Arabidopsis CNGC Family Members Contribute to Heavy Metal Ion Uptake in Plants
Abstract
: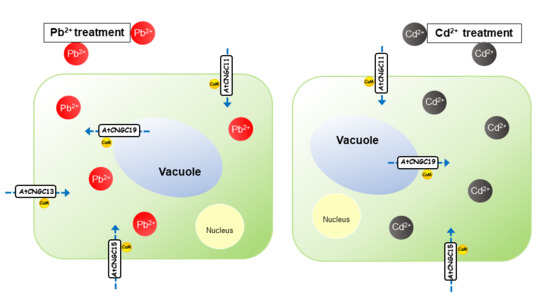
Graphical Abstract
1. Introduction
2. Results
2.1. Primary Root Growth Analysis in Pb2+ Stressed atcngc Mutant Plants
2.2. Primary Root Growth Analysis in Cd2+ Stressed atcngc Mutant Plants
2.3. Quantitative Assessment of Pb2+ Levels in atcngc Mutant Plants
2.4. Quantitative Assessment of Cd2+ Levels in atcngc Mutant Plants
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Condition
4.2. Root Growth Assay
4.3. Ion Content
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CNGC | Cyclic Nucleotide-Gated Channel |
| CN | Cyclic Nucleotide |
| cAMP | cyclic Adenosine MonoPhosphate |
| cGMP | cyclic Guanosine MonoPhosphate |
| CNBD | CN binding domains |
| CaM | Ca2+-activated calmodulin |
| ROS | Reactive Oxygen Species |
References
- Alloway, B.J. Sources of heavy metals and metalloids in soils. In Heavy Metals in Soil, 3rd ed.; Alloway, B.J., Ed.; Springer: Dordrecht, The Netherlands, 2013; Volume 22, pp. 11–50. ISBN 978-94-007-4469-1. [Google Scholar]
- Wu, Q.; Zhou, H.; Tam, N.F.; Tian, Y.; Tan, Y.; Zhou, S.; Li, Q.; Chen, Y.; Leung, J.Y. Contamination, toxicity and speciation of heavy metals in an industrialized urban river: Implications for the dispersal of heavy metals. Mar. Pollut. Bull. 2016, 104, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cobbina, S.J.; Mao, G.; Xu, H.; Zhang, Z.; Yang, L. A review of toxicity and mechanisms of individual and mixtures of heavy metals in the environment. Environ. Sci. Pollut. Res. Int. 2016, 23, 8244–8259. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Pourrut, B.; Dumat, C.; Nadeem, M.; Aslam, M.; Pinelli, E. Heavy-metal-induced reactive oxygen species: Phytotoxicity and physicochemical changes in plants. Rev. Environ. Contam. Toxicol. 2014, 232, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol. 2014, 7, 60–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansch, R.; Mendel, R.R. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 2009, 12, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Kanter, U.; Hauser, A.; Michalke, B.; Draxl, S.; Schaffner, A.R. Caesium and strontium accumulation in shoots of Arabidopsis thaliana: Genetic and physiological aspects. J. Exp. Bot. 2010, 61, 3995–4009. [Google Scholar] [CrossRef] [PubMed]
- White, P.J. The pathways of calcium movement to the xylem. J. Exp. Bot. 2001, 52, 891–899. [Google Scholar] [CrossRef] [Green Version]
- White, P.J.; Davenport, R.J. The voltage-independent cation channel in the plasma membrane of wheat roots is permeable to divalent cations and may be involved in cytosolic Ca2+ homeostasis. Plant Physiol. 2002, 130, 1386–1395. [Google Scholar] [CrossRef]
- White, P.J.; Broadley, M.R. Mechanisms of caesium uptake by plants. New Phytol. 2000, 147, 241–256. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Smolders, E. Plant uptake of radiocaesium: A review of mechanisms, regulation and application. J. Exp. Bot. 2000, 51, 1635–1645. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Moeder, W.; Yoshioka, K. Opening the gates: Insights into cyclic nucleotide-gated channel-mediated signaling. Trends Plant Sci. 2016, 21, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, B.; Sherman, T.; Fromm, H. Cyclic nucleotide-gated channels in plants. FEBS Lett. 2007, 581, 2237–2246. [Google Scholar] [CrossRef] [Green Version]
- Moeder, W.; Urquhart, W.; Ung, H.; Yoshioka, K. The role of cyclic nucleotide-gated ion channels in plant immunity. Mol. Plant 2011, 4, 442–452. [Google Scholar] [CrossRef]
- Talke, I.N.; Blaudez, D.; Maathuis, F.J.; Sanders, D. CNGCs: Prime targets of plant cyclic nucleotide signalling? Trends Plant Sci. 2003, 8, 286–293. [Google Scholar] [CrossRef]
