sorghum leaves. FEBS Lett. 2007, 581, 3468–3472. [Google Scholar] [CrossRef] [PubMed]
- Fukayama, H.; Tsuchida, H.; Agarie, S.; Nomura, M.; Onodera, H.; Ono, K.; Lee, B.H.; Hirose, S.; Toki, S.; Ku, M.S.; et al. Significant accumulation of C4-specific pyruvate, orthophosphate dikinase in a C3 plant, rice. Plant Physiol. 2001, 127, 1136–1146. [Google Scholar]
- Ku, M.S.B.; Agarie, S.; Nomura, M.; Fukayama, H.; Tsuchida, H.; Ono, K.; Hirose, S.; Toki, S.; Miyao, M.; Matsuoka, M. High-level expression of maize phosphoenolpyruvate carboxylasein transgenic rice plants. Nat. Biotechnol. 1999, 17, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Han, Y.; Liu, G.; An, B.; Yang, J.; Yang, G.; Li, Y.; Zhu, Y. Overexpression of sedoheptulose-1,7-bisphosphatase enhances photosynthesis and growth under salt stress in transgenic rice plants. Funct. Plant Biol. 2007, 34, 822–834. [Google Scholar] [CrossRef]
- Zhang, X.; Rerksiri, W.; Liu, A.; Zhou, X.; Xiong, H.; Xiang, J.; Chen, X.; Xiong, X. Transcriptome profile reveals heat response mechanism at molecular and metabolic levels in rice flag leaf. Gene 2013, 530, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.-L.; Lu, Y.; Li, Y.; Yang, C.; Peng, X.-X. Overexpression of glycolate oxidase confers improved photosynthesis under high light and high temperature in rice. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, Y.; Yang, Q.; Zhang, Z.; Chen, Y.; Zhang, S.; Peng, X. Suppression of glycolate oxidase causes glyoxylate accumulation that inhibits photosynthesis through deactivating Rubisco in rice. Physiol. Plant. 2014, 150, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Timm, S.; Florian, A.; Arrivault, S.; Stitt, M.; Fernie, A.R.; Bauwe, H. Glycine decarboxylase controls photosynthesis and plant growth. FEBS Lett. 2012, 586, 3692–3697. [Google Scholar] [CrossRef] [PubMed]
- Bunney, T.D.; van Walraven, H.S.; de Boer, A.H. 14-3-3 protein is a regulator of the mitochondrial and chloroplast ATP synthase. Proc. Natl. Acad. Sci. USA 2001, 98, 4249–4254. [Google Scholar] [CrossRef] [PubMed]
- Molik, S.; Karnauchov, I.; Weidlich, C.; Herrmann, R.G.; Klosgen, R.B. The Rieske Fe/S protein of the cytochromeb 6 /f complex in chloroplasts. J. Biol. Chem. 2001, 276, 42761–42766. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Kumar, V.; Khare, T.; Sharma, M.; Wani, S.H. Engineering crops for future: A phosphoproteomics approach. Curr. Protein Pept. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]


| Plant Species | Tissue/Organ | Variety | Treatment Conditions | Platforms | Protein Species (a) | Unique Proteins (b) | Reference |
|---|---|---|---|---|---|---|---|
| Agave americana | leaf chloroplast | nd | 55 °C; 4 h | 2DE, ESI-Q-Trap MS | 58 | 58(26/32) | [19] |
