Molecular Biology of Prune Dwarf Virus—A Lesser Known Member of the Bromoviridae but a Vital Component in the Dynamic Virus–Host Cell Interaction Network
Abstract
:1. Introduction
2. Genome Organization of Prune Dwarf Virus (PDV)
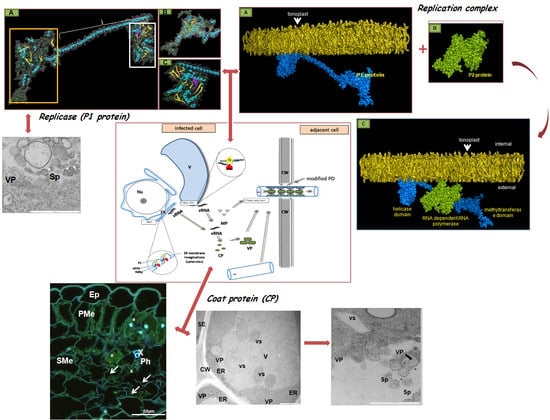
3. The Crucial Functions of Proteins Coded by PDV RNA
4. PDV Infection Cycle Based on the Bromoviridae Family Model
5. The Specificity of Cell-to-Cell Transport in Different Plant Viruses as Compared to PDV
6. Systemic Transport of PDV and Other Bromoviridae
7. Plant Response to Infections with PDV and Other Bromoviridae
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bujarski, J.J.; Figlerowicz, M.; Gallitelli, D.; Roossinck, M.J.; Scott, S.W. Family Bromoviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses-Ninth Report of the International Committee on Taxonomy of Viruses, 1st ed.; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: San Diego, CA, USA, 2012; pp. 972–976. ISBN 978-0123846846. [Google Scholar]
- Brunt, H.A.; Crabtree, K.; Dallawitz, M.J.; Gibs, A.J.; Watson, L. Viruses of Plants, 1st ed.; CAB International UK: Wallingford, UK, 1996; ISBN 978-0851987941. [Google Scholar]
- Codoñer, F.M.; Cuevas, J.M.; Sanchez-Navarro, J.A.; Pallas, V.; Elena, S.F. Molecular evolution of the plant virus family Bromoviridae based on RNA3-encoded proteins. J. Mol. Evol. 2005, 61, 697–705. [Google Scholar] [CrossRef]
- Codoñer, F.M.; Fares, M.A.; Elena, S.F. Adaptive covariation between the coat and movement proteins of Prunus necrotic ringspot virus. J. Virol. 2006, 80, 5833–5840. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV) Official Website. Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/251/bromoviridae (accessed on 16 November 2017).
- Fulton, R.W. Prune dwarf virus. C.M.I/A.A.B. Descr. Plant Viruses 1970, 1. Unavailable online. [Google Scholar]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Amari, K.; Sanchez-Pina, M.A.; Myrta, A.; Sanchez-Navarro, J.A. Ilarviruses of Prunus spp.: A continued concern for fruit trees. Phytopathology 2012, 102, 1108–1120. [Google Scholar] [CrossRef] [PubMed]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Sanchez-Navarro, J.A.; Scott, S.W. The molecular biology of ilarviruses. Adv. Virus Res. 2013, 87, 139–183. [Google Scholar] [CrossRef] [PubMed]
- Faquet, C.M.; Mayo, M.A.; Maniloff, J.; Desselberger, U.; Ball, L.A. Virus Taxonomy-Eight Report of the International Committee on Taxonomy of Viruses, 1st ed.; Elsevier Academic Press: London, UK, 2005; pp. 200–236. ISBN 978-0122499517. [Google Scholar]
- Lee, S.; Shin, Y.G. Development and practical use of RT-PCR for seed-transmitted Prune dwarf virus in quarantine. Plant Pathol. J. 2014, 30, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991, 5, 2108–2116. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solińska, J.; Bujarski, J.J. Insights into the single-cell reproduction cycle of members of the family Bromoviridae: Lessons from the use of protoplast systems. J. Virol. 2008, 82, 10330–10340. [Google Scholar] [CrossRef]
- Fulton, R.W. Ilavirus group. C.M.I/A.A.B. Descr. Plant Viruses 1983, 274. Unavailable online. [Google Scholar]
- Kajati, I. Metody ossledovanija ekomomičeskowo značenija virusnych zabolevanij plodovich kultur. (Methods of invastigation on the economic importance of the virus diseses of fruit trees). In Proceedings of the Konferencjia Stran-Tslenov SZEV po Zaščite i Karantenu Rastenij, Budapest, Hungary, 16–19 October 1976; pp. 119–120. [Google Scholar]
- Kalinowska, E.; Mroczkowska, K.; Paduch-Cichal, E.; Chodorska, M. Genetic variability among coat protein of Prune dwarf virus variants from different countries and different Prunus species. Eur. J. Plant Pathol. 2014, 4, 863–868. [Google Scholar] [CrossRef]
- Nemeth, M. Interferencja vizsgălatok a csonthĕjas gyümöcsfăk gyürüsfoltossăg (ringspot) virusavial. Növĕnyvĕdelem 1972, 8, 64–71. [Google Scholar]
