The Antagonist Effect of Arachidonic Acid on GLUT4 Gene Expression by Nuclear Receptor Type II Regulation
Abstract
:1. Introduction
2. Results
2.1. Anthropometric and Biochemical Parameters of Lean and Morbidly Obese Individuals
2.2. GLUT4 and LXR-α are Downregulated in Visceral Adipose Tissue in Morbidly Obese Individuals
2.3. The Fatty Acid Profile of the Visceral Adipose Tissue in the Non-Obese and Morbidly Obese Subjects
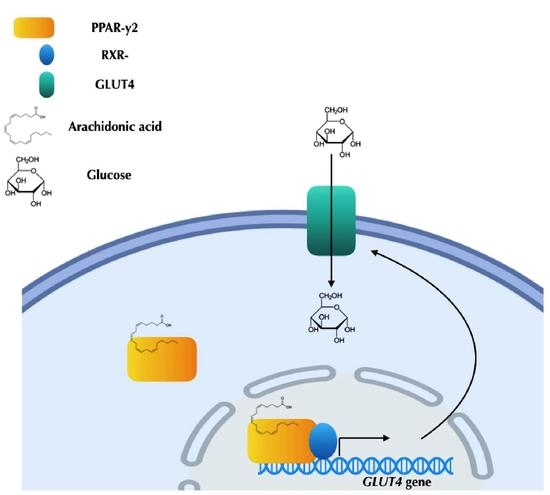
2.4. Arachidonic Acid Binds to PPAR-γ2 and Their Ligands’ Actions
2.5. Arachidonic Acid Mediates Repression Actions of GLUT4 mRNA Expression via PPAR-γ2 Activation
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Laboratory Measurements
4.3. Isolation of RNA and Quantitative RT-PCR
4.4. Western and Ligand Immunoblot Analysis
4.5. Visceral Adipose Tissue Fatty Acid Composition
4.6. Plasmids and DNA Constructs
4.6.1. Protein Expression Vectors
4.6.2. GST Fusion Protein Constructs
4.6.3. Reporter Gene Constructs
4.6.4. In Vitro Translation and Bacterial Overexpression of Proteins
4.7. GST Pull-Down Assays
4.8. Transfection and Luciferase Assays
4.9. Gene Silencing (siRNA)
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| NRs | Nuclear receptors |
| RXR | Retinoid X receptor |
| LXR-α | Liver X receptor type alpha |
| LXRE | LXR response elements |
| PPAR-g2 | Peroxisome proliferator-activated receptor type gamma 2 |
| GLUT4 | Glucose transporter type 4 |
| C/EBP-α | CCAAT-enhancer-binding proteins type alpha |
| TZD | Thiazolidinediones |
| EPA | Eicosapentaenoic acid |
| PUFA | Polyunsaturated fatty acid |
| MUFA | Monounsaturated fatty acid |
| DHA | Docosahexaenoic acid |
| AA | Arachidonic acid |
| LA | Linoleic acid |
| OA | Oleic acid |
| VAT | Visceral adipose tissue |
| LIR-MO | Low insulin resistance morbidly obese |
| HIR-MO | High insulin resistance morbidly obese |
| FFA | Free fatty acids |
| FA | Fatty acids |
| HOMA-IR | Homeostasis model of insulin resistance |
| DMSO | Dimethyl sulfoxide |
| SRC1 | Human steroid receptor co-activator 1 |
| HepG2 | Liver hepatocellular carcinoma cell line |
| Tk | Thymidine kinase |
| SFA | Saturated fatty acid |
| BMI | Body Mass index |
| SFA | Saturated fatty acids |
References
- Karelis, A.D. Metabolically healthy but obese individuals. Lancet 2008, 372, 1281–1283. [Google Scholar] [CrossRef]
- Sims, E.A.H. Are there persons who are obese, but metabolically healthy? Metab. Clin. Exp. 2001, 50, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- MacIas-Gonzalez, M.; Moreno-Santos, I.; García-Almeida, J.M.; Tinahones, F.J.; Garcia-Fuentes, E. PPARγ2 protects against obesity by means of a mechanism that mediates insulin resistance. Eur. J. Clin. Investig. 2009, 39, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Macias-Gonzalez, M.; Cardona, F.; Queipo-Ortuño, M.; Bernal, R.; Martin, M.; Tinahones, F.J. PPARgamma mRNA expression is reduced in peripheral blood mononuclear cells after fat overload in patients with metabolic syndrome. J. Nutr. 2008, 174–175. [Google Scholar] [CrossRef]