| Agrostis scabra/Agrostis stolonifera | root | nd; Penncross | 20 °C, 30 °C, 40 °C; 2 d, 10 d | 2DE, MALDI-TOF/TOF MS | 70 | 67(23/44) | [20] |
| Agrostis scabra/Agrostis stolonifera | leaf | nd; Penncross | 40 °C/35 °C day/night; 2 d, 10 d | 2DE-DIGE | 71 | 71(57/14) | [21] |
| Arabidopsis thaliana | leaf | Columbia (Col-0) | 40 °C; 6 h | 2DE, MALDI-TOF MS, ESI-Q-Trap MS | 37 | 33(12/21) | [22] |
| Avena sativa | seed | nd | 35 °C, 45 °C, 50 °C; 24 h, 2 d | 2DE, ESI-Orbitrap MS | 21 | 21(2/19) | [23] |
| Brassica oleracea | leaf | TSS-AVRDC-2; B-75 | 40 °C; 3 d | 2DE, MALDI-TOF MS | 24 | 24(10/13/1) | [24] |
| Carissa spinarum | leaf | nd | 42 °C/35 °C day/night; 48 h, 120 h | 2DE, MALDI-TOF/TOF MS | 49 | 26(13/13) | [25] |
| Glycine max | leaf, stem, root | Enrei | 40 °C; 6, 12, 24 h | 2DE, MALDI-TOF MSESI-Orbitrap MS | 150 | 150(122/28) | [26] |
| Glycine max | leaf | Surge; Davison | 40 °C/35 °C day/night; 6 d | 2DE-DIGE, MALDI-TOF MS | 88 | 44(19/25) | [27] |
| Glycine max | seed | Ningzhen No. 1 | 40 °C/30 °C day/night; 24, 96, 168 h | 2DE, MALDI-TOF MS | 42 | 42(22/20) | [28] |
| Hordeum spontaneum | leaf | nd | 42 °C, 2 h | 2DE-DIGE, MALDI-TOF/TOF MS | 20 | 20(12/8) | [29] |
| Hordeum vulgare | leaf | Arta; Keel | 36 °C/32 °C day/night; 7 d | 2DE, MALDI-TOF/TOF MS | 99 | 99(67/32) | [30] |
| Medicago sativa | leaf | Huaiyin | 36 °C; 24, 48, 72 h | 2DE, MALDI-TOF/TOF MS | 81 | 81 | [31] |
| Miscanthus sinensis | leaf | Kosung | 42 °C; 24, 48 h | 2DE, MALDI-TOF MS, 2DE, MALDI-TOF/TOF MS | 55 | 55(30/25) | [32] |
| Oryza meridionalis | leaf | nd | 45 °C; 24 h | 2DE, ESI-Q-Trap MS | 50 | 38(22/16) | [33] |
| Oryza sativa | leaf | Dongjin | 40 °C; 12, 24 h | 2DE, MALDI-TOF MS | 73 | 56(47/9) | [34] |
| Oryza sativa | leaf | nd | 35, 40, 45 °C; 48 h | 2DE, MALDI-TOF MS | 63 | 52(28/24) | [35] |
| Oryza sativa | leaf | N22 | 42 °C/32 °C day/night; 24 h | 2DE, MALDI-TOF MS | 111 | 52(37/15) | [36] |
| Oryza sativa | leaf, spikelet | N22; Gharib | 28 °C; 12 h | 2DE, MALDI-TOF MS | 36 | 36 | [37] |
| Oryza sativa | cell suspension cultures | Doongara | 44 °C; 3 d | 1DE, ESI-Q-Trap MS | 139 | 139 | [38] |
| Oryza sativa | grain | Khao Dawk Mali 105 | 40 °C; 3 d | ESI-Orbitap MS | 822 | 822 | [39] |
| Oryza sativa | leaf | Nipponbare | 42 °C, 12 h, 24 h | 2DE, MALDI-TOF/TOF MS | 12 | 12(9/3) | [40] * |
| Pinellia ternata | leaf | nd | 38 °C; 24 h | 2DE, MALDI-TOF/TOF MS | 27 | 24(17/7) | [41] |
| Portulaca oleracea | leaf | nd | 35 °C; 6h, 12 h, 24 h | 2D, MALDI-TOF/TOF MS | 154 | 51(36/15) | [42] |
| Prunus persica | mesocarp | nd | 39 °C; 3 d | 2DE, MALDI-TOF/TOF MS | 44 | 33(15/18) | [43] |
| Raphanus sativus | leaf | NAU-08Hr-10 | 40 °C; 0 h, 12 h, 24 h | 2DE, MALDI-TOF/TOF MS | 11 | 11(4/1/6) | [44] |