- Nemeth, M. Virus, Mycoplasma and Rikettsia Diseases of Fruit Trees, 1st ed.; Springer: Budapest, Hungary, 1986; pp. 600–650. ISBN 978-90-247-2868-8. [Google Scholar]
- Ramptish, C.; Estewell, K.C. The complete nucleotide sequence of prune dwarf ilarvirus RNA-1. Arch. Virol. 1997, 142, 1911–1918. [Google Scholar] [CrossRef]
- Scott, S.W.; Zimmerman, M.T.; Xin, G.; Mackenzie, D.J. The coat proteins and putative movement proteins of isolates of Prunus necrotic ringspot virus from different host species and geographic origins are extensively conserved are extensively conserved. Eur. J. Plant Pathol. 1998, 104, 155–161. [Google Scholar] [CrossRef]
- Bachman, E.J.; Scott, S.W.; Xin, G.; Vance, V.B. The complete nucleotide sequence of prune dwarf ilarvirus RNA 3: Implications for coat protein activation of genome replication in ilarviruses. Virology 1994, 201, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Ramptish, C.; Estewell, K.C.; Hall, J. Setting confidence limits for the detection of prune dwarf virus in Prunus avium with a monoclonal antibody-based triple antibody-sandwich ELISA. Ann. Appl. Biol. 1995, 126, 485–491. [Google Scholar] [CrossRef]
- Paduch-Cichal, E. Characterization of PNRSV and PDV. Associate Professor Thesis, Warsaw University of Life Sciences, Warsaw, Poland, 2000. [Google Scholar]
- Vaskova, D.; Petrzik, K.; Ŝpak, J. Molecular variability of the capsid protein of the Prune dwarf virus. Eur. J. Plant Pathol. 2000, 106, 573–580. [Google Scholar] [CrossRef]
- Ulubas-Serce, C.; Ertunc, F.; Özturk, A. Identification and genomic variability of Prune dwarf virus variants infecting stone fruit trees in Turkey. J. Phytopathol. 2009, 157, 298–305. [Google Scholar] [CrossRef]
- Loesch, L.S.; Fulton, R.W. Prunus necrotic ringspot virus as a multicomponent system. Virology 1975, 68, 71–78. [Google Scholar] [CrossRef]
- Rozanov, M.N.; Koonin, E.V.; Gorbalenya, A.E. Conservation of the putative methylotransferase domain: A hallmark of the ‘Sindbis like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 1992, 73, 2129–2134. [Google Scholar] [CrossRef] [PubMed]
- Korolev, S.; Hsieh, J.; Gauss, G.H.; Lohman, T.M.; Waksman, G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep-helicase bound to single stranded DNA and ADP. Cell 1997, 90, 635–647. [Google Scholar] [CrossRef]
- Bol, J.F. Replication of alfamo- and ilarviruses: Role of the coat protein. Annu. Rev. Phytopathol. 2005, 43, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Westler, W.M.; den Boon, G.; Wang, X.; Diaz, A.; Steinberg, H.A.; Alquist, P. An amphipathic alpha-helix controls multiple roles of Brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 2009, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Van der Hejden, M.W.; Carette, J.E.; Reinhoud, P.J.; Haegi, A.; Bol, J.F. Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J. Virol. 2001, 75, 1879–1887. [Google Scholar] [CrossRef] [PubMed]
- Cillo, F.; Roberts, I.M.; Palukaitis, P. In situ localization and tissue distribution of the replication-associated proteins of cucumber mosaic virus in tobacco and cucumber. J. Virol. 2002, 76, 10654–10664. [Google Scholar] [CrossRef] [PubMed]
- Kozieł, E.; Otulak, K.; Lockhart, B.E.L.; Garbaczewska, G. Subcelullar localization of proteins associated with Prune dwarf virus replication. Eur. J. Plant Pathol. 2017, 149, 653–668. [Google Scholar] [CrossRef]
- Dinant, S.; Janda, M.; Kroner, P.A.; Alquist, P. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J. Virol. 1993, 67, 7181–7189. [Google Scholar] [PubMed]
- Melcher, U. The “30K” superfamily of viral movement proteins. J. Gen. Virol. 2000, 81, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Kasteel, D.T.J.; van der Wel, N.N.; Jansen, K.A.J.; Goldbach, R.W.; van Lent, J.W.M. Tubule-forming capacity of the movement proteins of alfalfa mosaic virus and brome mosaic virus. J. Gen. Virol. 1997, 78, 2089–2093. [Google Scholar] [CrossRef] [PubMed]
