PsCor413pm2, a Plasma Membrane-Localized, Cold-Regulated Protein from Phlox subulata, Confers Low Temperature Tolerance in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Sequence Analysis of PsCor413pm2
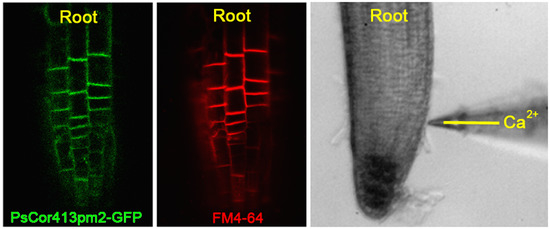
2.2. Subcellular Localization of PsCor413pm2
2.3. Overexpression of PsCor413pm2 in Arabidopsis Enhances Tolerance to Low Temperature
2.4. Overexpression of PsCor413pm2 in Arabidopsis Affects Ca2+ Flux and Stress-Related Genes Expression
3. Discussion
4. Materials and Methods
4.1. Identification of PsCor413pm2 and Sequence Analysis
4.2. Plant Material and Growth Conditions
4.3. RNA Extraction and Real-Time Quantitative PCR (qPCR) Analyses
4.4. Vector Construction and Plant Transformation
4.5. Confocal Laser Scanning Microscopy
4.6. Low Temperature and Osmosis Tolerance Assay
4.7. Net Ca2+ Flux Measurements
4.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Winfield, M.O.; Lu, C.; Wilson, I.D.; Coghill, J.A.; Edwards, K.J. Plant responses to cold: Transcriptome analysis of wheat. Plant Biotechnol. J. 2010, 8, 749–771. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.Q.; Shen, C.; Wu, L.H.; Tang, K.X.; Lin, J. CBF-dependent signaling pathway: A key responder to low temperature stress in plants. Crit. Rev. Biotechnol. 2011, 31, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Dong, C.H.; Zhu, J.K. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 2007, 10, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhong, H.; Ren, F.; Guo, Q.Q.; Hu, X.P.; Li, X.B. A novel cold-regulated gene, COR25, of Brassica napus is involved in plant response and tolerance to cold stress. Plant Cell Rep. 2011, 30, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Wathugala, D.L.; Richards, S.A.; Knight, H.; Knight, M.R. OsSFR6 is a functional rice orthologue of SENSITIVE TO FREEZING-6 and can act as a regulator of COR gene expression, osmotic stress and freezing tolerance in Arabidopsis. New Phytol. 2011, 191, 984–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, F.; Pan, Y.; Li, J.; Chen, X.; Pan, Y.; Wang, Y.; Tian, S.; Zhang, X. Heterologous expression of Arabidopsis C-repeat binding factor 3 (AtCBF3) and cold-regulated 15A (AtCOR15A) enhanced chilling tolerance in transgenic eggplant (Solanum melongena L.). Plant Cell Rep. 2014, 33, 1951–1961. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.M.; Sheng, Y.; Chen, W.J.; Lu, Y.Q.; Tang, X.; Ou-Yang, M.; Huang, X. Overexpression of Hevea brasiliensis HbICE1 Enhances Cold Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1462. [Google Scholar] [CrossRef] [PubMed]
- Steponkus, P.L.; Uemura, M.; Joseph, R.A.; Gilmour, S.J.; Thomashow, M.F. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1998, 95, 14570–14575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomashow, M.F. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 1998, 118, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Breton, G.; Danyluk, J.; Charron, J.B.; Sarhan, F. Expression profiling and bioinformatic analyses of a novel stress-regulated multispanning transmembrane protein family from cereals and Arabidopsis. Plant Physiol. 2003, 132, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Okawa, K.; Kakizaki, T.; Honma, T.; Itoh, H.; Inaba, T. Arabidopsis Cor15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiol. 2007, 144, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Thalhammer, A.; Hundertmark, M.; Popova, A.V.; Seckler, R.; Hincha, D.K. Interaction of two intrinsically disordered plant stress proteins (COR15A and COR15B) with lipid membranes in the dry state. BBA-Biomembr. 2010, 1798, 1812–1820. [Google Scholar] [CrossRef] [PubMed]
