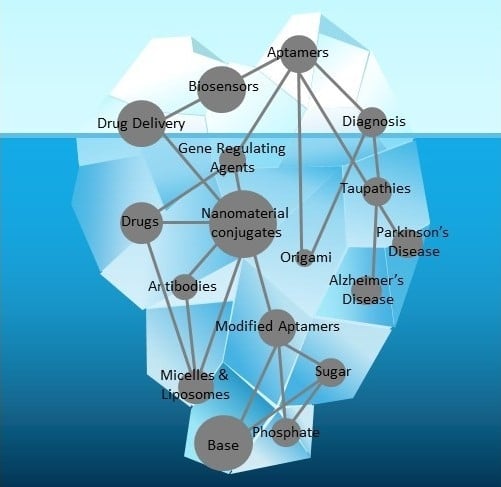
Nucleic Acid Aptamers: Emerging Applications in Medical Imaging, Nanotechnology, Neurosciences, and Drug Delivery
Abstract
:1. Introduction
2. Medical Imaging (MRI and PET)
2.1. Aptamers and MRI
2.2. Aptamers and PET Imaging
3. Aptamers for the Treatment and Diagnostics of Neurological Diseases
3.1. Aptamers and Neurotransmitters
3.2. Aptamers and Tauopathies
3.2.1. Alzheimer’s Disease
3.2.2. Parkinson’s Disease
4. Aptamers as Key Components in the Fabrication of Smart DNA Origami Objects
5. Aptamers as Drug Carriers and Gene Regulating Agents
5.1. Nanomaterials Conjugated Aptamers
5.2. Micelles and Liposomes Conjugated Aptamers
5.3. Aptamer-Drug Conjugates
5.4. Aptamer-Antibody Conjugate
5.5. Aptamers and Gene Regulating Agents
6. Recent Chemical Modifications of Aptamers
6.1. Base Modified Aptamers
6.2. Aptamers with an Extended Genetic Alphabet
6.3. Sugar Modified Aptamers
6.4. Phosphate Modified Aptamers
7. Conclusions and Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CTC | Circulating Cancer Cells |
| GO | Graphene oxide |
| AFM | Atomic Force Microscopy |
| SELEX | Systematic Evolution of Ligands by Exponential Enrichment |
| PET | Positron Emission Tomography |
| MRI | Magnetic Resonance Imaging |
| DOTA | 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid |
| KD | Dissociation constant |
| XNA | Xeno nucleic acid |
| SPIONs | Superparamagnetic iron oxide nanoparticles |
| NOTA | 1,4,7-triazacyclononane-triacetic acid |
| DNAzyme | DNA enzyme or catalytic DNA |
| SPECT | Single Photon Emission Computed Tomography |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| DOX | Doxorubicin |
| QD | Quantum Dot |
| PEG | Polyethyleneglycol |
| siRNA | Small interfering RNA |
| miRNA | Micro RNA |
| sgRNA | Single guide RNA |
| pRNA | Packaging RNA |
| 3WJ | Three-way junction |
| GABA | γ-Aminobutyric acid |
| BSA | Bovine Serum Albumin |
| Aβ | Amyloid β |
References
- Goyenvalle, A.; Griffith, G.; Babbs, A.; El Andaloussi, S.; Ezzat, K.; Avril, A.; Dugovic, B.; Chaussenot, R.; Ferry, A.; Voit, T.; et al. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 2015, 21, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Watts, J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017, 35, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.; Ke, Y. The beauty and utility of DNA origami. Chem 2017, 2, 359–382. [Google Scholar] [CrossRef]
- Hollenstein, M. DNA catalysis: The chemical repertoire of DNAzymes. Molecules 2015, 20, 20777–20804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverman, S.K. Catalyic DNA: Scope, applications, and biochemistry of deoxyribozymes. Trends Biochem. Sci. 2016, 41, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Rioz-Martinez, A.; Roelfes, G. DNA-based hybrid catalysis. Curr. Opin. Chem. Biol. 2015, 25, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.Y.; Liang, C.; Lv, Q.X.; Li, D.F.; Xu, X.G.; Liu, B.Q.; Lu, A.P.; Zhang, G. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int. J. Mol. Sci. 2016, 17, 358. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Q.; Wan, S.; Jiang, Y.; Wang, Y.Y.; Fu, T.; Liu, Q.L.; Cao, Z.J.; Qiu, L.P.; Tan, W.H. Molecular elucidation of disease biomarkers at the interface of chemistry and biology. J. Am. Chem. Soc. 2017, 139, 2532–2540. [Google Scholar] [CrossRef] [PubMed]
- Sedlyarova, N.; Rescheneder, P.; Magan, A.; Popitsch, N.; Rziha, N.; Bilusic, I.; Epshtein, V.; Zimmermann, B.; Lybecker, M.; Sedlyarov, V.; et al. Natural RNA polymerase aptamers regulate transcription in E. coli. Mol. Cell 2017, 67, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F. Forty years of in vitro evolution. Angew. Chem. Int. Ed. 2007, 46, 6420–6436. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, F.; Mayer, G. Selection and biosensor application of aptamers for small molecules. Front. Chem. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded-DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yu, Y.Y.; Jiang, F.; Zhou, J.W.; Li, Y.S.; Liang, C.; Dang, L.; Lu, A.P.; Zhang, G. Development of cell-SELEX technology and its application in cancer diagnosis and therapy. Int. J. Mol. Sci. 2016, 17, 2079. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Shangguan, D.; Xiong, X.L.; O’Donoghue, M.B.; Tan, W.H. Development of DNA aptamers using cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Li, Y.; Tang, Z.W.; Cao, Z.H.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.Y.J.; Tan, W.H. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.F.; Hesselberth, J.R.; Meyers, L.A.; Ellington, A.D. Aptamer database. Nucleic Acids Res. 2004, 32, D95–D100. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Toledo, J.; McKeague, M.; Zhang, X.R.; Giamberardino, A.; McConnell, E.; Francis, T.; DeRosa, M.C.; Dumontier, M. Aptamer base: A collaborative knowledge base to describe aptamers and SELEX experiments. Database 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.X.; Kwon, Y.J. Aptamers: The “evolution” of SELEX. Methods 2016, 106, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Hirota, M.; Waugh, S.M.; Murakami, I.; Suzuki, T.; Muraguchi, M.; Shibamori, M.; Ishikawa, Y.; Jarvis, T.C.; Carter, J.D.; et al. Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J. Biol. Chem. 2014, 289, 8706–8719. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, P.; Kurniawan, H.; Byrne, M.E.; Wower, J. Therapeutic RNA aptamers in clinical trials. Eur. J. Pharm. Sci. 2013, 48, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.H.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.M.; Liu, H.; Kuai, H.L.; Peng, R.Z.; Mo, L.T.; Zhang, X.B. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem. Soc. Rev. 2016, 45, 2583–2602. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.H.; Zhang, T.T.; Luo, H.; Yen, T.M.; Chen, P.W.; Han, Y.Y.; Lo, Y.H. Nucleic acid aptamers: An emerging tool for biotechnology and biomedical sensing. Sensors 2015, 15, 16281–16313. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed]
- Bunka, D.H.J.; Stockley, P.G. Aptamers come of age—At last. Nat. Rev. Microbiol. 2006, 4, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M.; Hartig, J.S.; Mayer, G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007, 107, 3715–3743. [Google Scholar] [CrossRef] [PubMed]
- Forier, C.; Boschetti, E.; Ouhammouch, M.; Cibiel, A.; Duconge, F.; Nogre, M.; Tellier, M.; Bataille, D.; Bihoreau, N.; Santambien, P.; et al. DNA aptamer affinity ligands for highly selective purification of human plasma-related proteins from multiple sources. J. Chromatogr. A 2017, 1489, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Z.; Huang, P.J.J.; Ding, J.S.; Liu, J. Aptamer-based biosensors for biomedical diagnostics. Analyst 2014, 139, 2627–2640. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.; Ban, C. Aptamer–nanoparticle complexes as powerful diagnostic and therapeutic tools. Exp. Mol. Med. 2016, 48, e230. [Google Scholar] [CrossRef] [PubMed]
- Uzawa, T.; Tada, S.; Wang, W. Expansion of the aptamer library from a “natural soup” to an “unnatural soup”. Chem. Commun. 2013, 49, 1786–1795. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Z.; Farokhzad, O.C. Current progress of aptamer-based molecular imaging. J. Nucl. Med. 2014, 55, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Boros, E.; Gale, E.M.; Caravan, P. MR imaging probes: Design and applications. Dalton Trans. 2015, 44, 4804–4818. [Google Scholar] [CrossRef] [PubMed]
- Hingorani, D.V.; Bernstein, A.S.; Pagel, M.D. A review of responsive MRI contrast agents: 2005–2014. Contrast Media Mol. Imaging 2015, 10, 245–265. [Google Scholar] [CrossRef] [PubMed]
- Que, E.L.; Chang, C.J. Responsive magnetic resonance imaging contrast agents as chemical sensors for metals in biology and medicine. Chem. Soc. Rev. 2010, 39, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Gale, E.M.; Jones, C.M.; Ramsay, I.; Farrar, C.T.; Caravan, P. A janus chelator enables biochemically responsive MRI contrast with exceptional dynamic range. J. Am. Chem. Soc. 2016, 138, 15861–15864. [Google Scholar] [CrossRef] [PubMed]
- Yigit, M.V.; Mazumdar, D.; Kim, H.K.; Lee, J.H.; Dintsov, B.; Lu, Y. Smart “turn-on” magnetic resonance contrast agents based on aptamer-functionalized superparamagnetic iron oxide nanoparticles. ChemBioChem 2007, 8, 1675–1678. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Park, I.K.; Jeong, Y.Y. Magnetic iron oxide nanoparticles for multimodal imaging and therapy of cancer. Int. J. Mol. Sci. 2013, 14, 15910–15930. [Google Scholar] [CrossRef] [PubMed]