- Kozieł, E.; Otulak, K.; Garbaczewska, G. Phylogenetic analysis of PDV movement protein compared to Bromoviridae members as justification of possible intercellular movement. Acta Biol. Crac. Ser. Bot. 2015, 57, 19–31. [Google Scholar] [CrossRef]
- Sanchez-Navarro, J.A.; Pallas, V. Evolutionary relationships in the ilarviruses: Nucleotide sequence of Prunus necrotic ringspot virus RNA 3. Arch. Virol. 1997, 142, 749–763. [Google Scholar] [CrossRef] [PubMed]
- Ansel-McKinney, P.; Scott, S.W.; Swanson, M.; Ge, X.; Gehrke, L. A plant viral coat protein RNA binding consensus sequence contains a crucial arginine. EMBO J. 1996, 15, 5077–5084. [Google Scholar] [PubMed]
- Aparicio, F.; Vilar, M.; Perez-Paya, E.; Pallas, V. The coat protein of Prunus necrotic ringspot virus specifically binds to and regulates the conformation of its genomic RNA. Virology 2003, 313, 213–223. [Google Scholar] [CrossRef]
- Aparicio, F.; Sánchez-Pina, M.A.; Sánchez-Navarro, J.A.; Pallás, V. Location of prunus necrotic ringspot ilarvirus within pollen grains of infected nectarine trees: Evidence from RT-PCR, dot-blot and in situ hybridisation. Eur. J. Plant Pathol. 1999, 105, 623–627. [Google Scholar] [CrossRef]
- Aparicio, F.; Sánchez-Navarro, J.A.; Pallás, V. Implication of the C terminus of the Prunus necrotic ringspot virus movement protein in cell-to-cell transport and in its interaction with the coat protein. J. Gen. Virol. 2010, 91, 1865–1870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neeleman, L.; Bol, J.F. Cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology 1999, 254, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.L.N. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 2006, 44, 61–87. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Tereso, S.; Nolasco, G.; Oliveira, M.M. Cellular location of Prune dwarf virus in almond sections by in situ reverse transcription-polymerase chain reaction. Phytopathology 2003, 93, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Öztürk, Y.; Çevik, B. Genetic diversity in the coat protein genes of Prune dwarf virus isolates from sweet cherry in Turkey. Plant Pathol. J. 2015, 31, 41–49. [Google Scholar] [CrossRef]
- Roenhorst, J.W.; van Lent, J.W.; Verduin, B.J. Binding of cowpea chlorotic mottle virus to cowpea protoplasts and relation of binding to virus entry and infection. Virology 1988, 164, 91–98. [Google Scholar] [CrossRef]
- Burgess, J.; Motoyoshi, F.; Fleming, E.N. The mechanism of infection of plant protoplasts by viruses. Planta 1973, 112, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Greber, R.S.; Teakle, D.S.; Mink, G.I. Thrips-facilitated transmission of prune dwarf and prunus necrotic ringspot viruses from cherry pollen to cucumber. Plant Dis. 1972, 76, 1039–1041. [Google Scholar] [CrossRef]
- Gallie, D.R.; Kobayashi, M. The role of the 3′-untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene 1994, 142, 152–165. [Google Scholar] [CrossRef]
- Ibrahim, A.; Hutchens, H.M.; Berg, R.H.; Loesch-Fries, S. Alfalfa mosaic virus replicase proteins, P1 and P2, localize to the tonoplast in the presence of virus RNA. Virology 2012, 433, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Jongejan, L.; Zheng, H.; Zhang, L.; Bol, J.F. Intracellular localization and movement phenotypes of Alfalfa mosaic virus movement protein mutants. Mol. Plant Microbe Interact. 2001, 14, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Houwing, C.J.; van de Putte, P.; Jaspars, E.M.J. Regulation of single strand RNA synthesis of Alfalfa mosaic virus in non-transgenic cowpea protoplasts by the viral coat protein. Arch. Virol. 1998, 143, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Olsthoorn, R.C.; Mertens, S.; Brederode, F.T.; Bol, J.F. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 1999, 18, 4856–4864. [Google Scholar] [CrossRef] [PubMed]