- Cukkemane, A.; Seifert, R.; Kaupp, U.B. Cooperative and uncooperative cyclic-nucleotide-gated ion channels. Trends Biochem. Sci. 2011, 36, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Matulef, K.; Zagotta, W.N. Cyclic nucleotide-gated ion channels. Annu. Rev. Cell Dev. Biol. 2003, 19, 23–44. [Google Scholar] [CrossRef] [PubMed]
- Spalding, E.P.; Harper, J.F. The ins and outs of cellular Ca2+ transport. Curr. Opin. Plant Biol. 2011, 14, 715–720. [Google Scholar] [CrossRef]
- Arazi, T.; Kaplan, B.; Fromm, H. A high-affinity calmodulin-binding site in a tobacco plasma-membrane channel protein coincides with a characteristic element of cyclic nucleotide-binding domains. Plant Mol. Biol. 2000, 42, 591–601. [Google Scholar] [CrossRef]
- Pilot, G.; Pratelli, R.; Gaymard, F.; Meyer, Y.; Sentenac, H. Five-group distribution of the Shaker-like K+ channel family in higher plants. J. Mol. Evol. 2003, 56, 418–434. [Google Scholar] [CrossRef]
- Leng, Q.; Mercier, R.W.; Hua, B.G.; Fromm, H.; Berkowitz, G.A. Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiol. 2002, 128, 400–410. [Google Scholar] [CrossRef]
- Leng, Q.; Mercier, R.W.; Yao, W.; Berkowitz, G.A. Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 1999, 121, 753–761. [Google Scholar] [CrossRef]
- Schuurink, R.C.; Shartzer, S.F.; Fath, A.; Jones, R.L. Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc. Natl. Acad. Sci. USA 1998, 95, 1944–1949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, J.M.; Maser, P.; Schroeder, J.I. Plant ion channels: Gene families, physiology, and functional genomics analyses. Annu Rev. Physiol. 2009, 71, 59–82. [Google Scholar] [CrossRef] [PubMed]
- Arazi, T.; Kaplan, B.; Sunkar, R.; Fromm, H. Cyclic-nucleotide- and Ca2+/calmodulin-regulated channels in plants: Targets for manipulating heavy-metal tolerance, and possible physiological roles. Biochem. Soc. Trans. 2000, 28, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Arazi, T.; Sunkar, R.; Kaplan, B.; Fromm, H. A tobacco plasma membrane calmodulin-binding transporter confers Ni2+ tolerance and Pb2+ hypersensitivity in transgenic plants. Plant J. 1999, 20, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Kohler, C.; Merkle, T.; Neuhaus, G. Characterisation of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. Plant J. 1999, 18, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Kohler, C.; Neuhaus, G. Characterisation of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Lett. 2000, 471, 133–136. [Google Scholar] [CrossRef]
- Maser, P.; Thomine, S.; Schroeder, J.I.; Ward, J.M.; Hirschi, K.; Sze, H.; Talke, I.N.; Amtmann, A.; Maathuis, F.J.; Sanders, D.; et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001, 126, 1646–1667. [Google Scholar] [CrossRef]
- Urquhart, W.; Chin, K.; Ung, H.; Moeder, W.; Yoshioka, K. The cyclic nucleotide-gated channels AtCNGC11 and 12 are involved in multiple Ca2+-dependent physiological responses and act in a synergistic manner. J. Exp. Bot. 2011, 62, 3671–3682. [Google Scholar] [CrossRef]
- Balague, C.; Lin, B.; Alcon, C.; Flottes, G.; Malmstrom, S.; Kohler, C.; Neuhaus, G.; Pelletier, G.; Gaymard, F.; Roby, D. HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 2003, 15, 365–379. [Google Scholar] [CrossRef]