- Thalhammer, A.; Hincha, D.K. A mechanistic model of COR15 protein function in plant freezing tolerance: Integration of structural and functional characteristics. Plant Signal. Behav. 2014, 9, e977722. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Msanne, J.; Lin, J.; Stone, J.M.; Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 2011, 234, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Boon, N.J.; Webb, A.A.; Tanaka, R.J. Synergistic Activation of RD29A via integration of salinity stress and abscisic acid in Arabidopsis thaliana. Plant Cell Physiol. 2016, 57, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Okawa, K.; Nakayama, K.; Kakizaki, T.; Yamashita, T.; Inaba, T. Identification and characterization of Cor413im proteins as novel components of the chloroplast inner envelope. Plant Cell Environ. 2008, 31, 1470–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okawa, K.; Inoue, H.; Adachi, F.; Nakayama, K.; Ito-Inaba, Y.; Schnell, D.J.; Uehara, S.; Inaba, T. Targeting of a polytopic membrane protein to the inner envelope membrane of chloroplasts in vivo involves multiple transmembrane segments. J. Exp. Bot. 2014, 65, 5257–5265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, A.; Sun, H.; Feng, S.; Zhou, M.; Gong, S.; Wang, J.; Zhang, S. A novel cold-regulated gene from Phlox subulata, PsCor413im1, enhances low temperature tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 495, 1688–1694. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Lissarre, M.; Ohta, M.; Sato, A.; Miura, K. Cold-responsive gene regulation during cold acclimation in plants. Plant Signal. Behav. 2010, 5, 948–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Gong, Z. One SNP in COLD1 determines cold tolerance during rice domestication. J. Genet. Genomics 2015, 42, 133–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z.; Creelman, R.A.; Zhu, J.K. From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol. 2004, 135, 615–621. [Google Scholar] [CrossRef] [PubMed]
- TMHMM Server v. 2.0. Available online: http://www.cbs.dtu.dk/ services/TMHMM/ (accessed on 25 January 2018).
- Molecular Evolutionary Genetics Analysis. Available online: http://www.megasoftware.net/ (accessed on 25 January 2018).
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, Y.; Du, Y.; Zhao, Z.; Zhu, X.; Jiang, X.; Shu, Z.; Yin, Y.; Li, X. Overexpression of Camellia sinensis H1 histone gene confers abiotic stress tolerance in transgenic tobacco. Plant Cell Rep. 2014, 33, 1829–1841. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Liu, E.; Ma, H.; Feng, S.; Gong, S.; Wang, J. NaCl-induced expression of AtVHA-c5 gene in the roots plays a role in response of Arabidopsis to salt stress. Plant Cell Rep. 2018, 37, 443–452. [Google Scholar] [CrossRef] [PubMed]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, A.; Liu, E.; Li, H.; Li, Y.; Feng, S.; Gong, S.; Wang, J. PsCor413pm2, a Plasma Membrane-Localized, Cold-Regulated Protein from Phlox subulata, Confers Low Temperature Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2579. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092579
Zhou A, Liu E, Li H, Li Y, Feng S, Gong S, Wang J. PsCor413pm2, a Plasma Membrane-Localized, Cold-Regulated Protein from Phlox subulata, Confers Low Temperature Tolerance in Arabidopsis. International Journal of Molecular Sciences. 2018; 19(9):2579. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092579
Chicago/Turabian StyleZhou, Aimin, Enhui Liu, He Li, Yang Li, Shuang Feng, Shufang Gong, and Jingang Wang. 2018. "PsCor413pm2, a Plasma Membrane-Localized, Cold-Regulated Protein from Phlox subulata, Confers Low Temperature Tolerance in Arabidopsis" International Journal of Molecular Sciences 19, no. 9: 2579. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092579