- Sivakumaran, K.; Chen, M.H.; Rosssinck, M.J.; Kao, C.C. Core promoter for initiation of cucumber mosaic virus subgenomic RNA4A. Mol. Plant Pathol. 2002, 3, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, I.; Rao, A.L.N. Deletions in the conserved amino-terminal basic arm of cucumber mosaic virus coat protein disrupt virion assembly but do not abolish infectivity and cell-to-cell movement. Virology 1998, 248, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solińska, J.; Dzianott, A.; Bujarski, J.J. Recombination of 5′ subgenomic RNA3a with genomic RNA3 of Brome mosaic bromovirus in vitro and in vivo. Virology 2011, 410, 129–141. [Google Scholar] [CrossRef]
- Schoelz, J.E.; Harries, P.A.; Nelson, R.S. Intracellular transport of plant viruses: Finding the door out of the cell. Mol. Plant 2011, 4, 813–831. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Heinlein, M. Cellular pathways for viral transport through plasmodesmata. Protoplasma 2011, 248, 75–99. [Google Scholar] [CrossRef] [PubMed]
- Otulak, K.; Garbaczewska, G. Ultrastructural events during hypersensitive response of potato cv. Rywal infected with necrotic strains of potato virus Y. Acta Physiol. Plant. 2010, 32, 635–644. [Google Scholar] [CrossRef]
- Otulak, K.; Kozieł, E.; Garbaczewska, G. Seeing is believing. The use of light, fluorescent and transmission electron microscopy in the observation of pathological changes during different plant—Virus interactions. In Microscopy: Advances in Scientific Research and Education, 6th ed.; Mendez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2014; Volume 1, pp. 367–376. ISBN 978-84-942134-3-4. [Google Scholar]
- Carrington, J.C.; Kasschau, K.D.; Mahajan, S.K.; Schaad, M.C. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 1996, 8, 1669–1681. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, T.V.; Peiro, A.; Pallas, V.; Sanchez-Navarro, J. Systemic transport of alfalfa mosaic virus can be mediated by the movement proteins of several viruses assigned to five genera of the 30K family. J. Gen. Virol. 2013, 94, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Otulak, K.; Garbaczewska, G. Cell-to-cell movement of three genera (+) ssRNA plant viruses. Acta Physiol. Plant. 2011, 33, 249–260. [Google Scholar] [CrossRef]
- Garbaczewska, G.; Chouda, M.; Otulak, K. Ultrastructural studies of plasmodesmatal and vascular translocation of tobacco rattle virus (TRV) in tobacco and potato. Acta Physiol. Plant. 2012, 34, 1229–1238. [Google Scholar] [CrossRef]
- Otulak, K.; Kozieł, E.; Garbaczewska, G. Ultrastructural impact of tobacco rattle virus on tobacco and pepper ovary and anther tissues. J. Phytopathol. 2015, 164, 217–289. [Google Scholar] [CrossRef]
- Leisner, S.; Turgeon, R. Movement of virus and photo-assimilate in the phloem: A comparative analysis. Bioessays 1993, 15, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Lucas, W.J.; Ding, B.; Van der Schoot, C. Plasmodesmata and the supracellular nature of plants. New Phytol. 1993, 125, 435–476. [Google Scholar] [CrossRef]
- Herranz, M.C.; Sanchez-Navarro, J.A.; Sauri, A.; Mingarro, I.; Pallas, V. Mutational analysis of the RNA-binding domain of the prunus necrotic ringspot virus (PNRSV) movement protein reveals its requirement for cell-to-cell movement. Virology 2005, 339, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Tomenius, K.; Clapham, D.; Meshi, T. Localization by immunogold cytochemistry of the virus coded 30K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology 1987, 160, 363–371. [Google Scholar] [CrossRef]
- Solovyev, A.G.; Savenkov, E.L. Factors involved in the systemic transport of plant RNA viruses: The emerging role of the nucleus. J. Exp. Bot. 2014, 65, 1689–1697. [Google Scholar] [CrossRef] [PubMed]