- Huizenga, D.E.; Szostak, J.W. A DNA aptamer that binds adenosine and ATP. Biochemistry 1995, 34, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Tasset, D.M.; Kubik, M.F.; Steiner, W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997, 272, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Yigit, M.V.; Mazumdar, D.; Lu, Y. MRI detection of thrombin with aptamer functionalized superparamagnetic iron oxide nanoparticles. Bioconjug. Chem. 2008, 19, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.C.; Lu, Y. A smart magnetic resonance imaging contrast agent responsive to adenosine based on a DNA aptamer-conjugated gadolinium complex. Chem. Commun. 2011, 47, 4998–5000. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.C.; Xing, H.; Lu, Y. A smart T-1-weighted MRI contrast agent for uranyl cations based on a DNAzyme-gadolinium conjugate. Analyst 2013, 138, 6266–6269. [Google Scholar] [CrossRef] [PubMed]
- Artemov, D.; Mori, N.; Ravi, R.; Bhujwalla, Z.M. Magnetic resonance molecular imaging of the Her-2/neu receptor. Cancer Res. 2003, 63, 2723–2727. [Google Scholar] [PubMed]
- Zhou, Z.X.; Lu, Z.R. Gadolinium-based contrast agents for magnetic resonance cancer imaging. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Hicke, B.J.; Stephens, A.W.; Gould, T.; Chang, Y.F.; Lynott, C.K.; Heil, J.; Borkowski, S.; Hilger, C.S.; Cook, G.; Warren, S.; et al. Tumor targeting by an aptamer. J. Nucl. Med. 2006, 47, 668–678. [Google Scholar] [PubMed]
- Bernard, E.D.; Beking, M.A.; Rajamanickam, K.; Tsai, E.C.; DeRosa, M.C. Target binding improves relaxivity in aptamer-gadolinium conjugates. J. Biol. Inorg. Chem. 2012, 17, 1159–1175. [Google Scholar] [CrossRef] [PubMed]
- Caravan, P. Protein-targeted gadolinium-based magnetic resonance imaging (MRI) contrast agents: Design and mechanism of action. Acc. Chem. Res. 2009, 42, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Lupold, S.E.; Hicke, B.J.; Lin, Y.; Coffey, D.S. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002, 62, 4029–4033. [Google Scholar] [PubMed]
- Wang, A.Z.; Bagalkot, V.; Vasilliou, C.C.; Gu, F.; Alexis, F.; Zhang, L.; Shaikh, M.; Yuet, K.; Cima, M.J.; Langer, R.; et al. Superparamagnetic iron oxide nanoparticle-aptamer bioconjugates for combined prostate cancer imaging and therapy. Chem. Med. Chem. 2008, 3, 1311–1315. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Cheng, J.J.; Teply, B.A.; Sherifi, I.; Jon, S.; Kantoff, P.W.; Richie, J.P.; Langer, R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 6315–6320. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.K.; Kim, D.; Lee, I.H.; So, J.S.; Jeong, Y.Y.; Jon, S. Image-guided prostate cancer therapy using aptamer-functionalized thermally cross-linked superparamagnetic iron oxide nanoparticles. Small 2011, 7, 2241–2249. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; You, J.; Dai, Y.; Shi, M.L.; Han, C.P.; Xu, K. Gadolinium oxide nanoparticles and aptamer-functionalized silver nanoclusters-based multimodal molecular imaging nanoprobe for optical/magnetic resonance cancer cell imaging. Anal. Chem. 2014, 86, 11306–11311. [Google Scholar] [CrossRef] [PubMed]
- Ji, K.L.; Lim, W.S.; Li, S.F.Y.; Bhakoo, K. A two-step stimulus-response cell-SELEX method to generate a DNA aptamer to recognize inflamed human aortic endothelial cells as a potential in vivo molecular probe for atherosclerosis plaque detection. Anal. Bioanal. Chem. 2013, 405, 6853–6861. [Google Scholar] [CrossRef] [PubMed]
- Ametamey, S.M.; Honer, M.; Schubiger, P.A. Molecular imaging with PET. Chem. Rev. 2008, 108, 1501–1516. [Google Scholar] [CrossRef] [PubMed]
- Perrin, D.M. [18F]-Organotrifluoroborates as radioprosthetic groups for PET imaging: From design principles to preclinical applications. Acc. Chem. Res. 2016, 49, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Bernard-Gauthier, V.; Bailey, J.J.; Liu, Z.B.; Wangler, B.; Wangler, C.; Jurkschat, K.; Perrin, D.M.; Schirrmacher, R. From unorthodox to established: The current status of F-18-trifluoroborate- and F-18-SiFA-based radiopharmaceuticals in PET nuclear imaging. Bioconjug. Chem. 2016, 27, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.W.; VanBrocklin, H.F.; Taylor, S.E. Photoconjugation of 3-azido-5-nitrobenzyl-[18F] fluoride to an oligonucleotide aptamer. J. Label. Compd. Radiopharm. 2002, 45, 257–268. [Google Scholar] [CrossRef]
- Jacobson, O.; Weiss, I.D.; Wang, L.; Wang, Z.; Yang, X.Y.; Dewhurst, A.; Ma, Y.; Zhu, G.Z.; Niu, G.; Kiesewetter, D.O.; et al. 18F-Labeled single-stranded DNA aptamer for PET imaging of protein tyrosine kinase-7 expression. J. Nucl. Med. 2015, 56, 1780–1785. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jacobson, O.; Avdic, D.; Rotstein, B.H.; Weiss, I.D.; Collier, L.; Chen, X.Y.; Vasdev, N.; Liang, S.H. Ortho-stabilized 18F-Azido click agents and their application in PET imaging with single-stranded DNA aptamers. Angew. Chem. Int. Ed. 2015, 54, 12777–12781. [Google Scholar] [CrossRef] [PubMed]
- Daniels, D.A.; Chen, H.; Hicke, B.J.; Swiderek, K.M.; Gold, L. A tenascin-c aptamer identified by tumor cell-SELEX: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA 2003, 100, 15416–15421. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, O.; Yan, X.F.; Niu, G.; Weiss, I.D.; Ma, Y.; Szajek, L.P.; Shen, B.Z.; Kiesewetter, D.O.; Chen, X.Y. PET imaging of tenascin-c with a radio labeled single-stranded DNA aptamer. J. Nucl. Med. 2015, 56, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.Z.; Zhang, H.M.; Jacobson, O.; Wang, Z.T.; Chen, H.J.; Yang, X.Y.; Niu, G.; Chen, X.Y. Combinatorial screening of DNA aptamers for molecular imaging of her2 in cancer. Bioconjug. Chem. 2017, 28, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lee, T.S.; Song, I.H.; Cho, Y.L.; Chae, J.R.; Yun, M.; Kang, H.; Lee, J.H.; Lim, J.H.; Cho, W.G.; et al. Hybridization-based aptamer labeling using complementary oligonucleotide platform for PET and optical imaging. Biomaterials 2016, 100, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Schulz, J.; Vimont, D.; Bordenave, T.; James, D.; Escudier, J.M.; Allard, M.; Szlosek-Pinaud, M.; Fouquet, E. Silicon-based chemistry: An original and efficient one-step approach to F-18 -nucleosides and F-18 -oligonucleotides for PET imaging. Chem. Eur. J. 2011, 17, 3096–3100. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Escudier, J.M.; Amigues, E.; Schulz, J.; Vitry, C.; Bordenave, T.; Szlosek-Pinaud, M.; Fouquet, E. A ‘click chemistry’ approach to the efficient synthesis of modified nucleosides and oligonucleotides for PET imaging. Tetrahedron Lett. 2010, 51, 1230–1232. [Google Scholar] [CrossRef]
- Li, Y.; Schaffer, P.; Perrin, D.M. Dual isotope labeling: Conjugation of P-32-oligonucleotides with F-18-aryltrifluoroborate via copper(I) catalyzed cycloaddition. Bioorg. Med. Chem. Lett. 2013, 23, 6313–6316. [Google Scholar] [CrossRef] [PubMed]
- Kuhnast, B.; de Bruin, A.; Hinnen, F.; Tavitian, B.; Dolle, F. Design and synthesis of a new F-18 fluoropyridine-based haloacetamide reagent for the labeling of oligonucleotides: 2-bromo-N-3-(2-F-18-fluoropyridin-3-yloxy)propyl acetamide. Bioconjug. Chem. 2004, 15, 617–627. [Google Scholar] [CrossRef] [PubMed]
- James, M.L.; Gambhir, S.S. A molecular imaging primer: Modalities, imaging agents, and applications. Physiol. Rev. 2012, 92, 897–965. [Google Scholar] [CrossRef] [PubMed]
- Gomes, S.D.; Miguel, J.; Azema, L.; Eimer, S.; Ries, C.; Dausse, E.; Loiseau, H.; Allard, M.; Toulme, J.J. Tc-99m-MAG3-aptamer for imaging human tumors associated with high level of matrix metalloprotease-9. Bioconjug. Chem. 2012, 23, 2192–2200. [Google Scholar] [CrossRef] [PubMed]
- Kryza, D.; Debordeaux, F.; Azema, L.; Hassan, A.; Paurelle, O.; Schulz, J.; Savona-Baron, C.; Charignon, E.; Bonazza, P.; Taleb, J.; et al. Ex vivo and in vivo imaging and biodistribution of aptamers targeting the human matrix metalloprotease-9 in melanomas. PLoS ONE 2016, 11, e0149387. [Google Scholar] [CrossRef] [PubMed]
- Macedo, B.; Cordeiro, Y. Unraveling prion protein interactions with aptamers and other PrP-binding nucleic acids. Int. J. Mol. Sci. 2017, 18, 1023. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Yu, S.Q.; Zheng, Y.; Zheng, Y.; Yang, H.; Zhang, J.L. Aptamer and its applications in neurodegenerative diseases. Cell. Mol. Life Sci. 2017, 74, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Wolter, O.; Mayer, G. Aptamers as valuable molecular tools in neurosciences. J. Neurosci. 2017, 37, 2517–2523. [Google Scholar] [CrossRef] [PubMed]