| Triticum aestivum | endosperm | Récital | 34 °C/10 °C day/night | 2DE, MALDI-TOF MS | 37 | 23(22/1) | [45] |
| Triticum aestivum | non-prolamins | Thésée | 34 °C/10 °C day/night | 2DE, MALDI-TOF MS | 42 | 24(16/8) | [46] |
| Triticum aestivum | seed | Svevo | 37 °C/17 °C day/night; 5 d | 2DE, MALDI-TOF/TOF MS | 47 | 47(37/10) | [47] |
| Triticum aestivum | spikelet | Vinjett | 32 °C/24 °C day/night; 10 d | 2DE, MALDI-TOF/TOF MS | 57 | 57(36/21) | [48] |
| Triticum aestivum | leaf | 810; 1039 | 35 °C/26 °C day/night; 5 d | 2DE, MALDI-TOF MS | 49 | 49(32/11, 12/21) | [49] |
| Vitis vinifera | leaf | Cabernet Sauvignon | 43 °C; 6 h | iTRAQ, ESI-Q-TOF MS | 113 | 113(48/65) | [50] |
| Zea mays | leaf | Zhengdan 958 | heat from 28 to 42 °C, total 8 h | iTRAQ, ESI-Orbitrap MS | 172 | 172(77/95) | [51] * |
| Protein Name | Abbreviation | Plant Species |
|---|---|---|
| 1. Photoreaction | ||
| Magnesium chelatase subunit | CHLI | Po; Gm; Vv |
| Chlorophyll a/b binding protein | LHC | Ta; Pt; Os; Vv; MS |
| Oxygen-evolving enhancer protein 1 | OEE1 | Gm; Aa; Ta; Os; Cs; Asc; Ast; At |
| Oxygen-evolving enhancer protein 2 | OEE2 | Asc; Rs; Gm; Os; Gm; At; Ms |
| Photosystem I PsaA subunit | PsaA | Aa; Vv |
| Photosystem I PsaB subunit | PsaB | Aa |
| Photosystem I PsaD subunit | PsaD | Aa; At; Vv; Ms |
| Photosystem I PsaE subunit | PsaE | Asc; Ast |
| Photosystem I PsaN subunit | PsaN | Aa; Asc; Vv |
| Photosystem I PsaL subunit | PsaL | Vv |
| Photosystem I PsaF subunit | PsaF | Vv |
| Photosystem I PsaH subunit | PsaH | Vv |
| Photosystem II PsbA subunit | PsbA | Vv |
| Photosystem II PsbP subunit | PsbP | Os |
| Photosystem II PsbO subunit | PsbO | Os |
| Photosystem II PsbS subunit | PsbS | Vv |
| Photosystem II PsbR subunit | PsbR | Vv |
| 2. Calvin cycle | ||
| RuBisCO activase | RCA | Os; Ta; Om; Asc; Ast; Os; At; Vv; Ms |
| RuBisCO large subunit | Rubisco LS | Gm; Os; Bo; Aa; Ta; Po; Cs; Cs; Om; Asc; Ast; At; Vv; Ms |
| RuBisCO small subunit | Rubisco SS | Ta; Gm; Os; Bo; Asc; Ast; At; Vv; Ms |
| Phosphoribulokinase | PRK | Os; Om; Asc; Ast; Vv |
| Transketolase | TK | Os; Om |
| Sedoheptulose-1,7-bisphosphatase | SBPase | Gm; Ta; Om |
| Chloroplast fructose-bisphosphate aldolase | FBPA | At; Asc; Ast; Gm; Ta; Vv |
| C4-specific pyruvate orthophosphate dikinase | PPDK | Ta |
| Carbonic anhydrase | CA | Asc; Ast; Gm; Vv |
| Phosphoglycerate kinase | PGK | Om |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | At; Ast; Asc; Ta; Ms Vv, |
| 3. Photorespiration | ||
| Glycolate oxidase | GO | Po |
| Glycine dehydrogenase | GLDC | Os; Om |
| Glycine decarboxylase | GDC | Ta; Os |
| Glutamine synthetase | GS | Os |
| 4. ATP synthesis and electrical transport chain | ||
| ATP synthase CF (0) b subunit | CF0 | Vv |