- Citovsky, V.; Wong, M.L.; Shaw, A.L.; Prasad, P.V.; Zambryski, P. Visualization and characterization of tobacco mosaic virus movement protein binding to single-stranded nucleic acids. Plant Cell 1992, 4, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Citovsky, V. Probing plasmodesmal transport with plant viruses. Plant Physiol. 1993, 102, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Heinlein, M.; Epel, B.L.; Padgett, H.S.; Beachy, R.N. Interaction of Tobamovirus movement proteins with the plant cytoskeleton. Science 1995, 270, 1983–1985. [Google Scholar] [CrossRef] [PubMed]
- McLean, B.G.; Zupan, J.; Zambryski, P.C. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 1995, 7, 2101–2114. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, C.; Niehl, A.; Sambade, A.; Steinmetz, A.; Heinlein, M. Inhibition of tobacco mosaic virus movement by expression of an actin-binding protein. Plant Physiol. 2009, 149, 1810–1823. [Google Scholar] [CrossRef] [PubMed]
- Vale, R.D. Intracellular transport using microtubule-based motors. Annu. Rev. Cell Biol. 1987, 3, 347–378. [Google Scholar] [CrossRef] [PubMed]
- St Johnston, D. The intracellular localization of messenger RNAs. Cell 1995, 81, 161–170. [Google Scholar] [CrossRef]
- Harries, P.A.; Schoelz, J.E.; Nelson, R.S. Intracellular transport of viruses and their components: Utilizing the cytoskeleton and membrane high-ways. Mol. Plant Microbe Interact. 2010, 23, 1381–1393. [Google Scholar] [CrossRef] [PubMed]
- Boyko, V.; Ferralli, J.; Ashby, J.; Schellenbaum, P.; Heinlein, M. Function of microtubules in intercellular transport of plant virus RNA. Nat. Cell Biol. 2000, 2, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Boyko, V.; Ferralli, J.; Heinlein, M. Cell-to-cell movement of TMV RNA is temperature-dependent and corresponds to the association of movement protein with microtubules. Plant J. 2000, 22, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Boyko, V.; Hu, Q.; Seemanpillai, M.; Ashby, J.; Heinlein, M. Validation of microtubule-associated to tobacco mosaic virus RNA movement and involvement of microtubule-aligned particle trafficking. Plant J. 2007, 51, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Sambade, A.; Brandner, K.; Hofmann, C.; Seemanpillai, M.; Mutterer, J.; Heinlein, M. Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata. Traffic 2008, 9, 2073–2088. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Gil, L.; Sanchez-Navarro, J.A.; Cruz, A.; Pallas, V.; Perez-Gil, J.; Mingarro, I. Plant virus cell-to-cell movement is not dependent on the transmembrane disposition of its movement protein. J. Virol. 2009, 83, 5535–5543. [Google Scholar] [CrossRef] [PubMed]
- Hipper, C.; Brault, V.; Ziegeler-Graf, V.; Revers, F. Viral and cellular factors involved in phloem transport of plant viruses. Front. Plant Sci. 2013, 4, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.L.N. Molecular studies on Bromovirus capsid protein. III. Analysis of cell-to-cell movement competence of coat protein defective variants of cowpea chlorotic mottle virus. Virology 1997, 232, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, S.; Watanabe, Y.; Beachy, R.N. Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. USA 2004, 101, 6291–6296. [Google Scholar] [CrossRef] [PubMed]
- Blackman, L.M.; Boevink, P.; Santa Cruz, S.; Palukaitis, P.; Oparka, K.J. The movement protein of cucumber mosaic virus traffics into sieve elements in minor veins of Nicotiana clevelandii. Plant Cell 1998, 10, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Canto, T.; Palukaitis, P. Are tubules generated by the 3a protein necessary for cucumber mosaic virus movement? Mol. Plant Microbe Interact. 1999, 12, 985–993. [Google Scholar] [CrossRef]
- Su, S.; Liu, Z.; Chen, C.; Zhang, Y.; Wang, X.; Zhu, L.; Miao, L.; Wang, X.C.; Yuan, M. Cucumber mosaic virus movement protein severs actin filaments to increase the plasmodesmal size exclusion limit in tobacco. Plant Cell 2010, 22, 1373–1387. [Google Scholar] [CrossRef] [PubMed]