- Nedergaard, M.; Takano, T.; Hansen, A.J. Beyond the role of glutamate as a neurotransmitter. Nat. Rev. Neurosci. 2002, 3, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Perry, M.; Li, Q.; Kennedy, R.T. Review of recent advances in analytical techniques for the determination of neurotransmitters. Anal. Chim. Acta 2009, 653, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Hokfelt, T.; Bartfai, T.; Bloom, F. Neuropeptides: Opportunities for drug discovery. Lancet Neurol. 2003, 2, 463–472. [Google Scholar] [CrossRef]
- Li, B.R.; Hsieh, Y.J.; Chen, Y.X.; Chung, Y.T.; Pan, C.Y.; Chen, Y.T. An ultrasensitive nanowire-transistor biosensor for detecting dopamine release from living pc12 cells under hypoxic stimulation. J. Am. Chem. Soc. 2013, 135, 16034–16037. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, A.; Ekavali. A review on Alzheimer’s disease pathophysiology and its management: An update. Pharmacol. Rep. 2015, 67, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Castren, E. Is mood chemistry? Nat. Rev. Neurosci. 2005, 6, 241–246. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.M.; Holahan, M.R.; DeRosa, M.C. Aptamers as promising molecular recognition elements for diagnostics and therapeutics in the central nervous system. Nucleic Acid Ther. 2014, 24, 388–404. [Google Scholar] [CrossRef] [PubMed]
- Mannironi, C.; DiNardo, A.; Fruscoloni, P.; TocchiniValentini, G.P. In vitro selection of dopamine RNA ligands. Biochemistry 1997, 36, 9726–9734. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, Y.; Yang, X.R. Aptamer-based colorimetric biosensing of dopamine using unmodified gold nanoparticles. Sens. Actuator B Chem. 2011, 156, 95–99. [Google Scholar] [CrossRef]
- Farjami, E.; Campos, R.; Nielsen, J.S.; Gothelf, K.V.; Kjems, J.; Ferapontova, E.E. RNA aptamer-based electrochemical biosensor for selective and label-free analysis of dopamine. Anal. Chem. 2013, 85, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.; DeRosa, M.C. Retention of function in the DNA homolog of the RNA dopamine aptamer. Biochem. Biophys. Res. Commun. 2009, 388, 732–735. [Google Scholar] [CrossRef] [PubMed]
- Holahan, M.R.; Madularu, D.; McConnell, E.M.; Walsh, R.; DeRosa, M.C. Intra-accumbens injection of a dopamine aptamer abates MK-801-induced cognitive dysfunction in a model of schizophrenia. PLoS ONE 2011, 6, e22239. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Martos, I.; Ferapontova, E.E. A DNA sequence obtained by replacement of the dopamine RNA aptamer bases is not an aptamer. Biochem. Biophys. Res. Commun. 2017, 489, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Kammer, M.N.; Olmsted, I.R.; Kussrow, A.K.; Morris, M.J.; Jackson, G.W.; Bornhop, D.J. Characterizing aptamer small molecule interactions with backscattering interferometry. Analyst 2014, 139, 5879–5884. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Carrillo, M.P.; Phillips, T.; King, B. Development of DNA aptamers for cytochemical detection of acetylcholine. In Vitro Cell. Dev. Biol. Anim. 2008, 44, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Chavez, J.L.; Hagen, J.A.; Kelley-Loughnane, N. Fast and selective plasmonic serotonin detection with aptamer-gold nanoparticle conjugates. Sensors 2017, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Tatemoto, K. Neuropeptide Y: Complete amino-acid-sequence of the brain peptide. Proc. Natl. Acad. Sci. USA 1982, 79, 5485–5489. [Google Scholar] [CrossRef] [PubMed]
- Van den Pol, A.N. Neuropeptide transmission in brain circuits. Neuron 2012, 76, 98–115. [Google Scholar] [CrossRef] [PubMed]
- Mendonsa, S.D.; Bowser, M.T. In vitro selection of aptamers with affinity for neuropeptide Y using capillary electrophoresis. J. Am. Chem. Soc. 2005, 127, 9382–9383. [Google Scholar] [CrossRef] [PubMed]
- Proske, D.; Hofliger, M.; Soll, R.M.; Beck-Sickinger, A.G.; Famulok, M. A Y2 receptor mimetic aptamer directed against neuropeptide Y. J. Biol. Chem. 2002, 277, 11416–11422. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, R.E.; Sanghavi, B.J.; Farmehini, V.; Chavez, J.L.; Hagen, J.; Kelley-Loughnane, N.; Chou, C.F.; Swami, N.S. Aptamer-functionalized graphene-gold nanocomposites for label-free detection of dielectrophoretic-enriched neuropeptide Y. Electrochem. Commun. 2016, 72, 144–147. [Google Scholar] [CrossRef]
- Banerjee, S.; Hsieh, Y.J.; Liu, C.R.; Yeh, N.H.; Hung, H.H.; Lai, Y.S.; Chou, A.C.; Chen, Y.T.; Pan, C.Y. Differential releases of dopamine and neuropeptide Y from histamine-stimulated pc12 cells detected by an aptamer-modified nanowire transistor. Small 2016, 12, 5524–5529. [Google Scholar] [CrossRef] [PubMed]
- Eulberg, D.; Buchner, K.; Maasch, C.; Klussmann, S. Development of an automated in vitro selection protocol to obtain RNA-based aptamers: Identification of a biostable substance P antagonist. Nucleic Acids Res. 2005, 33, e45. [Google Scholar] [CrossRef] [PubMed]
- Vater, A.; Klussmann, S. Turning mirror-image oligonucleotides into drugs: The evolution of spiegelmer therapeutics. Drug Discov. Today 2015, 20, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Faulhammer, D.; Eschgfaller, B.; Stark, S.; Burgstaller, P.; Englberger, W.; Erfurth, J.; Kleinjung, F.; Rupp, J.; Vulcu, S.D.; Schroder, W.; et al. Biostable aptamers with antagonistic properties to the neuropeptide nociceptin/orphanin FQ. RNA 2004, 10, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Amino, T.; Miyachi, Y.; Ogino, C.; Kondo, A. Screening and evaluation of aptamers against somatostatin, and sandwich-like monitoring of somatostatin based on atomic force microscopy. Sens. Actuator B Chem. 2017, 252, 813–821. [Google Scholar] [CrossRef]
- Kobelt, P.; Helmling, S.; Stengel, A.; Wlotzka, B.; Andresen, V.; Klapp, B.F.; Wiedenmann, B.; Klussmann, S.; Monnikes, H. Anti-ghrelin spiegelmer NOX-B11 inhibits neurostimulatory and orexigenic effects of peripheral ghrelin in rats. Gut 2006, 55, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Vater, A.; Sell, S.; Kaczmarek, P.; Maasch, C.; Buchner, K.; Pruszynska-Oszmalek, E.; Kolodziejski, P.; Purschke, W.G.; Nowak, K.W.; Strowski, M.Z.; et al. A mixed mirror-image DNA/RNA aptamer inhibits glucagon and acutely improves glucose tolerance in models of type 1 and type 2 diabetes. J. Biol. Chem. 2013, 288, 21136–21147. [Google Scholar] [CrossRef] [PubMed]
- Heiat, M.; Ranjbar, R.; Latifi, A.M.; Rasaee, M.J. Selection of a high-affinity and in vivo bioactive ssDNA aptamer against angiotensin II peptide. Peptides 2016, 82, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Vater, A.; Jarosch, F.; Buchner, K.; Klussmann, S. Short bioactive spiegelmers to migraine-associated calcitonin gene-related peptide rapidly identified by a novel approach: Tailored-SELEX. Nucleic Acids Res. 2003, 31, e130. [Google Scholar] [CrossRef] [PubMed]
- Kahsai, A.W.; Wisler, J.W.; Lee, J.; Ahn, S.; Cahill, T.J.; Dennison, S.M.; Staus, D.P.; Thomsen, A.R.B.; Anasti, K.M.; Pani, B.; et al. Conformationally selective RNA aptamers allosterically modulate the β(2)-adrenoceptor. Nat. Chem. Biol. 2016, 12, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Daniels, D.A.; Sohal, A.K.; Rees, S.; Grisshammer, R. Generation of RNA aptamers to the G-protein-coupled receptor for neurotensin, NTS-1. Anal. Biochem. 2002, 305, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Clawson, G.A.; Abraham, T.; Pan, W.H.; Tang, X.M.; Linton, S.S.; McGovern, C.O.; Loc, W.S.; Smith, J.P.; Butler, P.J.; Kester, M.; et al. A cholecystokinin B receptor-specific DNA aptamer for targeting pancreatic ductal adenocarcinoma. Nucleic Acid Ther. 2017, 27, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Zurzolo, C. The cell biology of prion-like spread of protein aggregates: Mechanisms and implication in neurodegeneration. Biochem. J. 2013, 452, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Verwilst, P.; Kim, H.-R.; Seo, J.; Sohn, N.-W.; Cha, S.-Y.; Kim, Y.; Maeng, S.; Shin, J.-W.; Kwak, J.H.; Kang, C.; et al. Rational design of in vivo tau tangle-selective near-infrared fluorophores: Expanding the BODIPY universe. J. Am. Chem. Soc. 2017, 139, 13393–13403. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.L.; Lee, V.M.Y. Seeding of normal tau by pathological tau conformers drives pathogenesis of Alzheimer-like tangles. J. Biol. Chem. 2011, 286, 15317–15331. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, E.; Choi, W.H.; Lee, J.; Lee, J.H.; Lee, H.; Kim, D.E.; Suh, Y.H.; Lee, M.J. Inhibitory RNA aptamers of tau oligomerization and their neuroprotective roles against proteotoxic stress. Mol. Pharm. 2016, 13, 2039–2048. [Google Scholar] [CrossRef] [PubMed]