- Shulman, A.I.; Mangelsdorf, D.J. Retinoid X Receptor Heterodimers in the Metabolic Syndrome. N. Engl. J. Med. 2005, 353, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Laurencikiene, J.; Rydén, M. Liver X receptors and fat cell metabolism. Int. J. Obes. 2012, 36, 1494. [Google Scholar] [CrossRef] [PubMed]
- Peet, D.J.; Turley, S.D.; Ma, W.; Janowski, B.A.; Lobaccaro, J.M.; Hammer, R.E.; Mangelsdorf, D.J. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell 1998, 93, 693–704. [Google Scholar] [CrossRef]
- Steffensen, K.R.; Schuster, G.U.; Parini, P.; Holter, E.; Sadek, C.M.; Cassel, T.; Eskild, W.; Gustafsson, J.A. Different regulation of the LXRalpha promoter activity by isoforms of CCAAT/enhancer-binding proteins. Biochem. Biophys. Res. Commun. 2002, 293, 1333–1340. [Google Scholar] [CrossRef]
- Hong, C.; Tontonoz, P. Liver X receptors in lipid metabolism: Opportunities for drug discovery. Nat. Rev. Drug Discov. 2014, 13, 433. [Google Scholar] [CrossRef]
- Chawla, A.; Boisvert, W.A.; Lee, C.H.; Laffitte, B.A.; Barak, Y.; Joseph, S.B.; Liao, D.; Nagy, L.; Edwards, P.A.; Curtiss, L.K.; et al. A PPARγ-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell 2001, 7, 161–171. [Google Scholar] [CrossRef]
- Laffitte, B.A.; Chao, L.C.; Li, J.; Walczak, R.; Hummasti, S.; Joseph, S.B.; Castrillo, A.; Wilpitz, D.C.; Mangelsdorf, D.J.; Collins, J.L.; et al. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc. Natl. Acad. Sci. USA 2003, 100, 5419–5424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, S.E.; Erickson, R.L.; Gerin, I.; DeRose, P.M.; Bajnok, L.; Longo, K.A.; Misek, D.E.; Kuick, R.; Hanash, S.M.; Atkins, K.B.; et al. Microarray Analyses during Adipogenesis: Understanding the Effects of Wnt Signaling on Adipogenesis and the Roles of Liver X Receptor in Adipocyte Metabolism. Mol. Cell. Biol. 2002, 22, 5989–5999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, J.B.; Moon, H.M.; Kim, W.S.; Lee, Y.S.; Jeong, H.W.; Yoo, E.J.; Ham, J.; Kang, H.; Park, M.G.; Steffensen, K.R.; et al. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor $$ expression. Mol. Cell. Biol. 2004, 24, 3430–3444. [Google Scholar] [CrossRef]
- Dalen, K.T.; Ulven, S.M.; Bamberg, K.; Gustafsson, J.Å.; Nebb, H.I. Expression of the Insulin-responsive Glucose Transporter GLUT4 in Adipocytes Is Dependent on Liver X Receptor α. J. Biol. Chem. 2003, 278, 48283–48291. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.R.; Tu, H.; Luk, A.; Repa, J.J.; Medina, J.C.; Li, L.; Schwendner, S.; Wang, S.; Thoolen, M.; Mangelsdorf, D.J.; et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000, 14, 2831–2838. [Google Scholar] [CrossRef] [Green Version]
- Chandra, V.; Huang, P.; Hamuro, Y.; Raghuram, S.; Wang, Y.; Burris, T.P.; Rastinejad, F. Structure of the intact PPAR-γ-RXR-α nuclear receptor complex on DNA. Nature 2008, 456, 350. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Ye, F.; Gui, C.; Luo, H.; Cai, J.; Shen, J.; Chen, K.; Shen, X.; Jiang, H. Ligand-binding regulation of LXR/RXR and LXR/PPAR heterodimerizations: SPR technology-based kinetic analysis correlated with molecular dynamics simulation. Protein Sci. 2005, 14, 812–822. [Google Scholar] [CrossRef] [Green Version]
- Li, A.C.; Glass, C.K. PPAR- and LXR-dependent pathways controlling lipid metabolism and the development of atherosclerosis. J. Lipid Res. 2004, 45, 2161–2173. [Google Scholar] [CrossRef] [Green Version]
