1. Introduction
Cyanotis arachnoidea C. B. Clarke (Commelinaceae), a Chinese traditional medicine herb, has been use for treatment of limb numbness and rheumatoid arthritis, as well as for promoting blood circulation and muscle relaxing [
1]. Rich phytoecdysteroids, including 20-hydroxyecdysone (20E), dihydroxyrubrosterone, rubrosterone, poststerone, and cyanosterone B, were found in this plant [
1,
2]. 20E was reported to have agricultural applications in enhancing the synchronous development of
Bombyx mori, elevating silk yield [
3] and reducing the time of the molting cycle of
Alpheus heterochelis [
4]. 20E and its derivatives exhibited a wide variety of pharmacological effects, including anti-depression, antioxidation, anti-diabetes, and neuron protection [
5,
6]. 20E is primarily obtained from ecdysteroid-rich plants, such as
C. arachnoidea,
Ajuga turkestanica, and
Serratula wolffi [
6]. Dried roots and aerial parts from
C. arachnoidea have been reported to contain 20E at 5.50% and 0.52%, respectively [
7]. However, due to the limitation of the wild resources for
C. arachnoidea, the supply of 20E is in great shortage and its application is always restricted [
6].
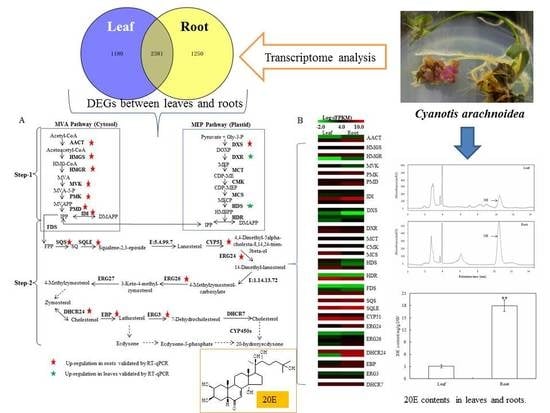
Originally, the ecdysteroid biosynthetic pathway has been described as a cytosolic pathway, starting from mevalonate (MVA) as a precursor [
8]. Subsequently, it has been proven that isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) required for the triterpenoid synthesis can be produced not only from the MVA pathway in cytosol but also the 2-
C-methyl-
d-erythritol 4-phosphate (MEP) pathway in plastids [
9]. However, the studies to date on phytoecdysteroid biosynthesis are limited to the parts mediated by the MVA pathway [
8]. In many plants, ecdysteroids were synthesized from cholesterol [
10,
11]. However, Adler and Grebenok [
12] identified lathosterol as a precursor of 20E in spinach. It is obviously controversial whether cholesterol or lathosterol is the preferred substrate for phytoecdysteriod biosynthesis [
8]. Conversion of a sterol to ecdysteroid requires several structural modifications, such as the hydroxylation of the sterol nucleus and side chain [
12]. In insects, cytochrome P-450 monooxygenases (CYP450s), which catalyze the bioreaction from cholesterol to 20E, have been cloned and identified. However, only one CYP450 enzyme in plants has been identified to be involved in 20E biosynthesis in the hairy roots of
Ajuga reptans [
13]. To date, the biosynthesis of 20E, especially the origin of the precursor IPP and its downstream pathway, remains largely unknown.
The accumulation of 20E in
C. arachnoidea was different among various tissues. Mu et al. [
7] reported that the 20E content in roots was 4–20 times higher than that in leaves. Biosynthesis and accumulation of secondary metabolites are often tissue-specific [
14]. Tomás et al. [
6] reported that the ecdysteroid content was related to organized structures of whole plants and in vitro propagated plantlets in
A. reptans, while calluses from leaves or roots could not produce ecdysteroids [
15]. The cultured plantlets of
A. reptans produced seven ecdysteroids with the tissue differentiation. Tomas et al. has revealed that ecdysteroid production is root-specific in
A. reptans [
16]. In recent years, comparative transcriptome analysis has been applied to investigate the biosynthesis of secondary metabolites. Yang et al. compared the transcriptome of leaves and roots from
Salvia miltiorrhiza and identified candidate genes involved in the biosynthesis of tanshinones [
17]. Biosynthesis of withanolide A, a medicinal component synthesized specifically in roots of
Withania somnifrea, was also investigated by comparative transcriptome analysis [
18]. Despite the important pharmacological value of 20E, there is little genetic information revealing the biosynthesis of 20E in
C. arachnoidea. Thus, as a follow-up to our previous works on the biotechnological production of 20E [
19] and cloning of key enzyme genes in 20E biosynthesis [
20], we conducted a comparative transcriptome analysis of the leaves and roots of
C. arachnoidea to identify genes putatively involved in 20E biosynthesis. As we know, this study reported the de novo sequencing of
C. arachnoidea for the first time. Meanwhile, transcription factors (TFs), which have been found to regulate the secondary metabolism in plants, were also searched for in the transcriptome database. Furthermore, we provided valuable information for developing the important molecular marker of simple sequence repeats (SSRs). This study was designed to characterize the transcriptome of