- Krylova, S.M.; Musheev, M.; Nutiu, R.; Li, Y.F.; Lee, G.; Krylov, S.N. Tau protein binds single-stranded DNA sequence specifically—The proof obtained in vitro with non-equilibrium capillary electrophoresis of equilibrium mixtures. FEBS Lett. 2005, 579, 1371–1375. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Wark, A.W.; Lee, H.J. Femtomolar detection of tau proteins in undiluted plasma using surface plasmon resonance. Anal. Chem. 2016, 88, 7793–7799. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. The amyloid β peptide: A chemist’s perspective. Role in Alzheimer’s and fibrillization. Chem. Rev. 2012, 112, 5147–5192. [Google Scholar] [CrossRef] [PubMed]
- Lauren, J.; Gimbel, D.A.; Nygaard, H.B.; Gilbert, J.W.; Strittmatter, S.M. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature 2009, 457, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Balducci, C.; Beeg, M.; Stravalaci, M.; Bastone, A.; Sclip, A.; Biasini, E.; Tapella, L.; Colombo, L.; Manzoni, C.; Borsello, T.; et al. Synthetic amyloid-β oligomers impair long-term memory independently of cellular prion protein. Proc. Natl. Acad. Sci. USA 2010, 107, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Kessels, H.W.; Nguyen, L.N.; Nabavi, S.; Malinow, R. The prion protein as a receptor for amyloid-β. Nature 2010, 466, E3–E4. [Google Scholar] [CrossRef] [PubMed]
- Benilova, I.; De Strooper, B. Prion protein in alzheimer’s pathogenesis: A hot and controversial issue. EMBO Mol. Med. 2010, 2, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.Y.; Shi, Y.S.; Kou, Z.W.; Peng, Y.H.; Chen, W.J.; Li, X.W.; Li, S.J.; Wang, Y.; Wang, F.; Zhang, X.M. Inhibition of bace1 activity by a DNA aptamer in an Alzheimer’s disease cell model. PLoS ONE 2015, 10, e0140733. [Google Scholar] [CrossRef] [PubMed]
- Eremeeva, E.; Abramov, M.; Margamuljana, L.; Rozenski, J.; Pezo, V.; Marliere, P.; Herdewijn, P. Chemical morphing of DNA containing four noncanonical bases. Angew. Chem. Int. Ed. 2016, 55, 7515–7519. [Google Scholar] [CrossRef] [PubMed]
- Gasse, C.; Zaarour, M.; Noppen, S.; Abramov, M.; Marliere, P.; Liekens, S.; De Strooper, B.; Herdewijn, P. Modulation of BACE1 activity by chemically modified aptamers. Chembiochem 2017. submitted. [Google Scholar]
- Rentmeister, A.; Bill, A.; Wahle, T.; Walter, J.; Famulok, M. RNA aptamers selectively modulate protein recruitment to the cytoplasmic domain of β-secretase BACE1 in vitro. RNA 2006, 12, 1650–1660. [Google Scholar] [CrossRef] [PubMed]
- Ylera, F.; Lurz, R.; Erdmann, V.A.; Furste, J.P. Selection of RNA aptamers to the Alzheimer’s disease amyloid peptide. Biochem. Biophys. Res. Commun. 2002, 290, 1583–1588. [Google Scholar] [CrossRef] [PubMed]
- Farrar, C.T.; William, C.M.; Hudry, E.; Hashimoto, T.; Hyman, B.T. RNA aptamer probes as optical imaging agents for the detection of amyloid plaques. PLoS ONE 2014, 9, e89901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahimi, F.; Murakami, K.; Summers, J.L.; Chen, C.-H.B.; Bitan, G. RNA aptamers generated against oligomeric aβ40 recognize common amyloid aptatopes with low specificity but high sensitivity. PLoS ONE 2009, 4, e7694. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Tada, K.; Mihara, H. RNA aptamers selected against amyloid β-peptide (Aβ) inhibit the aggregation of ab. Mol. Biosyst. 2009, 5, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Nishikawa, F.; Noda, K.; Yokoyama, T.; Nishikawa, S. Anti-bovine prion protein RNA aptamer containing tandem gga repeat interacts both with recombinant bovine prion protein and its β isoform with high affinity. Prion 2008, 2, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, D.; Hasegawa, H.; Kaneko, K.; Sode, K.; Ikebukuro, K. Screening of DNA aptamer against mouse prion protein by competitive selection. Prion 2007, 1, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Proske, D.; Gilch, S.; Wopfner, F.; Schatzl, H.M.; Winnacker, E.L.; Famulok, M. Prion-protein-specific aptamer reduces PrPSc formation. ChemBioChem 2002, 3, 717–725. [Google Scholar] [CrossRef]
- Wang, P.; Hatcher, K.L.; Bartz, J.C.; Chen, S.G.; Skinner, P.; Richt, J.; Liu, H.; Sreevatsan, S. Selection and characterization of DNA aptamers against PrPSc. Exp. Biol. Med. 2011, 236, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. Alpha-synuclein and neurodegenerative diseases. Nat. Rev. Neurosci. 2001, 2, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.Y.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef] [PubMed]
- Tsukakoshi, K.; Harada, R.; Sode, K.; Ikebukuro, K. Screening of DNA aptamer which binds to alpha-synuclein. Biotechnol. Lett. 2010, 32, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Sode, K.; Ikebukuro, K. Selection of DNA aptamers against VEGF(165) using a protein competitor and the aptamer blotting method. Biotechnol. Lett. 2008, 30, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Tsukakoshi, K.; Abe, K.; Sode, K.; Ikebukuro, K. Selection of DNA aptamers that recognize alpha-synuclein oligomers using a competitive screening method. Anal. Chem. 2012, 84, 5542–5547. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Xia, N.; Zhao, L.J.; Liu, K.; Hou, W.J.; Liu, L. Aptasensors for the selective detection of alpha-synuclein oligomer by colorimetry, surface plasmon resonance and electrochemical impedance spectroscopy. Sens. Actuator B Chem. 2017, 245, 87–94. [Google Scholar] [CrossRef]
- Liu, L.; Chang, Y.; Yu, J.; Jiang, M.S.; Xia, N. Two-in-one polydopamine nanospheres for fluorescent determination of β-amyloid oligomers and inhibition of β-amyloid aggregation. Sens. Actuator B Chem. 2017, 251, 359–365. [Google Scholar] [CrossRef]
- Zhou, Y.L.; Zhang, H.Q.; Liu, L.T.; Li, C.M.; Chang, Z.; Zhu, X.; Ye, B.X.; Xu, M.T. Fabrication of an antibody-aptamer sandwich assay for electrochemical evaluation of levels of β-amyloid oligomers. Sci. Rep. 2016, 6, 35186. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.F.; Chen, B.C.; Chen, B.; Li, X.J.; Liao, H.L.; Huang, H.M.; Guo, Z.J.; Zhang, W.Y.; Wu, L. Detection of ab oligomers based on magnetic-field-assisted separation of aptamer-functionalized Fe3O4 magnetic nanoparticles and bayf5:Yb,er nanoparticles as upconversion fluorescence labels. Talanta 2017, 170, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.L.; Zhang, J.Y.; Wang, F.Y.; Wang, Y.; Lu, L.L.; Feng, C.C.; Xu, Z.A.; Zhang, W. Selective amyloid β oligomer assay based on abasic site-containing molecular beacon and enzyme-free amplification. Biosens. Bioelectron. 2016, 78, 206–212. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, C.K.; Hamblin, G.D.; Sleiman, H.F. Supramolecular DNA assembly. Chem. Soc. Rev. 2011, 40, 5647–5656. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Yang, Y.; Sugiyama, H. DNA origami technology for biomaterials applications. Biomater. Sci. 2013, 1, 347–360. [Google Scholar] [CrossRef]
- Chhabra, R.; Sharma, J.; Ke, Y.G.; Liu, Y.; Rinker, S.; Lindsay, S.; Yan, H. Spatially addressable multiprotein nanoarrays templated by aptamer-tagged DNA nanoarchitectures. J. Am. Chem. Soc. 2007, 129, 10304–10305. [Google Scholar] [CrossRef] [PubMed]
- Rinker, S.; Ke, Y.G.; Liu, Y.; Chhabra, R.; Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat. Nanotechnol. 2008, 3, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Godonoga, M.; Lin, T.Y.; Oshima, A.; Sumitomo, K.; Tang, M.S.L.; Cheung, Y.W.; Kinghorn, A.B.; Dirkzwager, R.M.; Zhou, C.S.; Kuzuya, A.; et al. A DNA aptamer recognising a malaria protein biomarker can function as part of a DNA origami assembly. Sci. Rep. 2016, 6, 21266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walter, H.K.; Bauer, J.; Steinmeyer, J.; Kuzuya, A.; Niemeyer, C.M.; Wagenknecht, H.A. “DNA origami traffic lights” with a split aptamer sensor for a bicolor fluorescence readout. Nano Lett. 2017, 17, 2467–2472. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.R.; Sha, R.J.; Seeman, N.C. Modifying the surface features of two-dimensional DNA crystals. J. Am. Chem. Soc. 1999, 121, 917–922. [Google Scholar] [CrossRef]
- Green, L.S.; Jellinek, D.; Jenison, R.; Ostman, A.; Heldin, C.H.; Janjic, N. Inhibitory DNA ligands to platelet-derived growth factor B-Chain. Biochemistry 1996, 35, 14413–14424. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.G.; Lindsay, S.; Chang, Y.; Liu, Y.; Yan, H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science 2008, 319, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.W.; Kwok, J.; Law, A.W.L.; Watt, R.M.; Kotaka, M.; Tanner, J.A. Structural basis for discriminatory recognition of plasmodium lactate dehydrogenase by a DNA aptamer. Proc. Natl. Acad. Sci. USA 2013, 110, 15967–15972. [Google Scholar] [CrossRef] [PubMed]