- Escher, P.; Braissant, O.; Basu-Modak, S.; Michalik, L.; Wahli, W.; Desvergne, B. Rat PPARs: Quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology 2001, 142, 4195–4202. [Google Scholar] [CrossRef]
- Sugii, S.; Olson, P.; Sears, D.D.; Saberi, M.; Atkins, A.R.; Barish, G.D.; Hong, S.H.; Castro, G.L.; Yin, Y.Q.; Nelson, M.C.; et al. PPAR activation in adipocytes is sufficient for systemic insulin sensitization. Proc. Natl. Acad. Sci. USA 2009, 106, 22504–22509. [Google Scholar] [CrossRef]
- He, W.; Barak, Y.; Hevener, A.; Olson, P.; Liao, D.; Le, J.; Nelson, M.; Ong, E.; Olefsky, J.M.; Evans, R.M. Adipose-specific peroxisome proliferator-activated receptor knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. USA 2003, 100, 15712–15717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fajas, L.; Schoonjans, K.; Gelman, L.; Kim, J.B.; Najib, J.; Martin, G.; Fruchart, J.C.; Briggs, M.; Spiegelman, B.M.; Auwerx, J. Regulation of Peroxisome Proliferator-Activated Receptor γ Expression by Adipocyte Differentiation and Determination Factor 1/Sterol Regulatory Element Binding Protein 1: Implications for Adipocyte Differentiation and Metabolism. Mol. Cell. Biol. 1999, 19, 5495–5503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Rosen, E.D.; Brun, R.; Hauser, S.; Adelmant, G.; Troy, A.E.; McKeon, C.; Darlington, G.J.; Spiegelman, B.M. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 1999, 3, 151–158. [Google Scholar] [CrossRef]
- Giorgino, F.; Leonardini, A.; Laviola, L.; Perrini, S.; Natalicchio, A. Cross-Talk between PPARγ and insulin signaling and modulation of Insulin Sensitivity. Ppar Res. 2009, 2009, 818945. [Google Scholar]
- Gross, B.; Staels, B. PPAR agonists: Multimodal drugs for the treatment of type-2 diabetes. Best Pract. Res. Clin. Endocrinol. Metab. 2007, 21, 687–710. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, A.B.; Viswanad, B.; Ramarao, P. PPARγ agonists partially restores hyperglycemia induced aggravation of vascular dysfunction to angiotensin II in thoracic aorta isolated from rats with insulin resistance. Pharmacol. Res. 2007, 55, 400–407. [Google Scholar] [CrossRef]
- Akiyama, T.E.; Meinke, P.T.; Berger, J.P. PPAR ligands: Potential therapies for metabolic syndrome. Curr. Diabetes Rep. 2005, 5, 45–52. [Google Scholar] [CrossRef]
- Wu, Z.; Xie, Y.; Morrison, R.F.; Bucher, N.L.R.; Farmer, S.R. PPARγ induces the insulin-dependent glucose transporter GLUT4 in the absence of C/EBPα during the conversion of 3T3 fibroblasts into adipocytes. J. Clin. Investig. 1998. [Google Scholar] [CrossRef]
- Rieusset, J.; Auwerx, J.; Vidal, H. Regulation of gene expression by activation of the peroxisome proliferator-activated receptor γ with rosiglitazone (BRL 49653) in human adipocytes. Biochem. Biophys. Res. Commun. 1999, 265, 265–271. [Google Scholar] [CrossRef]
- Furuta, M.; Yano, Y.; Gabazza, E.C.; Araki-Sasaki, R.; Tanaka, T.; Katsuki, A.; Hori, Y.; Nakatani, K.; Sumida, Y.; Adachi, Y. Troglitazone improves GLUT4 expression in adipose tissue in an animal model of obese type 2 diabetes mellitus. Diabetes Res. Clin. Pract. 2002, 56, 159–171. [Google Scholar] [CrossRef]