C. arachnoidea and provide a valuable basis for further elucidating the biosynthetic pathway of 20E.
3. Materials and Methods
3.1. Plant Materials
C. arachnoidea seeds were collected in September 2011 from the suburbs of Luquan County, Yunnan Province of China with its voucher specimen (SCU-110923), identified by C.Y. Liu. They were deposited in the herbarium of Soochow University. Germination and transplantation were performed in the horticultural nursery of Soochow University, Suzhou, China. One month after germination, the seedlings were transplanted into a plastic pot (35 × 25 × 7 cm in length, width, and height, respectively) containing sand and vermiculite in a 1:2 (v/v) ratio. Cultures were maintained in a growth chamber at 25 ± 2 °C under a 14/10-h light/dark cycle photoperiod with white fluorescent light at 1500 lux. Leaf and root tissues of four-month old C. arachnildea were collected separately, frozen in liquid nitrogen, and stored at −80 °C until use. At least three biological replicates were used for subsequent studies, and each replicate contained leaf or root tissues from at least 15 seedlings.
3.2. 20E Extraction and Analysis
The extraction and quantification of 20E in
C. arachnoidea leaves and roots were conducted as described by our previous report [
20] with slight modifications. Dry leaves or roots (0.5 g) were ground into a powder and extracted with 30 mL methanol under sonication for 90 min. The extract was then evaporated to dryness and dissolved in 1 mL methanol. High performance liquid chromatography (HPLC) conditions are as follows: an Aglient 1280 HPLC system equipped with 250 × 4.6 mm Aglient HC-C18 column, samples were eluted with 20:80 (
v/
v) acetonitrile/water at a flow rate 1 mL/min, and monitored at 242 nm. 20E was quantified with a genuine standard (Sigma, St. Louis, CA, USA).
Figure 1A presented a typical chromatogram of 20E in
C. arachnoidea leaves and roots under the condition.
3.3. cDNA Library Construction, Sequencing and Quality Control
For cDNA library construction, total RNA was firstly extracted from leaf or root tissues of four-month old C. arachnildea using a mirVana™ RNA isolation kit (Applied Biosystems, Foster City, CA, USA) and then treated with DNase I for 30 min at 37 °C. RNA integrity and purity was confirmed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). For each pool, total RNA (10 μg) was prepared for the cDNA library. The mRNA was purified from total RNA using magnetic beads with Oligo (dT). Subsequently, the mRNA was sheared into small fragments for cDNA synthesis. Double-stranded cDNA was synthesized using random hexamers. These cDNA fragments were subjected to an end repair process, the ligation of adapters, and were enriched by PCR to create the final libraries. Paired-end sequencing at 125 bp was performed using a HiSeq™2500 platform (Illumina, San Diego, CA, USA). High-quality reads were obtained by removing adaptor fragments, reads containing more than 5% ambiguous bases, and low-quality reads containing more than 20% of bases with a Q value ≤ 20.
3.4. De Novo Assembly and Sequence Annotation
3.5. Identification of DEGs
3.6. Real-Time Quantitative PCR Analysis
The expression levels of selected unigenes were analyzed through RT-qPCR. Total RNA of
C. arachnoidea leaves and roots were isolated with the RNAprep Pure Plant Kit (Tiangen, Beijing, China). The first cDNA strand was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, San Jose, CA, USA) according to the manufacturer’s instructions. RT-qPCR was performed using the CFX96™ Real-Time System (Bio-Red, Hercules, CA, USA). The reaction mixture included 2 μL five-fold diluted cDNA template, 1 μL of 10 mM forward primer, 1 μL of 10 mM reverse primer, 10 μL FS Universal SYBR Green Master (Roche, Indianapolis, IN, USA), and 6 μL ddH
2O. Amplification conditions were 94 °C for 4 min, and then 40 cycles of 94 °C for 1 min, 56 °C for 30 s, and 72 °C for 15 s. All the primers used were listed in
Table S1.
3.7. Identification of Simple Sequence Repeats
The identification of SSRs in the
C. arachnoidea transcriptome was conducted using a microsatellite program (MISA) (
http://pgrc.ipk-gatersleben.de/misa/). SSRs from the mononucleotides to the hexa-nucleotides were searched for in all unigenes. Both perfect and compound repeats were identified.
3.8. Statistical Analysis
To examine significant differences statistically between the means of two groups, we used Microsoft Excel software to conduct Student’s t-test. Values are reported as the mean ± SD from three independent experiments. Asterisks represented significant differences: * p < 0.05 and ** p < 0.01.