- Kuzuya, A.; Sakai, Y.; Yamazaki, T.; Xu, Y.; Komiyama, M. Nanomechanical DNA origami ‘single-molecule beacons’ directly imaged by atomic force microscopy. Nat. Commun. 2011, 2, 449. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.L.; Yan, M.M.; Yang, S.M. Split aptamers and their applications in sandwich aptasensors. Trac-Trends Anal. Chem. 2016, 80, 581–593. [Google Scholar] [CrossRef]
- Walter, H.K.; Bohlander, P.R.; Wagenknecht, H.A. Development of a wavelength-shifting fluorescent module for the adenosine aptamer using photostable cyanine dyes. Chemistry 2015, 4, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Holzhauser, C.; Wagenknecht, H.A. DNA and RNA “traffic lights”: Synthetic wavelength-shifting fluorescent probes based on nucleic acid base substitutes for molecular imaging. J. Org. Chem. 2013, 78, 7373–7379. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, N.; Wu, J.; Xu, X.Y.; Kamaly, N.; Farokhzad, O.C. Cancer nanotechnology: The impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 2014, 66, 2–25. [Google Scholar] [CrossRef] [PubMed]
- Strebhardt, K.; Ullrich, A. Paul Ehrlich’s magic bullet concept: 100 years of progress. Nat. Rev. Cancer 2008, 8, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.L.; Jin, C.; Wang, Y.Y.; Fang, X.H.; Zhang, X.B.; Chen, Z.; Tan, W.H. Aptamer-conjugated nanomaterials for specific cancer cell recognition and targeted cancer therapy. NPG Asia Mater. 2014, 6, e95. [Google Scholar] [CrossRef]
- Nair, R.R.; Wu, H.A.; Jayaram, P.N.; Grigorieva, I.V.; Geim, A.K. Unimpeded permeation of water through helium-leak-tight graphene-based membranes. Science 2012, 335, 442–444. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, S.J.; Min, D.H. Emerging approaches for graphene oxide biosensor. Anal. Chem. 2017, 89, 232–248. [Google Scholar] [CrossRef] [PubMed]
- Nellore, B.P.V.; Kanchanapally, R.; Pramanik, A.; Sinha, S.S.; Chavva, S.R.; Hamme, A.; Ray, P.C. Aptamer-conjugated graphene oxide membranes for highly efficient capture and accurate identification of multiple types of circulating tumor cells. Bioconjug. Chem. 2015, 26, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Bahreyni, A.; Yazdian-Robati, R.; Hashemitabar, S.; Ramezani, M.; Ramezani, P.; Abnous, K.; Taghdisi, S.M. A new chemotherapy agent-free theranostic system composed of graphene oxide nano-complex and aptamers for treatment of cancer cells. Int. J. Pharm. 2017, 526, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Hu, H.; Zhang, M.G.; Song, J.; Nie, L.; Wang, S.; Niu, G.; Huang, P.; Lu, G.; Chen, X. An aptamer-targeting photoresponsive drug delivery system using “off-on” graphene oxide wrapped mesoporous silica nanoparticles. Nanoscale 2015, 7, 6304–6310. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, X.B.; Ye, M.; Jiang, J.H.; Yang, R.H.; Fu, T.; Chen, Y.; Wang, K.M.; Liu, C.; Tan, W.H. Aptamer-conjugated nanomaterials and their applications. Adv. Drug Deliv. Rev. 2011, 63, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Chen, X.; Tan, W.; Veige, A.S. N-Heterocyclic Carbene–Gold(I) complexes conjugated to a Leukemia-Specific DNA aptamer for targeted drug delivery. Angew. Chem. Int. Ed. 2016, 55, 8889–8893. [Google Scholar] [CrossRef] [PubMed]
- Siafaka, P.I.; Okur, N.U.; Karavas, E.; Bikiaris, D.N. Surface modified multifunctional and stimuli responsive nanoparticles for drug targeting: Current status and uses. Int. J. Mol. Sci. 2016, 17, 1440. [Google Scholar] [CrossRef] [PubMed]
- Latorre, A.; Posch, C.; Garcimartin, Y.; Celli, A.; Sanlorenzo, M.; Vujic, I.; Ma, J.; Zekhtser, M.; Rappersberger, K.; Ortiz-Urda, S.; et al. DNA and aptamer stabilized gold nanoparticles for targeted delivery of anticancer therapeutics. Nanoscale 2014, 6, 7436–7442. [Google Scholar] [CrossRef] [PubMed]
- Massich, M.D.; Giljohann, D.A.; Schmucker, A.L.; Patel, P.C.; Mirkin, C.A. Cellular response of polyvalent oligonucleotide-gold nanoparticle conjugates. ACS Nano 2010, 4, 5641–5646. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-S.; Wei, S.-C.; Chang, H.-T.; Lin, H.-J.; Huang, C.-C. Gold nanoparticles modified with self-assembled hybrid monolayer of triblock aptamers as a photoreversible anticoagulant. J. Control. Release 2016, 221, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Swift, B.J.F.; Shadish, J.A.; DeForest, C.A.; Baneyx, F. Streamlined synthesis and assembly of a hybrid sensing architecture with solid binding proteins and click chemistry. J. Am. Chem. Soc. 2017, 139, 3958–3961. [Google Scholar] [CrossRef] [PubMed]
- Geissler, D.; Linden, S.; Liermann, K.; Wegner, K.D.; Charbonniere, L.J.; Hildebrandt, N. Lanthanides and quantum dots as forster resonance energy transfer agents for diagnostics and cellular imaging. Inorg. Chem. 2014, 53, 1824–1838. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.J. Quantum dot-nucleic acid/aptamer bioconjugate-based fluorimetric biosensors. Biochem. Soc. Trans. 2012, 40, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Elgqvist, J. Nanoparticles as theranostic vehicles in experimental and clinical applications-focus on prostate and breast cancer. Int. J. Mol. Sci. 2017, 18, 1102. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Ma, Q.; Fei, X.; Zhang, H.; Su, X. A novel aptamer functionalized cuins2 quantum dots probe for daunorubicin sensing and near infrared imaging of prostate cancer cells. Anal. Chim. Acta 2014, 818, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.R.; Sefah, K.; Liu, H.P.; Wang, R.W.; Tan, W.H. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc. Natl. Acad. Sci. USA 2010, 107, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.P.; Zhu, Z.; Kang, H.Z.; Wu, Y.R.; Sefan, K.; Tan, W.H. DNA-based micelles: Synthesis, micellar properties and size-dependent cell permeability. Chem. Eur. J. 2010, 16, 3791–3797. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, D.; Lee, J.B. DNA aptamer-based carrier for loading proteins and enhancing the enzymatic activity. RSC Adv. 2017, 7, 1643–1645. [Google Scholar] [CrossRef]
- Xiong, X.L.; Liu, H.P.; Zhao, Z.L.; Altman, M.B.; Lopez-Colon, D.; Yang, C.J.; Chang, L.J.; Liu, C.; Tan, W.H. DNA aptamer-mediated cell targeting. Angew. Chem. Int. Ed. 2013, 52, 1472–1476. [Google Scholar] [CrossRef] [PubMed]
- Lale, S.V.; Aswathy, R.G.; Aravind, A.; Kumar, D.S.; Koul, V. AS1411 aptamer and folic acid functionalized pH-responsive atrp fabricated pPEGMA-PCL-pPEGMA polymeric nanoparticles for targeted drug delivery in cancer therapy. Biomacromolecules 2014, 15, 1737–1752. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.C.; Zhang, N.; Tang, Y.F.; Yang, Y.N.; Chu, X.; Zhao, Y.X. Sl2b aptamer and folic acid dual-targeting DNA nanostructures for synergic biological effect with chemotherapy to combat colorectal cancer. Int. J. Nanomed. 2017, 12, 2657–2672. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Hu, Y.; Duan, J.; Yang, X.-D. Aptamer-guided DNA tetrahedron as a novel targeted drug delivery system for muc1-expressing breast cancer cells in vitro. Oncotarget 2016, 7, 38257–38269. [Google Scholar] [CrossRef] [PubMed]
- Stiriba, S.E.; Frey, H.; Haag, R. Dendritic polymers in biomedical applications: From potential to clinical use in diagnostics and therapy. Angew. Chem. Int. Ed. 2002, 41, 1329–1334. [Google Scholar] [CrossRef]
- MacEwan, S.R.; Chilkoti, A. From composition to cure: A systems engineering approach to anticancer drug carriers. Angew. Chem. Int. Ed. 2017, 56, 6712–6733. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Zhang, L.; Teply, B.A.; Mann, N.; Wang, A.; Radovic-Moreno, A.F.; Langer, R.; Farokhzad, O.C. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc. Natl. Acad. Sci. USA 2008, 105, 2586–2591. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Gu, F.X.; Langer, R.; Farokhzad, O.C.; Lippard, S.J. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc. Natl. Acad. Sci. USA 2008, 105, 17356–17361. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Kolishetti, N.; Lippard, S.J.; Farokhzad, O.C. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 1850–1855. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.Y.; Deng, H.P.; Su, Y.; He, L.; Wang, R.B.; Tong, G.S.; He, D.N.; Zhu, X.Y. Aptamer-functionalized and backbone redox-responsive hyperbranched polymer for targeted drug delivery in cancer therapy. Biomacromolecules 2016, 17, 2050–2062. [Google Scholar] [CrossRef] [PubMed]