- Young, P.W.; Cawthorne, M.A.; Coyle, P.J.; Holder, J.C.; Holman, G.D.; Kozka, I.J.; Kirkham, D.M.; Lister, C.A.; Smith, S.A. Repeat treatment of obese mice with BRL 49653, a new and potent insulin sensitizer, enhances insulin action in white adipocytes: Association with increased insulin binding and cell-surface GLUT4 as measured by photoaffinity labeling. Diabetes 1995, 44, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.A.; Yoshizaki, T.; Favelyukis, S.; Patsouris, D.; Imamura, T.; Verma, I.M.; Olefsky, J.M. Suppression of PPAR- attenuates insulin-stimulated glucose uptake by affecting both GLUT1 and GLUT4 in 3T3-L1 adipocytes. AJP Endocrinol. Metab. 2007. [Google Scholar] [CrossRef]
- Lehmann, J.M.; Lenhard, J.M.; Oliver, B.B.; Ringold, G.M.; Kliewer, S.A. Peroxisome proliferator-activated receptors α and γ are activated by indomethacin and other non-steroidal anti-inflammatory drugs. J. Biol. Chem. 1997, 272, 3406–3410. [Google Scholar] [CrossRef] [PubMed]
- Kletzien, R.F.; Clarke, S.D.; Ulrich, R.G. Enhancement of adipocyte differentiation by an insulin-sensitizing agent. Mol. Pharm. 1992, 41, 393–398. [Google Scholar]
- Forman, B.M.; Chen, J.; Evans, R.M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors and. Proc. Natl. Acad. Sci. USA 1997, 94, 4312–4317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimomura, I.; Hammer, R.E.; Richardson, J.A.; Ikemoto, S.; Bashmakov, Y.; Goldstein, J.L.; Brown, M.S. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: Model for congenital generalized lipodystrophy. Genes Dev. 1998, 12, 3182–3194. [Google Scholar] [CrossRef]
- Nadler, S.T.; Stoehr, J.P.; Schueler, K.L.; Tanimoto, G.; Yandell, B.S.; Attie, A.D. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc. Natl. Acad. Sci. USA 2000, 97, 11371–11376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moitra, J.; Mason, M.M.; Olive, M.; Krylov, D.; Gavrilova, O.; Marcus-Samuels, B.; Feigenbaum, L.; Lee, E.; Aoyama, T.; Eckhaus, M.; et al. Life without white fat: A transgenic mouse. Genes Dev. 1998, 12, 3168–3181. [Google Scholar] [CrossRef]
- Yu, Z.W.; Burén, J.; Enerbäck, S.; Nilsson, E.; Samuelsson, L.; Eriksson, J.W. Insulin can enhance GLUT4 gene expression in 3T3-F442A cells and this effect is mimicked by vanadate but counteracted by cAMP and high glucose—Potential implications for insulin resistance. Biochim. Biophys. Acta Mol. Basis Dis. 2001, 1535, 174–185. [Google Scholar] [CrossRef]
- Flores-Riveros, J.R.; McLenithan, J.C.; Ezaki, O.; Lane, M.D. Insulin down-regulates expression of the insulin-responsive glucose transporter (GLUT4) gene: Effects on transcription and mRNA turnover. Proc. Natl. Acad. Sci. USA 1993, 90, 512–516. [Google Scholar] [CrossRef]