- Chan, C.W.; Schorrak, L.M.; Smith, R.K., Jr.; Bent, A.F.; Sussman, M.R. A cyclic nucleotide-gated ion channel, CNGC2, is crucial for plant development and adaptation to calcium stress. Plant Physiol. 2003, 132, 728–731. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Ali, R.; Berkowitz, G.A. Characterization of plant phenotypes associated with loss-of-function of AtCNGC1, a plant cyclic nucleotide gated cation channel. Plant Physiol. Biochem. 2006, 44, 494–505. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Park, G.; Amtmann, A.; Sanders, D.; Maathuis, F.J. Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J. Exp. Bot. 2006, 57, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Borsics, T.; Harrington, H.M.; Christopher, D.A. Arabidopsis AtCNGC10 rescues potassium channel mutants of E. coli, yeast and Arabidopsis and is regulated by calcium/calmodulin and cyclic GMP in E. coli. Funct. Plant Biol. 2005, 32, 643–653. [Google Scholar] [CrossRef]
- Guo, K.M.; Babourina, O.; Christopher, D.A.; Borsic, T.; Rengel, Z. The cyclic nucleotide-gated channel AtCNGC10 transports Ca2+ and Mg2+ in Arabidopsis. Physiol. Plant. 2010, 139, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kaplan, B.; Bouche, N.; Arazi, T.; Dolev, D.; Talke, I.N.; Maathuis, F.J.; Sanders, D.; Bouchez, D.; Fromm, H. Expression of a truncated tobacco NtCBP4 channel in transgenic plants and disruption of the homologous Arabidopsis CNGC1 gene confer Pb2+ tolerance. Plant J. 2000, 24, 533–542. [Google Scholar] [CrossRef]
- Ohki, S.; Ikura, M.; Zhang, M. Identification of Mg2+-binding sites and the role of Mg2+ on target recognition by calmodulin. Biochemistry 1997, 36, 4309–4316. [Google Scholar] [CrossRef]
- Ouyang, H.; Vogel, H.J. Metal ion binding to calmodulin: NMR and fluorescence studies. Biometals 1998, 11, 213–222. [Google Scholar] [CrossRef]
- Simons, T.J.; Pocock, G. Lead enters bovine adrenal medullary cells through calcium channels. J. Neurochem. 1987, 48, 383–389. [Google Scholar] [CrossRef]
- Tomsig, J.L.; Suszkiw, J.B. Permeation of Pb2+ through calcium channels: Fura-2 measurements of voltage- and dihydropyridine-sensitive Pb2+ entry in isolated bovine chromaffin cells. Biochim. Biophys. Acta 1991, 1069, 197–200. [Google Scholar] [CrossRef]
- Genger, R.K.; Jurkowski, G.I.; McDowell, J.M.; Lu, H.; Jung, H.W.; Greenberg, J.T.; Bent, A.F. Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “defense, no death” mutants. Mol. Plant-Microbe Interact. 2008, 21, 1285–1296. [Google Scholar] [CrossRef] [PubMed]
- Yuen, C.Y.; Christopher, D.A. The group IV-A cyclic nucleotide-gated channels, CNGC19 and CNGC20, localize to the vacuole membrane in Arabidopsis thaliana. AoB Plants 2013, 5, plt012. [Google Scholar] [CrossRef]
- Sharma, S.S.; Dietz, K.J.; Mimura, T. Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants. Plant Cell Environ. 2016, 39, 1112–1126. [Google Scholar] [CrossRef] [PubMed]
- Kugler, A.; Kohler, B.; Palme, K.; Wolff, P.; Dietrich, P. Salt-dependent regulation of a CNG channel subfamily in Arabidopsis. BMC Plant Biol. 2009, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Yuen, C.Y.; Christopher, D.A. The role of cyclic nucleotide-gated channels in cation nutrition and abiotic stress. In Ion Channels and Plant Stress Responses; Demidchik, V., Maathuis, F., Eds.; Springer: Berlin, Germany, 2010; Volume 5, pp. 137–157. ISBN 978-3-642-10493-0. [Google Scholar]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, J.Y.; Belloeil, C.; Ianna, M.L.; Shin, R. Arabidopsis CNGC Family Members Contribute to Heavy Metal Ion Uptake in Plants. Int. J. Mol. Sci. 2019, 20, 413. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20020413
Moon JY, Belloeil C, Ianna ML, Shin R. Arabidopsis CNGC Family Members Contribute to Heavy Metal Ion Uptake in Plants. International Journal of Molecular Sciences. 2019; 20(2):413. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20020413
Chicago/Turabian StyleMoon, Ju Yeon, Célestine Belloeil, Madeline Louise Ianna, and Ryoung Shin. 2019. "Arabidopsis CNGC Family Members Contribute to Heavy Metal Ion Uptake in Plants" International Journal of Molecular Sciences 20, no. 2: 413. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20020413