- Lao, Y.H.; Phua, K.K.L.; Leong, K.W. Aptamer nanomedicine for cancer therapeutics: Barriers and potential for translation. ACS Nano 2015, 9, 2235–2254. [Google Scholar] [CrossRef] [PubMed]
- Taghavi, S.; Ramezani, M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. Chitosan-modified plga nanoparticles tagged with 5tr1 aptamer for in vivo tumor-targeted drug delivery. Cancer Lett. 2017, 400, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Coles, D.J.; Rolfe, B.E.; Boase, N.R.B.; Veedu, R.N.; Thurecht, K.J. Aptamer-targeted hyperbranched polymers: Towards greater specificity for tumours in vivo. Chem. Commun. 2013, 49, 3836–3838. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.R.; Dong, R.J.; Chen, J.X.; Chen, F.; Jiang, W.F.; Zhou, Y.F.; Zhu, X.Y.; Yan, D.Y. Synthesis and self-assembly of arnphiphilic aptamer-functionalized hyperbranched multiarm copolymers for targeted cancer imaging. Biomacromolecules 2014, 15, 1828–1836. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.J.; Siddiqui, I.A.; Nihal, M.; Pilla, S.; Rosenthal, K.; Mukhtar, H.; Gong, S.Q. Aptamer-conjugated and doxorubicin-loaded unimolecular micelles for targeted therapy of prostate cancer. Biomaterials 2013, 34, 5244–5253. [Google Scholar] [CrossRef] [PubMed]
- Torchilin, V.P. Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Discov. 2005, 4, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Huwyler, J.; Wu, D.; Pardridge, W.M. Brain drug delivery of small molecules using immunoliposomes. Proc. Natl. Acad. Sci. USA 1996, 93, 14164–14169. [Google Scholar] [CrossRef] [PubMed]
- Schnyder, A.; Huwyler, J. Drug transport to brain with targeted liposomes. NeuroRX 2005, 2, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Willis, M.C.; Collins, B.; Zhang, T.; Green, L.S.; Sebesta, D.P.; Bell, C.; Kellogg, E.; Gill, S.C.; Magallanez, A.; Knauer, S.; et al. Liposome anchored vascular endothelial growth factor aptamers. Bioconjug. Chem. 1998, 9, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Ara, M.N.; Matsuda, T.; Hyodo, M.; Sakurai, Y.; Hatakeyama, H.; Ohga, N.; Hida, K.; Harashima, H. An aptamer ligand based liposomal nanocarrier system that targets tumor endothelial cells. Biomaterials 2014, 35, 7110–7120. [Google Scholar] [CrossRef] [PubMed]
- Alshaer, W.; Hillaireau, H.; Vergnaud, J.; Ismail, S.; Fattal, E. Functionalizing liposomes with anti-CD44 aptamer for selective targeting of cancer cells. Bioconjug. Chem. 2015, 26, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Plourde, K.; Derbali, R.M.; Desrosiers, A.; Dubath, C.; Vallée-Bélisle, A.; Leblond, J. Aptamer-based liposomes improve specific drug loading and release. J. Control. Release 2017, 251, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Wochner, A.; Menger, M.; Orgel, D.; Cech, B.; Rimmele, M.; Erdmann, V.A.; Glokler, J. A DNA aptamer with high affinity and specificity for therapeutic anthracyclines. Anal. Biochem. 2008, 373, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Barenholz, Y. Doxil®-the first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Hah, S.S. Improved ligand binding by antibody—Aptamer pincers. Bioconjug. Chem. 2014, 25, 1421–1427. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.C.; Twu, K.Y.; Ellington, A.D.; Levy, M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006, 34, e73. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.W.; Zhu, G.Z.; Mei, L.; Xie, Y.; Ma, H.B.; Ye, M.; Qing, F.L.; Tan, W.H. Automated modular synthesis of aptamer-drug conjugates for targeted drug delivery. J. Am. Chem. Soc. 2014, 136, 2731–2734. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.Z.; Niu, G.; Chen, X.Y. Aptamer-drug conjugates. Bioconjug. Chem. 2015, 26, 2186–2197. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-F.; Shangguan, D.; Liu, H.; Phillips, J.A.; Zhang, X.; Chen, Y.; Tan, W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. ChemBioChem 2009, 10, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Zheng, J.; Song, E.; Donovan, M.; Zhang, K.; Liu, C.; Tan, W. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc. Natl. Acad. Sci. USA 2013, 110, 7998–8003. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.H.; Cao, Z.H.C.; Li, Y.; Tan, W.H. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin. Chem. 2007, 53, 1153–1155. [Google Scholar] [CrossRef] [PubMed]
- Mallikaratchy, P. Evolution of complex target SELEX to identify aptamers against mammalian cell-surface antigens. Molecules 2017, 22, 215. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Meng, L.; Ye, M.; Yang, L.; Sefah, K.; O’Donoghue, M.B.; Chen, Y.; Xiong, X.; Huang, J.; Song, E.; et al. Self-assembled aptamer-based drug carriers for bispecific cytotoxicity to cancer cells. Chem. Asian J. 2012, 7, 1630–1636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhao, N.X.; Zeng, Z.H.; Chang, C.C.; Zu, Y.L. Combination of an aptamer probe to CD4 and antibodies for multicolored cell phenotyping. Am. J. Clin. Pathol. 2010, 134, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Jo, N.; Mailhos, C.; Ju, M.H.; Cheung, E.; Bradley, J.; Nishijima, K.; Robinson, G.S.; Adarnis, A.P.; Shima, D.T. Inhibition of platelet-derived growth factor B signaling enhances the efficacy of anti-vascular enclothelial growth factor therapy in multiple models of ocular neovascularization. Am. J. Pathol. 2006, 168, 2036–2053. [Google Scholar] [CrossRef] [PubMed]
- Heo, K.; Min, S.-W.; Sung, H.J.; Kim, H.G.; Kim, H.J.; Kim, Y.H.; Choi, B.K.; Han, S.; Chung, S.; Lee, E.S.; et al. An aptamer-antibody complex (oligobody) as a novel delivery platform for targeted cancer therapies. J. Control. Release 2016, 229, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Chen, Z.C.; He, A.B.; Zhan, Y.H.; Li, J.F.; Liu, L.; Wu, H.W.; Zhuang, C.L.; Lin, J.H.; Zhang, Q.X.; et al. Targeting cellular mRNAs translation by CRISPR-Cas9. Sci. Rep. 2016, 6, 29652. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Su, J.H.; Zhang, F.; Zhuang, X.W. An RNA-aptamer-based two-color CRISPR labeling system. Sci. Rep. 2016, 6, 26857. [Google Scholar] [CrossRef] [PubMed]
- Dassie, J.P.; Giangrande, P.H. Current progress on aptamer-targeted oligonucleotide therapeutics. Ther. Deliv. 2013, 4, 1527–1546. [Google Scholar] [CrossRef] [PubMed]
- Kruspe, S.; Mittelberger, F.; Szameit, K.; Hahn, U. Aptamers as drug delivery vehicles. Chem. Med. Chem. 2014, 9, 1998–2011. [Google Scholar] [CrossRef] [PubMed]
- Kruspe, S.; Giangrande, P. Aptamer-siRNA chimeras: Discovery, progress, and future prospects. Biomedicines 2017, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.O.; Andrechek, E.R.; Wang, Y.; D Viles, K.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006, 24, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Dassie, J.P.; Liu, X.Y.; Thomas, G.S.; Whitaker, R.M.; Thiel, K.W.; Stockdale, K.R.; Meyerholz, D.K.; McCaffrey, A.P.; McNamara, J.O.; Giangrande, P.H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009, 27, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Yu, X.L.; Liu, H.T.; Wu, D.Q.; She, J.X. Co-targeting EGFR and survivin with a bivalent aptamer-dual siRNA chimera effectively suppresses prostate cancer. Sci. Rep. 2016, 6, 30346. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Gao, X.H. A universal protein tag for delivery of siRNA-aptamer chimeras. Sci. Rep. 2013, 3, 3129. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Lee, S.H.; Hwang, Y.; Yoo, H.; Jung, H.; Kim, S.H.; Mok, H. Multivalent aptamer-RNA conjugates for simple and efficient delivery of doxorubicin/siRNA into multidrug-resistant cells. Macromol. Biosci. 2017, 17, 1600343. [Google Scholar] [CrossRef] [PubMed]
- Wilner, S.E.; Wengerter, B.; Maier, K.; Magalhaes, M.D.B.; Del Amo, D.S.; Pai, S.; Opazo, F.; Rizzoli, S.O.; Yan, A.; Levy, M. An RNA alternative to human transferrin: A new tool for targeting human cells. Mol. Ther. Nucleic Acids 2012, 1, e21. [Google Scholar] [CrossRef] [PubMed]
- Breitz, H.B.; Weiden, P.L.; Beaumier, P.L.; Axworthy, D.B.; Seiler, C.; Su, F.M.; Graves, S.; Bryan, K.; Reno, J.M. Clinical optimization of pretargeted radioimmunotherapy with antibody-streptavidin conjugate and Y-90-DOTA-biotin. J. Nucl. Med. 2000, 41, 131–140. [Google Scholar] [PubMed]
- Binzel, D.W.; Shu, Y.; Li, H.; Sun, M.Y.; Zhang, Q.S.; Shu, D.; Guo, B.; Guo, P.X. Specific delivery of miRNA for high efficient inhibition of prostate cancer by RNA nanotechnology. Mol. Ther. 2016, 24, 1267–1277. [Google Scholar] [CrossRef] [PubMed]