- Sun, X.; Haas, M.E.; Miao, J.; Mehta, A.; Graham, M.J.; Crooke, R.M.; De Barros, J.P.; Wang, J.G.; Aikawa, M.; Masson, D.; et al. Insulin dissociates the effects of liver X receptor on lipogenesis, endoplasmic reticulum stress, and inflammation. J. Biol. Chem. 2016, 291, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xu, Z.; Zhang, Z.; Yin, X.; Lin, X.; Li, H.; Zheng, F. Impaired adipose expansion caused by liver X receptor activation is associated with insulin resistance in mice fed a high-fat diet. J. Mol. Endocrinol. 2017, 58, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Afman, L.A.; Müller, M. Human nutrigenomics of gene regulation by dietary fatty acids. Prog. Lipid Res. 2012, 51, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Petrus, P.; Edholm, D.; Rosqvist, F.; Dahlman, I.; Sundbom, M.; Arner, P.; Ryden, M.; Risérus, U. Depot-specific differences in fatty acid composition and distinct associations with lipogenic gene expression in abdominal adipose tissue of obese women. Int. J. Obes. 2017, 41, 1295. [Google Scholar] [CrossRef] [PubMed]
- Marinou, K.; Hodson, L.; Vasan, S.K.; Fielding, B.A.; Banerjee, R.; Brismar, K.; Koutsilieris, M.; Clark, A.; Neville, M.J.; Karpe, F. Structural and functional properties of deep abdominal subcutaneous adipose tissue explain its association with insulin resistance and cardiovascular risk in men. Diabetes Care 2014, 37, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Nugent, C.; Prins, J.B.; Whitehead, J.P.; Wentworth, J.M.; Chatterjee, V.K.K.; O’Rahilly, S. Arachidonic acid stimulates glucose uptake in 3T3-L1 adipocytes by increasing GLUT1 and GLUT4 levels at the plasma membrane: Evidence for involvement of lipoxygenase metabolites and peroxisome proliferator-activated receptor γ. J. Biol. Chem. 2001, 276, 9149–9157. [Google Scholar] [CrossRef] [PubMed]
- Grunfeld, C.; Baird, K.L.; Kahn, C.R. Maintenance of 3T3-L1 cells in culture media containing saturated fatty acids decreases insulin binding and insulin action. Biochem. Biophys. Res. Commun. 1981, 103, 219–226. [Google Scholar] [CrossRef]
- García-Serrano, S.; Moreno-Santos, I.; Garrido-Sánchez, L.; Gutierrez-Repiso, C.; García-Almeida, J.M.; García-Arnés, J.; Rivas-Marín, J.; Gallego-Perales, J.L.; García-Escobar, E.; Rojo-Martinez, G.; et al. Stearoyl-CoA desaturase-1 is associated with insulin resistance in morbidly obese subjects. Mol. Med. 2011, 17, 273–280. [Google Scholar] [CrossRef]
- Lepage, G.; Roy, C.C. Direct transesterification of all classes of lipids in a one-step reaction. J. Lipid Res. 1986, 27, 114–120. [Google Scholar]
- Soriguer, F.; García-Serrano, S.; García-Almeida, J.M.; Garrido-Sánchez, L.; García-Arnés, J.; Tinahones, F.J.; Cardona, I.; Rivas-Marín, J.; Gallego-Perales, J.L.; García-Fuentes, E. Changes in the serum composition of free-fatty acids during an intravenous glucose tolerance test. Obesity 2009, 17, 10–15. [Google Scholar] [CrossRef]



| Variables | Control (n = 11) | LIR-MO (n = 15) | HIR-MO (n = 19) |
|---|---|---|---|
| Sex (male/female) | 11 (4/7) | 15 (5/10) | 19 (7/12) |
| Age (years) | 44.6 ± 10.8 | 43.1 ± 11.2 | 41.1 ± 7.6 |
| Weight (kg) | 71.1 ± 12.6 a,b | 145.3 ± 30.7 | 156.4 ± 25.1 |
| BMI (kg/m2) | 23.0 ± 1.6 a,b | 53.8 ± 6.9 | 56.4 ± 6.8 |
| Waist circumference (cm) | 86.3 ± 4.0 a,b | 136.9 ± 19.7 | 146.8 ± 18.9 |
| Glucose (mg/dL) | 88.0 ± 5.7 b | 94.6 ± 9.7 c | 110.6 ± 17.6 |
| Insulin (µIU/mL) | 10.7 ± 3.1 a,b | 17.1 ± 4.6 c | 38.6 ± 14.5 |
| Total cholesterol (mg/dL) | 178.0 ± 44.1 | 195.9 ± 44.8 | 196.1 ± 28.6 |
| FFA (mmol/L) | 0.278 ± 0.133 a,b | 0.635 ± 0.323 | 0.663 ± 0.241 |
| Triglycerides (mg/dl) | 72.9 ± 37.0 a,b | 111.4 ± 46.1 c | 189.6 ± 79.9 |