| ADP, ATP carrier protein 1 | AAC | Aa |
| ATP synthase α subunit | α | Os; Ta |
| ATP synthase β subunit | β | Aa; Asc; Ast; Bo; Gm; Om; Os; Ta; Vv |
| ATP synthase γ subunit | γ | Asc; Ast; At; Vv |
| ATP synthase δ subunit | δ | Cs; Vv |
| Cytochrome c oxidase assembly protein | COX | Po |
| Rieske Fe/S protein of cytochrome b6/f complex | Fe/S | Pt |
| Ferredoxin-NADP(H) oxidoreductase | FNR | Om; Gm; Vv |
| Plastocyanin | PC | Ms |
| 5. Glycolysis | ||
| Triosephosphate isomerase | TPI | Gm; Ta; Cs; Os; At |
| Alcohol dehydrogenase | ADH | Gm; Po |
| Fructokinase | FK | Gm |
| Fructose-bisphosphate aldolase | FBPA | Gm; Asc; Ast; Os |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | Gm; Gm; Ta; Asc; Ast; Os; At |
| Phosphoglycerate kinase | PGK | Gm; Asc; Ast; Os |
| Phosphoglycerate mutase | PGM | Ta |
| Pyruvate kinase | PK | Ast |
| Enolase | Ta; Asc; Ast; Ms | |
| 6. Pentose phosphate pathway | ||
| Phosphogluconate dehydrogenase | PGD | Asc; Ast; Vv |
| Transketolase | TK | Os |
| 7. TCA cycle | ||
| Malate dehydrogenase | MDH | Os; Gm; Asc; Ast; Vv; Ms |
| Isocitrate dehydrogenase | IDH | Ta |
| Cytoplasmic aconitate hydratase | AH | Asc; Ast |
| Phosphoenolpyruvate carboxylase | PEPC | Po; Gm; Aa |
| Dihydrolipoyl dehydrogenase | DLDH | Ta |
| Pyruvate dehydrogenase | PDH | Os; Cs; Gm |
| Citrate synthase | CS | Ms; Vv |
| Fumarate hydratase | FH | Ms |
| 8. Signaling | ||
| Phospholipase C | PLC | Ta |
| GTP-binding protein | G protein | Po |
| Ras-related nuclear protein 1A | Ran 1A | Asc; Ast |
| Ras-related protein | Rab | Gm |
| Ca2+-transporting ATPase-like protein | Aa | |
| Calcium/calmodulin-dependent protein kinase | CDPK | Gm |
| Calcium-binding protein | CaB | Aa |
| BRI1-KD interacting protein 114 | Ta | |
| Nucleoside diphosphate kinase | NDPK | As; Gm; Ms; Os |
| WRKY transcriptional factor | WRKY | Po |
| MYB family transcription factor | MYB | Po |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Xu, C.; Cai, X.; Wang, Q.; Dai, S. Heat-Responsive Photosynthetic and Signaling Pathways in Plants: Insight from Proteomics. Int. J. Mol. Sci. 2017, 18, 2191. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102191
Wang X, Xu C, Cai X, Wang Q, Dai S. Heat-Responsive Photosynthetic and Signaling Pathways in Plants: Insight from Proteomics. International Journal of Molecular Sciences. 2017; 18(10):2191. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102191
Chicago/Turabian StyleWang, Xiaoli, Chenxi Xu, Xiaofeng Cai, Quanhua Wang, and Shaojun Dai. 2017. "Heat-Responsive Photosynthetic and Signaling Pathways in Plants: Insight from Proteomics" International Journal of Molecular Sciences 18, no. 10: 2191. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102191