- Serganov, A.; Patel, D.J. Ribozymes, riboswitches and beyond: Regulation of gene expression without proteins. Nat. Rev. Genet. 2007, 8, 776–790. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lopez, C.; Barroso-delJesus, A.; Puerta-Fernandez, E.; Berzal-Herranz, A. Interfering with hepatitis C virus ires activity using RNA molecules identified by a novel in vitro selection method. Biol. Chem. 2005, 386, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lopez, C.; Diaz-Gonzalez, R.; Barroso-DelJesus, A.; Berzal-Herranz, A. Inhibition of hepatitis C virus replication and internal ribosome entry site-dependent translation by an RNA molecule. J. Gen. Virol. 2009, 90, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lopez, C.; Lahlali, T.; Berzal-Herranz, B.; Berzal-Herranz, A. Development of optimized inhibitor RNAs allowing multisite-targeting of the HCV genome. Molecules 2017, 22, 861. [Google Scholar] [CrossRef] [PubMed]
- Travascio, P.; Li, Y.F.; Sen, D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998, 5, 505–517. [Google Scholar] [CrossRef]
- Golub, E.; Albada, H.B.; Liao, W.C.; Biniuri, Y.; Willner, I. Nucleoapzymes: Hemin/G-quadruplex DNAzyme-aptamer binding site conjugates with superior enzyme-like catalytic functions. J. Am. Chem. Soc. 2016, 138, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef] [PubMed]
- Diafa, S.; Hollenstein, M. Generation of aptamers with an expanded chemical repertoire. Molecules 2015, 20, 16643–16671. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Hongdilokkul, N.; Liu, Z.X.; Thirunavukarasu, D.; Romesberg, F.E. The expanding world of DNA and RNA. Curr. Opin. Chem. Biol. 2016, 34, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Dellafiore, M.A.; Montserrat, J.M.; Iribarren, A.M. Modified nucleoside triphosphates for in vitro selection techniques. Front. Chem. 2016, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Tolle, F.; Mayer, G. Dressed for success—Applying chemistry to modulate aptamer functionality. Chem. Sci. 2013, 4, 60–67. [Google Scholar] [CrossRef]
- Dunn, M.R.; Jimenez, R.M.; Chaput, J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017, 1, 76. [Google Scholar] [CrossRef]
- Gawande, B.N.; Rohloff, J.C.; Carter, J.D.; von Carlowitz, I.; Zhang, C.; Schneider, D.J.; Janjic, N. Selection of DNA aptamers with two modified bases. Proc. Natl. Acad. Sci. USA 2017, 114, 2898–2903. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.H.; Hipolito, C.J.; Hollenstein, M.; Perrin, D.M. A divalent metal-dependent self-cleaving DNAzyme with a tyrosine side chain. Org. Biomol. Chem. 2011, 9, 6949–6954. [Google Scholar] [CrossRef] [PubMed]
- Renders, M.; Miller, E.; Lam, C.H.; Perrin, D.M. Whole cell-SELEX of aptamers with a tyrosine-like side chain against live bacteria. Org. Biomol. Chem. 2017, 15, 1980–1989. [Google Scholar] [CrossRef] [PubMed]
- So, H.-M.; Park, D.-W.; Jeon, E.-K.; Kim, Y.-H.; Kim, B.S.; Lee, C.-K.; Choi, S.Y.; Kim, S.C.; Chang, H.; Lee, J.-O. Detection and Titer estimation of Escherichia coli using aptamer-functionalized single-walled carbon-nanotube field-effect transistors. Small 2008, 4, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Minagawa, H.; Onodera, K.; Fujita, H.; Sakamoto, T.; Akitomi, J.; Kaneko, N.; Shiratori, I.; Kuwahara, M.; Horii, K.; Waga, I. Selection, characterization and application of artificial DNA aptamer containing appended bases with sub-nanomolar affinity for a salivary biomarker. Sci. Rep. 2017, 7, 42716. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, Y.; Kasahara, Y.; Fujita, H.; Kitadume, S.; Ozaki, H.; Endoh, T.; Kuwahara, M.; Sugimoto, N. Efficacy of base-modification on target binding of small molecule DNA aptamers. J. Am. Chem. Soc. 2013, 135, 9412–9419. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.J.; Lee, J.-W.; Ellington, A.D. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. 2009, 2, 241–264. [Google Scholar] [CrossRef] [PubMed]
- Vaish, N.K.; Larralde, R.; Fraley, A.W.; Szostak, J.W.; McLaughlin, L.W. A novel, modification-dependent ATP-binding aptamer selected from an RNA library incorporating a cationic functionality. Biochemistry 2003, 42, 8842–8851. [Google Scholar] [CrossRef] [PubMed]
- Kabza, A.M.; Sczepanski, J.T. An L-RNA aptamer with expanded chemical functionality that inhibits microRNA biogenesis. ChemBioChem 2017, 18, 1824–1827. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.H.; Lei, Y.; Yeung, W.; Hili, R. Enzymatic synthesis of sequence-defined synthetic nucleic acid polymers with diverse functional groups. Angew. Chem. Int. Ed. 2016, 55, 13164–13168. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Kong, D.H.; Hili, R. A high-fidelity codon set for the T4 DNA ligase-catalyzed polymerization of modified oligonucleotides. ACS Comb. Sci. 2015, 17, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Yeung, W.; Hili, R. In vitro selection of diversely-functionalized aptamers. J. Am. Chem. Soc. 2017, 139, 13977–13980. [Google Scholar] [CrossRef] [PubMed]
- Feldman, A.W.; Romesberg, F.E. In vivo structure–activity relationships and optimization of an unnatural base pair for replication in a semi-synthetic organism. J. Am. Chem. Soc. 2017, 139, 11427–11433. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Yamashige, R.; Matsunaga, K.-I.; Yokoyama, S.; Hirao, I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013, 31, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Nakamura, M.; Hirao, I. Post-exSELEX stabilization of an unnatural-base DNA aptamer targeting VEGF(165) toward pharmaceutical applications. Nucleic Acids Res. 2016, 44, 7487–7494. [Google Scholar] [PubMed]
- Matsunaga, K.-I.; Kimoto, M.; Hirao, I. High-affinity DNA aptamer generation targeting von willebrand factor A1-domain by genetic alphabet expansion for systematic evolution of ligands by exponential enrichment using two types of libraries composed of five different bases. J. Am. Chem. Soc. 2017, 139, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I.; Kawai, G.; Yoshizawa, S.; Nishimura, Y.; Ishido, Y.; Watanabe, K.; Miura, K. Most compact hairpin-turn structure exerted by a short DNA fragment, d(GCGAAGC) in solution—An extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res. 1994, 22, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, K.; Kimoto, M.; Hanson, C.; Sanford, M.; Young, H.A.; Hirao, I. Architecture of high-affinity unnatural-base DNA aptamers toward pharmaceutical applications. Sci. Rep. 2015, 5, 18478. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I.; Kimoto, M.; Lee, K.H. DNA aptamer generation by exSELEX using genetic alphabet expansion with a mini-hairpin DNA stabilization method. Biochimie 2017. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Chen, F.; Chamberlin, S.G.; Benner, S.A. Expanded genetic alphabets in the polymerase chain reaction. Angew. Chem. Int. Ed. 2010, 49, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Benner, S.A.; Karalkar, N.B.; Hoshika, S.; Laos, R.; Shaw, R.W.; Matsuura, M.; Fajardo, D.; Moussatche, P. Alternative Watson-Crick synthetic genetic systems. Cold Spring Harb. Perspect. Biol. 2016, 8, a023770. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Yang, Z.; Bradley, K.M.; Hoshika, S.; Jiménez, E.; Zhang, L.; Zhu, G.; Shanker, S.; Yu, F.; Turek, D.; et al. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA 2014, 111, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, Z.; Le Trinh, T.; Teng, I.T.; Wang, S.; Bradley, K.M.; Hoshika, S.; Wu, Q.; Cansiz, S.; Rowold, D.J.; et al. Aptamers against cells overexpressing glypican 3 from expanded genetic systems combined with cell engineering and laboratory evolution. Angew. Chem. Int. Ed. 2016, 55, 12372–12375. [Google Scholar] [CrossRef] [PubMed]
- Biondi, E.; Lane, J.D.; Das, D.; Dasgupta, S.; Piccirilli, J.A.; Hoshika, S.; Bradley, K.M.; Krantz, B.A.; Benner, S.A. Laboratory evolution of artificially expanded DNA gives redesignable aptamers that target the toxic form of anthrax protective antigen. Nucleic Acids Res. 2016, 44, 9565–9577. [Google Scholar] [CrossRef] [PubMed]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. Qgrs mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef] [PubMed]
- Malyshev, D.A.; Dhami, K.; Lavergne, T.; Chen, T.J.; Dai, N.; Foster, J.M.; Correa, I.R.; Romesberg, F.E. A semi-synthetic organism with an expanded genetic alphabet. Nature 2014, 509, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lamb, B.M.; Feldman, A.W.; Zhou, A.X.; Lavergne, T.; Li, L.; Romesberg, F.E. A semisynthetic organism engineered for the stable expansion of the genetic alphabet. Proc. Natl. Acad. Sci. USA 2017, 114, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Rothlisberger, P.; Levi-Acobas, F.; Sarac, I.; Marliere, P.; Herdewijn, P.; Hollenstein, M. On the enzymatic incorporation of an imidazole nucleotide into DNA. Org. Biomol. Chem. 2017, 15, 4449–4455. [Google Scholar] [CrossRef] [PubMed]