- Van Lent, J.; Wellink, J.; Goldbach, R.W. Evidence for the involvement of the 58K and 48K proteins in the intracellular movement of cowpea mosaic virus. J. Gen. Virol. 1990, 71, 219–223. [Google Scholar] [CrossRef]
- Van Lent, J.; Storms, M.; Van der Meer, F.; Wellink, J.; Goldbach, R. Tubular structures involved in movement of cowpea mosaic virus are also formed in infected cowpea protoplasts. J. Gen. Virol. 1991, 72, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Flasinski, S.; Dzianott, A.; Pratt, S.; Bujarski, J. Mutational analysis of coat protein gene of brome mosaic virus: Effects on replication and movement protein in barley and on Chenopodium hybridum. Mol. Plant Microbe Interact. 1995, 8, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.L.; Grantham, G.L. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology 1995, 211, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Van der Vossen, E.A.; Neeleman, L.; Bol, J.F. Early and late functions of alfalfa mosaic virus coat protein can be mutated separately. Virology 1994, 202, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Van der Wel, N.N.; Goldbach, R.W.; van Lent, J. The movement protein and coat protein of alfalfa mosaic virus accumulate in structurally modified plasmodesmata. Virology 1998, 244, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Kaido, M.; Inoue, Y.; Takeda, Y.; Sugiyama, K.; Takeda, A.; Mori, M.; Tamai, A.; Meshi, T.; Okuno, T.; Mise, K. Downregulation of the NbNACa1 gene encoding a movement-protein-interacting protein reduces cell-to-cell movement of brome mosaic virus in Nicotiana benthamiana. Mol. Plant Microbe Interact. 2007, 20, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Navarro, J.A.; Herranz, M.C.; Pallas, V. Cell-to-cell movement of alfalfa mosaic virus can be mediated by the movement proteins of Ilar-, bromo-, cucumo-, tobamo- and comoviruses and does not require virion formation. Virology 2006, 346, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhang, L. Association of the movement protein of alfalfa mosaic virus with the endoplasmic reticulum and its trafficking in epidermal cells of onion bulb scales. Mol. Plant Microbe Interact. 1999, 12, 680–690. [Google Scholar] [CrossRef]
- Ueki, S.; Citovsky, V. Spread throughout the plant: Systemic transport of viruses. In Viral Transport in Plants, 1st ed.; Waigmann, E., Heinlein, M., Eds.; Springer: Berlin, Germany, 2007; pp. 85–118. ISBN 978-3-540-69967-5. [Google Scholar]
- Ding, B. Intercellular protein trafficking through plasmodesmata. In Plant Molecular Biology: Protein Trafficking in Plant Cells, 1st ed.; Soll, J., Ed.; Springer: Berlin, Germany, 1998; pp. 279–310. ISBN 978-94-011-5298-3. [Google Scholar]
- Oparka, K.J.; Santa Cruz, S. The great escape: Phloem transport and unloading of macromolecules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 323–347. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Yamanaka, K.; Okada, Y. Long-distance movement and viral assembly of tobacco mosaic virus mutants. Virology 1990, 176, 329–336. [Google Scholar] [CrossRef]
- Pallas, V.; Garcia, J.A. How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 2011, 92, 2691–2705. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.C.; Peng, N.I.; Hema, M.; Huang, X.; Dragnea, B. The coat protein leads the way: An update on basic and applied studies with the brome mosaic virus coat protein. Mol. Plant Pathol. 2011, 12, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Salánki, K.; Kiss, L.; Gellért, A.; Balázs, E. Identification a coat protein region of cucumber mosaic virus (CMV) essential for long-distance movement in cucumber. Arch. Virol. 2011, 156, 2279–2283. [Google Scholar] [CrossRef]