| Leptin (ng/mL) | 11.8 ± 2.7 a,b | 156.2 ± 90.9 | 149.6± 87.6 |
| Adiponectin (ng/mL) | 18.2 ± 5.9 a,b | 11.4 ± 4.0 | 8.5 ± 4.3 |
| HOMA-IR | 2.81 ± 1.22 a,b | 3.96 ± 1.08 c | 10.37 ± 5.07 |
| Fatty Acids | Control (n = 11) | LIR-MO (n = 15) | HIR-MO (n = 19) |
|---|---|---|---|
| 12:0 (lauric) | 0.37 ± 0.20 b | 0.46 ± 0.31 c | 0.21 ± 0.11 |
| 14:0 (myristic) | 1.75 ± 0.55 b | 1.91 ± 0.45 c | 1.36 ± 0.33 |
| 16:0 (palmitic) | 16.61 ± 2.13 a,b | 18.78 ± 2.58 | 18.51 ± 1.70 |
| 16.1:n7 (palmitoleic) | 3.21 ± 0.78 a,b | 4.48 ± 1.24 | 4.02 ± 0.88 |
| 18:0 (stearic) | 3.94 ± 1.26 b | 3.11 ± 1.01 | 2.79 ± 0.69 |
| 18:1n9 (oleic) | 51.64 ± 8.07 | 48.93 ± 5.18 | 50.33 ± 5.24 |
| 18:2n6 (linoleic) | 18.37 ± 6.18 | 17.83 ± 2.14 | 18.26 ± 4.38 |
| 18:3n6 (α-linolenic) | 0.43 ± 0.06 b | 0.33 ± 0.16 | 0.38 ± 0.06 |
| 18:3n3 (γ-linolenic) | 0.016 ± 0.0188 b | 0.036 ± 0.034 | 0.037 ± 0.025 |
| 20:0 (arachidic) | 0.24 ± 0.03 a,b | 0.11 ± 0.012 c | 0.14 ± 0.03 |
| 20:1n9 (eicosenoic) | 0.72 ± 0.18 | 0.67 ± 0.06 c | 0.75 ± 0.14 |
| 20:4n6 (arachidonic) | 0.25 ± 0.05 b | 0.28 ± 0.20c | 0.41 ± 0.06 |
| 20:5n3 (eicosatetraenoic) | 0.040 ± 0.031 | 0.045 ± 0.020 | 0.043 ± 0.010 |
| 22:4n6 (adrenic) | 0.14 ± 0.04 a,b | 0.24 ± 0.10 | 0.25 ± 0.08 |
| 22:5n3 (docosapentaenoic) | 0.15 ± 0.07 | 0.18 ± 0.08 | 0.16 ± 0.05 |
| 22:6n3 (docosahexaenoic) | 0.30 ± 0.08 b | 0.25 ± 0.06 c | 0.20 ± 0.04 |
| SFA | 25.47 ± 3.50 b | 24.39 ± 3.28 | 23.05 ± 2.42 |
| MUFA | 59.62 ± 4.28 | 56.14 ± 5.09 | 57.03 ± 5.68 |
| PUFA | 14.90 ± 1.17 a,b | 19.45 ± 2.25 | 19.91 ± 4.48 |
| n3 (Omega 3 fatty acid) | 0.51 ± 0.20 | 0.52 ± 0.14 | 0.45 ± 0.11 |
| n6 (Omega 6 fatty acid) | 14.38 ± 1.22 a,b | 18.93 ± 2.31 | 19.45 ± 4.39 |
| n3/n6 | 0.036 ± 0.016 b | 0.028 ± 0.010 c | 0.023 ± 0.003 |
| SFA/MUFA | 0.43 ± 0.08 | 0.44 ± 0.09 | 0.40 ± 0.07 |
| SFA/PUFA | 1.70 ± 0.19 a,b | 1.25 ± 0.13 | 1.19 ± 0.24 |
| Arachidonic/linoleic | 0.016 ± 0.008 b | 0.015 ± 0.010 c | 0.023 ± 0.006 |
| Docosapentaenoic/arachidonic | 0.15 ± 0.10 | 0.11 ± 0.05 | 0.10 ± 0.03 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-Santos, I.; Garcia-Serrano, S.; Boughanem, H.; Garrido-Sanchez, L.; Tinahones, F.J.; Garcia-Fuentes, E.; Macias-Gonzalez, M. The Antagonist Effect of Arachidonic Acid on GLUT4 Gene Expression by Nuclear Receptor Type II Regulation. Int. J. Mol. Sci. 2019, 20, 963. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20040963
Moreno-Santos I, Garcia-Serrano S, Boughanem H, Garrido-Sanchez L, Tinahones FJ, Garcia-Fuentes E, Macias-Gonzalez M. The Antagonist Effect of Arachidonic Acid on GLUT4 Gene Expression by Nuclear Receptor Type II Regulation. International Journal of Molecular Sciences. 2019; 20(4):963. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20040963
Chicago/Turabian StyleMoreno-Santos, Inmaculada, Sara Garcia-Serrano, Hatim Boughanem, Lourdes Garrido-Sanchez, Francisco José Tinahones, Eduardo Garcia-Fuentes, and Manuel Macias-Gonzalez. 2019. "The Antagonist Effect of Arachidonic Acid on GLUT4 Gene Expression by Nuclear Receptor Type II Regulation" International Journal of Molecular Sciences 20, no. 4: 963. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20040963