- Kaul, C.; Muller, M.; Wagner, M.; Schneider, S.; Carell, T. Reversible bond formation enables the replication and amplification of a crosslinking salen complex as an orthogonal base pair. Nat. Chem. 2011, 3, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Matyasovsky, J.; Perlikova, P.; Malnuit, V.; Pohl, R.; Hocek, M. 2-substituted dATP derivatives as building blocks for polymerase-catalyzed synthesis of DNA modified in the minor groove. Angew. Chem. Int. Ed. 2016, 55, 15856–15859. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M. Deoxynucleoside triphosphates bearing histamine, carboxylic acid, and hydroxyl residues—Synthesis and biochemical characterization. Org. Biomol. Chem. 2013, 11, 5162–5172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eremeeva, E.; Abramov, M.; Margamuljana, L.; Herdewijn, P. Base-modified nucleic acids as a powerful tool for synthetic biology and biotechnology. Chem. Eur. J. 2017, 23, 9560–9576. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M. Synthesis of deoxynucleoside triphosphates that include proline, urea, or sulfamide groups and their polymerase incorporation into DNA. Chem. Eur. J. 2012, 18, 13320–13330. [Google Scholar] [CrossRef] [PubMed]
- Röthlisberger, P.; Levi-Acobas, F.; Hollenstein, M. New synthetic route to ethynyl-dUTP: A means to avoid formation of acetyl and chloro vinyl base-modified triphosphates that could poison SELEX experiments. Bioorg. Med. Chem. Lett. 2017, 27, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Houlihan, G.; Arangundy-Franklin, S.; Holliger, P. Engineering and application of polymerases for synthetic genetics. Curr. Opin. Biotechnol. 2017, 48, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Hongdilokkul, N.; Liu, Z.; Adhikary, R.; Tsuen, S.S.; Romesberg, F.E. Evolution of thermophilic DNA polymerases for the recognition and amplification of C2′-modified DNA. Nat. Chem. 2016, 8, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Thirunavukarasu, D.; Chen, T.; Liu, Z.; Hongdilokkul, N.; Romesberg, F.E. Selection of 2′-fluoro-modified aptamers with optimized properties. J. Am. Chem. Soc. 2017, 139, 2892–2895. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, T.; Romesberg, F.E. Evolved polymerases facilitate selection of fully 2′-OMe-modified aptamers. Chem. Sci. 2017. [Google Scholar] [CrossRef]
- Pinheiro, V.B.; Holliger, P. The XNA world: Progress towards replication and evolution of synthetic genetic polymers. Curr. Opin. Chem. Biol. 2012, 16, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Bravo, I.A.; Cozens, C.; Holliger, P.; DeStefano, J.J. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015, 43, 9587–9599. [Google Scholar]
- Yu, H.Y.; Zhang, S.; Chaput, J.C. Darwinian evolution of an alternative genetic system provides support for tna as an RNA progenitor. Nat. Chem. 2012, 4, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Diafa, S.; Evéquoz, D.; Leumann, C.J.; Hollenstein, M. Enzymatic synthesis of 7′,5′-bicyclo-DNA oligonucleotides. Chem. Asian J. 2017, 12, 1347–1352. [Google Scholar] [CrossRef] [PubMed]
- Maiti, M.; Maiti, M.; Knies, C.; Dumbre, S.; Lescrinier, E.; Rosemeyer, H.; Ceulemans, A.; Herdewijn, P. Xylonucleic acid: Synthesis, structure, and orthogonal pairing properties. Nucleic Acids Res. 2015, 43, 7189–7200. [Google Scholar] [CrossRef] [PubMed]
- Siegmund, V.; Santner, T.; Micura, R.; Marx, A. Screening mutant libraries of T7 RNA polymerase for candidates with increased acceptance of 2′-modified nucleotides. Chem. Commun. 2012, 48, 9870–9872. [Google Scholar] [CrossRef] [PubMed]
- Inoue, N.; Shionoya, A.; Minakawa, N.; Kawakami, A.; Ogawa, N.; Matsuda, A. Amplification of 4′-thioDNA in the presence of 4′-thio-dTTP and 4′-thio-dCTP, and 4′-thioDNA-directed transcription in vitro and in mammalian cells. J. Am. Chem. Soc. 2007, 129, 15424–15425. [Google Scholar] [CrossRef] [PubMed]
- Kojima, T.; Furukawa, K.; Maruyama, H.; Inoue, N.; Tarashima, N.; Matsuda, A.; Minakawa, N. PCR amplification of 4′-thioDNA using 2′-deoxy-4′-thionucleoside 5′-triphosphates. ACS Synth. Biol. 2013, 2, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Houlihan, G.; Arangundy-Franklin, S.; Holliger, P. Exploring the chemistry of genetic information storage and propagation through polymerase engineering. Acc. Chem. Res. 2017, 50, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Ghadessy, F.J.; Ramsay, N.; Boudsocq, F.; Loakes, D.; Brown, A.; Iwai, S.; Vaisman, A.; Woodgate, R.; Holliger, P. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nat. Biotechnol. 2004, 22, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Higashimoto, Y.; Matsui, T.; Nishino, Y.; Taira, J.; Inoue, H.; Takeuchi, M.; Yamagishi, S. Blockade by phosphorothioate aptamers of advanced glycation end products-induced damage in cultured pericytes and endothelial cells. Microvasc. Res. 2013, 90, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Volk, D.; Lokesh, G. Development of phosphorothioate DNA and DNA thioaptamers. Biomedicines 2017, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhu, Y.; Wang, C.; Guan, Z.; Zhang, L.; Yang, Z. Alkylation of phosphorothioated thrombin binding aptamers improves the selectivity of inhibition of tumor cell proliferation upon anticoagulation. Biochim. Biophys. Acta 2017, 1861, 1864–1869. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, K.; Padmanabhan, K.P.; Ferrara, J.D.; Sadler, J.E.; Tulinsky, A. The structure of α-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993, 268, 17651–17654. [Google Scholar] [PubMed]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef] [PubMed]
- Lato, S.M.; Ozerova, N.D.S.; He, K.Z.; Sergueeva, Z.; Shaw, B.R.; Burke, D.H. Boron-containing aptamers to ATP. Nucleic Acids Res. 2002, 30, 1401–1407. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.S.; Chen, Y.H.; Lennox, K.A.; Behlke, M.A.; Davidson, B.L. In vivo SELEX for identification of brain-penetrating aptamers. Mol. Ther. Nucleic Acids 2013, 2, e67. [Google Scholar] [CrossRef] [PubMed]
- Monaco, I.; Camorani, S.; Colecchia, D.; Locatelli, E.; Calandro, P.; Oudin, A.; Niclou, S.; Arra, C.; Chiariello, M.; Cerchia, L.; et al. Aptamer functionalization of nanosystems for glioblastoma targeting through the blood-brain barrier. J. Med. Chem. 2017, 60, 4510–4516. [Google Scholar] [CrossRef] [PubMed]
- Temme, J.S.; Drzyzga, M.G.; MacPherson, I.S.; Krauss, I.J. Directed evolution of 2g12-targeted nonamannose glycoclusters by SELMA. Chem. Eur. J. 2013, 19, 17291–17295. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.C.; Dunn, M.R.; Hatch, A.; Sau, S.P.; Youngbull, C.; Chaput, J.C. A general strategy for expanding polymerase function by droplet microfluidics. Nat. Commun. 2016, 7, 11235. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, I.S.; Temme, J.S.; Krauss, I.J. DNA display of folded RNA libraries enabling RNA-SELEX without reverse transcription. Chem. Commun. 2017, 53, 2878–2881. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.C.; Hong, C.Y. Magnetic-assisted rapid aptamer selection (MARAS) for generating high-affinity DNA aptamer using rotating magnetic fields. ACS Comb. Sci. 2014, 16, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Renders, M.; Miller, E.; Hollenstein, M.; Perrin, D.M. A method for selecting modified DNAzymes without the use of modified DNA as a template in PCR. Chem. Commun. 2015, 51, 1360–1362. [Google Scholar] [CrossRef] [PubMed]









© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Röthlisberger, P.; Gasse, C.; Hollenstein, M. Nucleic Acid Aptamers: Emerging Applications in Medical Imaging, Nanotechnology, Neurosciences, and Drug Delivery. Int. J. Mol. Sci. 2017, 18, 2430. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18112430
Röthlisberger P, Gasse C, Hollenstein M. Nucleic Acid Aptamers: Emerging Applications in Medical Imaging, Nanotechnology, Neurosciences, and Drug Delivery. International Journal of Molecular Sciences. 2017; 18(11):2430. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18112430
Chicago/Turabian StyleRöthlisberger, Pascal, Cécile Gasse, and Marcel Hollenstein. 2017. "Nucleic Acid Aptamers: Emerging Applications in Medical Imaging, Nanotechnology, Neurosciences, and Drug Delivery" International Journal of Molecular Sciences 18, no. 11: 2430. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18112430