- Requena, A.; Simón-Buela, L.; Salcedo, G.; García-Arenal, F. Potential involvement of a cucumber homolog of phloem protein 1 in the long-distance movement of cucumber mosaic virus particles. Mol. Plant Microbe Interact. 2006, 19, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Bol, J.F. Alfalfa mosaic virus and ilarviruses: Involvement of coat protein in multiple steps of the replication cycle. J. Gen. Virol. 1999, 80, 1089–1102. [Google Scholar] [CrossRef] [PubMed]
- Tenllado, F.; Bol, J.F. Genetic dissection of the multiple functions of alfalfa mosaic virus coat protein in viral RNA replication, encapsidation, and movement. Virology 2000, 268, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Miller, J.; Nozaki, Y.; Sukamuto, J.; Takeda, M.; Shah, J.; Hase, S.; Ikegami, M.; Ehara, Y.; Dinesh-Kumar, S.P. RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J. 2002, 32, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Inaba, J.; Kim, B.M.; Shimura, H.; Masuta, C. Virus-induced necrosis is a consequence of direct protein-protein interaction between a viral RNA-silencing suppressor and a host catalase. Plant Physiol. 2011, 156, 2026–2036. [Google Scholar] [CrossRef] [PubMed]











| Ilarviruses | |
|---|---|
| Number | Name |
| Subgroup 1 | |
| 1 | Parietaria mottle virus, PMoV |
| 2 | Tobacco streak virus, TSV |
| Subgroup 2 | |
| 3 | Asparagus virus 2, AV-2 |
| 4 | Citrus leaf rugose virus, CiLRV |
| 5 | Citrus variegation virus, CVV |
| 6 | Elm mottle virus, EMoV |
| 7 | Lilac ring mottle virus, LiRMoV |
| 8 | Spinach latent virus, SpLV |
| 9 | Tulare apple mosaic virus, TaMV |
| Subgroup 3 | |
| 10 | Apple mosaic virus, ApMV |
| 11 | Blueberry shock virus, BlShV |
| 12 | Prunus necrotic ringspot virus, PNRSV |
| Subgroup 4 | |
| 13 | Fragaria chiloensis latent virus, FCILV |
| 14 | Prune dwarf virus, PDV |
| No relationships to other existing groups | |
| 15 | American plum line pattern virus, APLPV |
| 16 | Humulus japonicus latent virus, HJLV |
| Infected Species | Strain Name | Disease Name |
|---|---|---|
| Prunus avium, P. cerasifera, P. cerasus, P. domestica, P. mahaleb | Cherry chlorotic ringspot of Prune dwarf virus | Cherry chlorotic ringspot |
| Prunus avium, P. cerasifera, P. cerasus, P. domestica, P. mahaleb | Cherry chlorotic necrotic ringspot of Prune dwarf virus | Cherry chlorotic ringspot |
| Prunus avium, P. cerasus | Cherry ring mosaic of Prune dwarf virus | Cherry chlorotic necrotic ringspot |
| Prunus avium | Cherry ring mottleof Prune dwarf virus | Cherry ring mosaic |
| Prunus avium | Cherry yellow mosaic of Prune dwarf virus | Cherry ring mottle |
| Prunus serrulata cv. Amanogawa, Prunus serrulata cv. Kwanzan, P. avium var. plena, P fontanesiana, P.incisa, P. lannesiana | Cherry yellow mottle of Prune dwarf virus | Cherry yellow mosaic |
| Prunus domestica | Type strain of Prune dwarf virus | Cherry yellow mottle |
| Prunus armeniaca, P. avium, P. cerasus | Apricot gummosis of Prune dwarf virus | Chlorotic-necrotic ringspot |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozieł, E.; Bujarski, J.J.; Otulak, K. Molecular Biology of Prune Dwarf Virus—A Lesser Known Member of the Bromoviridae but a Vital Component in the Dynamic Virus–Host Cell Interaction Network. Int. J. Mol. Sci. 2017, 18, 2733. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18122733
Kozieł E, Bujarski JJ, Otulak K. Molecular Biology of Prune Dwarf Virus—A Lesser Known Member of the Bromoviridae but a Vital Component in the Dynamic Virus–Host Cell Interaction Network. International Journal of Molecular Sciences. 2017; 18(12):2733. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18122733
Chicago/Turabian StyleKozieł, Edmund, Józef J. Bujarski, and Katarzyna Otulak. 2017. "Molecular Biology of Prune Dwarf Virus—A Lesser Known Member of the Bromoviridae but a Vital Component in the Dynamic Virus–Host Cell Interaction Network" International Journal of Molecular Sciences 18, no. 12: 2733. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18122733